Genetic Diversity and Population Structure Analysis of Anatolian Kara Grapevine (Vitis vinifera L.) Germplasm Using Simple Sequence Repeats
Abstract
:1. Introduction
2. Materials and Methods
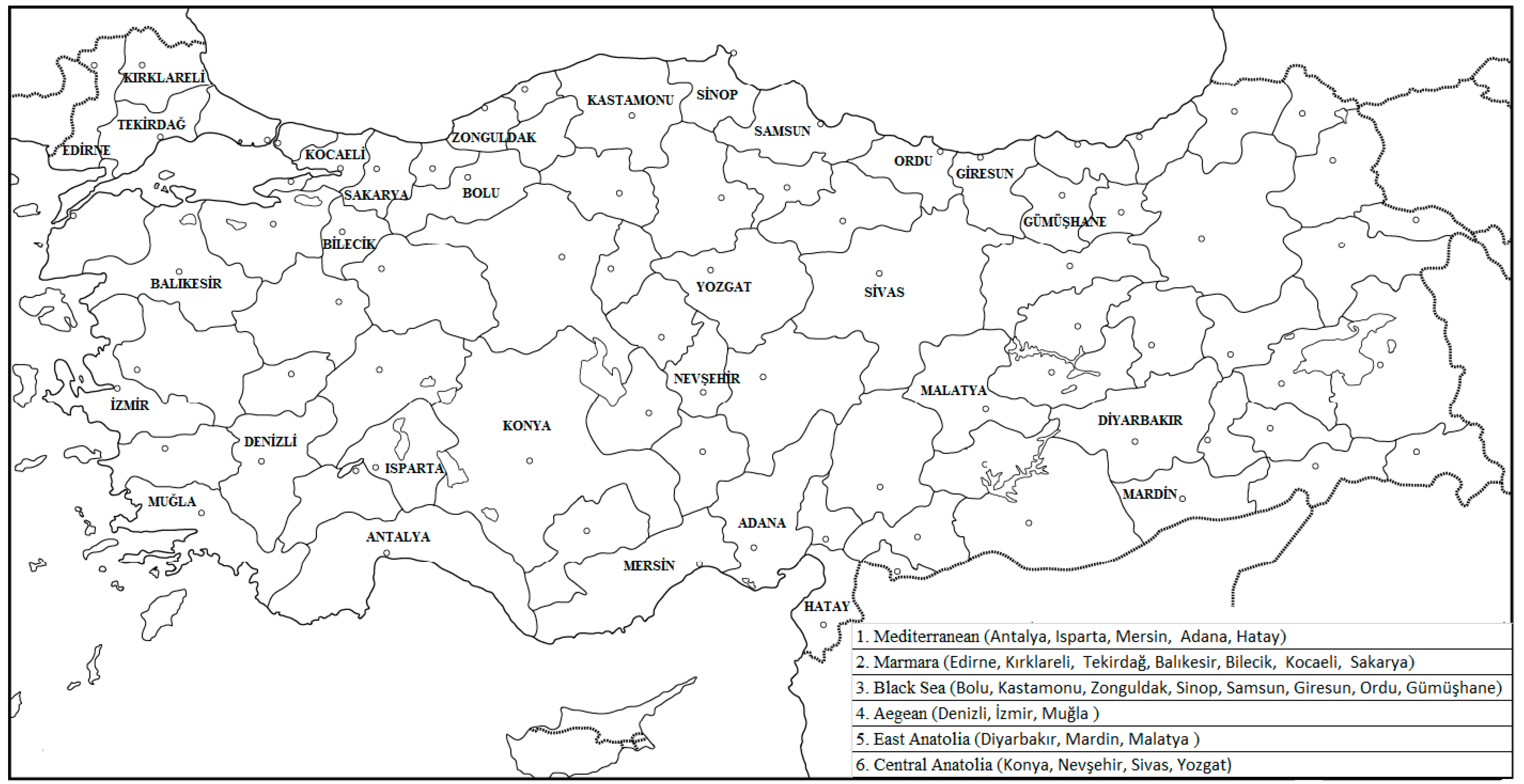
2.1. Plant Material
2.2. OIV Data Analysis
2.3. DNA Isolation
2.4. SSR Analysis and Capillary Electrophoresis
2.5. Genetic Analysis
2.5.1. SSR Analysis
2.5.2. Population Genetic Analysis
2.5.3. Clonal Analysis
3. Results
3.1. OIV Data Analysis
3.2. SSR Analysis
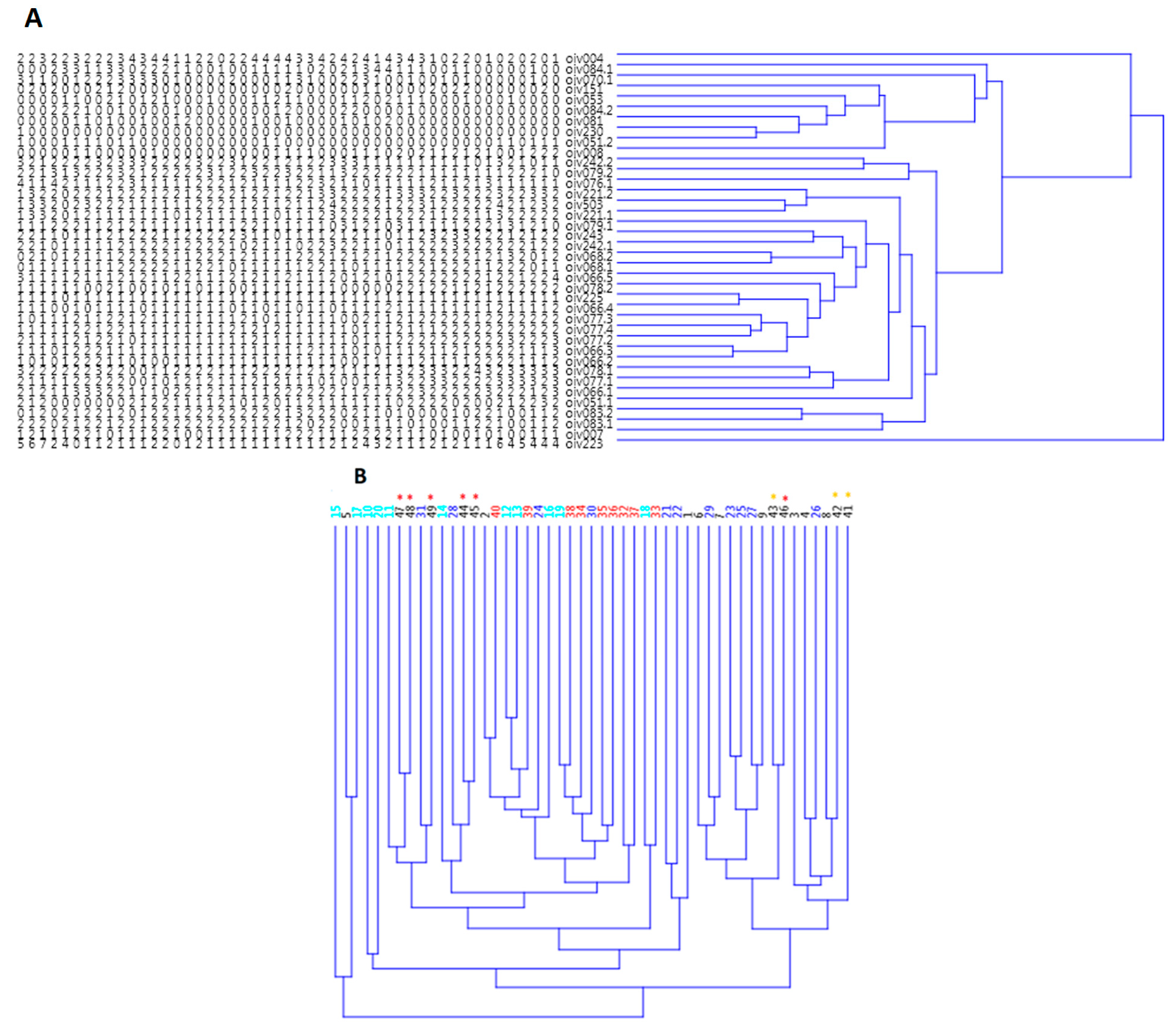
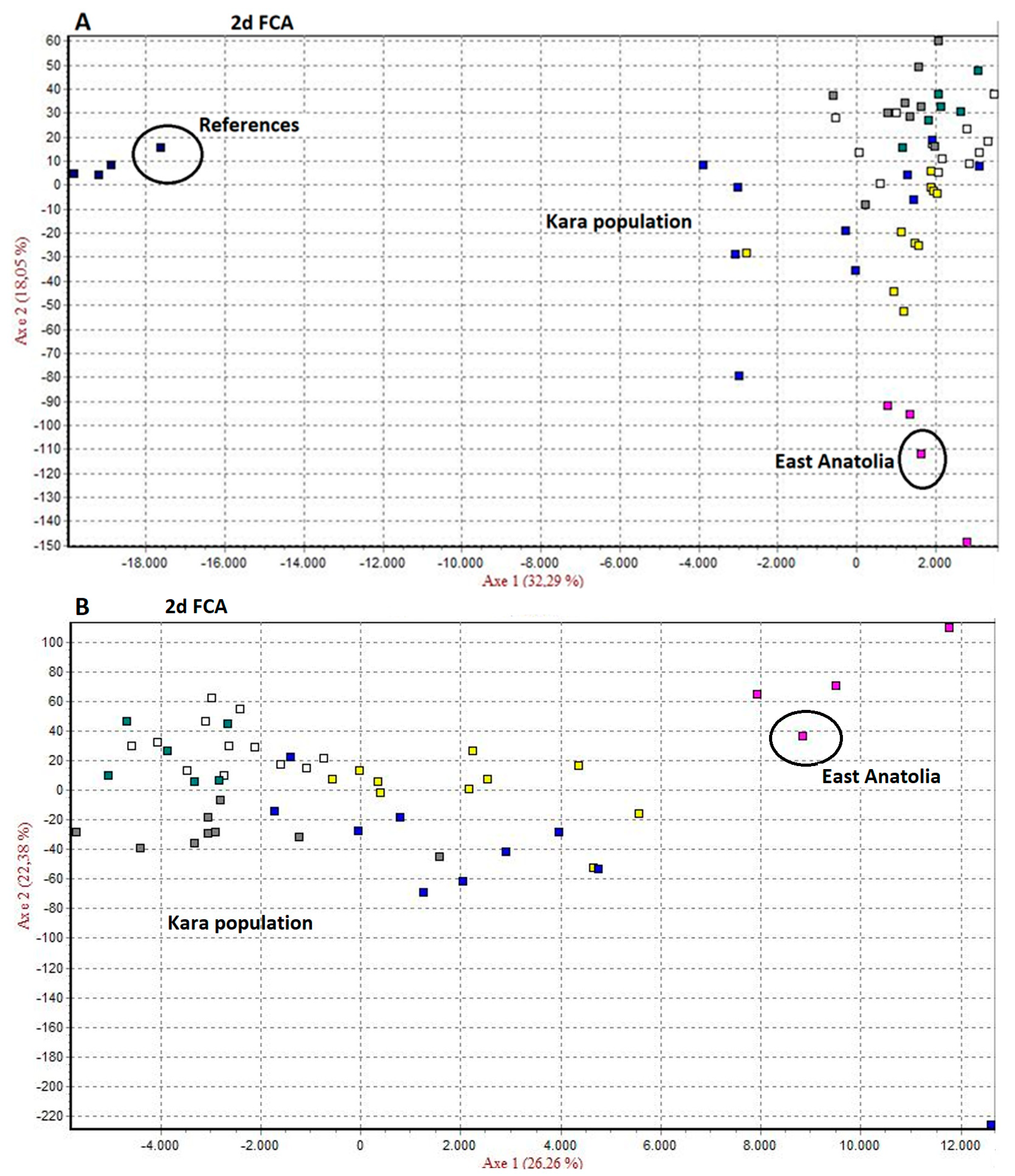
3.2.1. Genetic Relations among the Kara Grape Cultivars
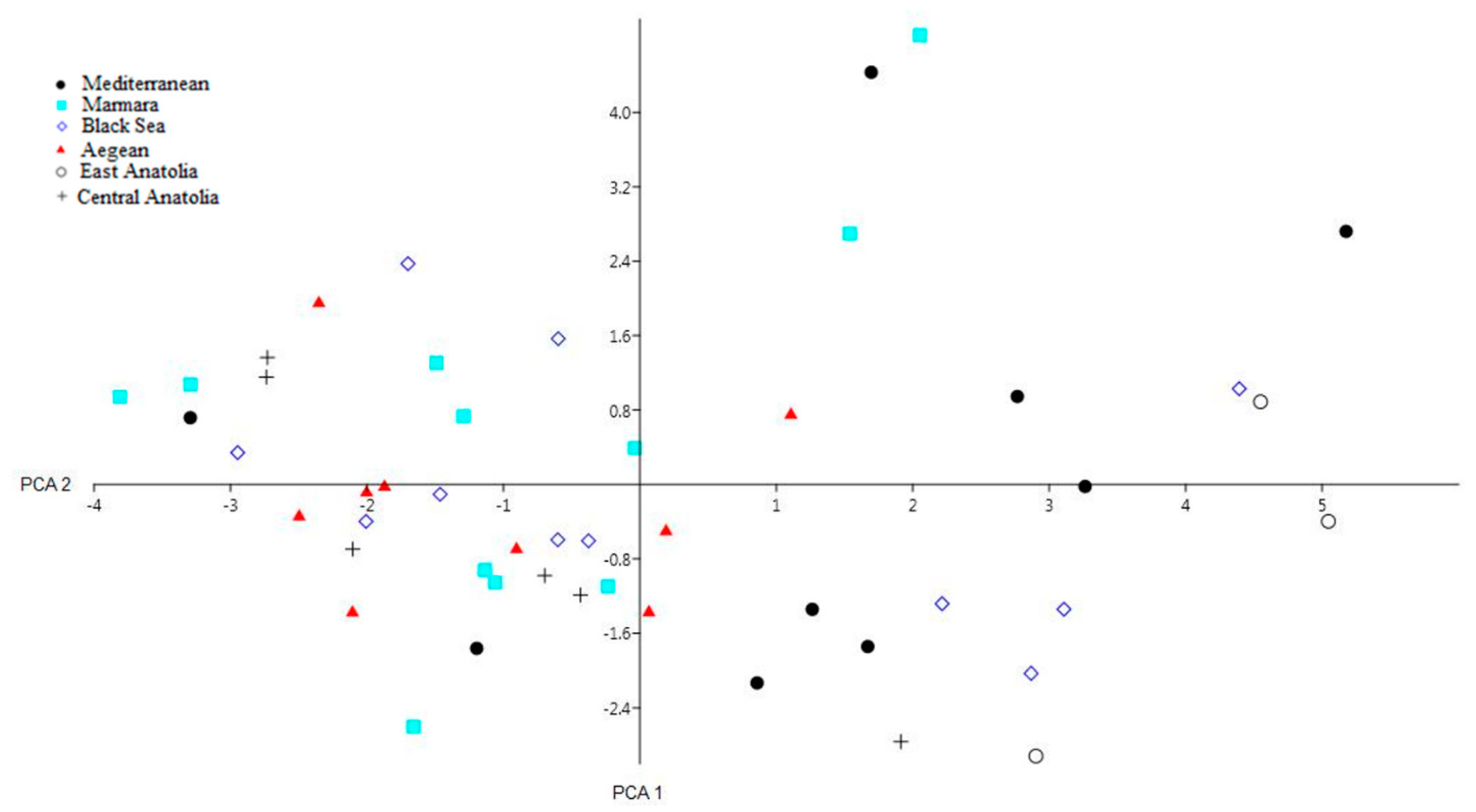
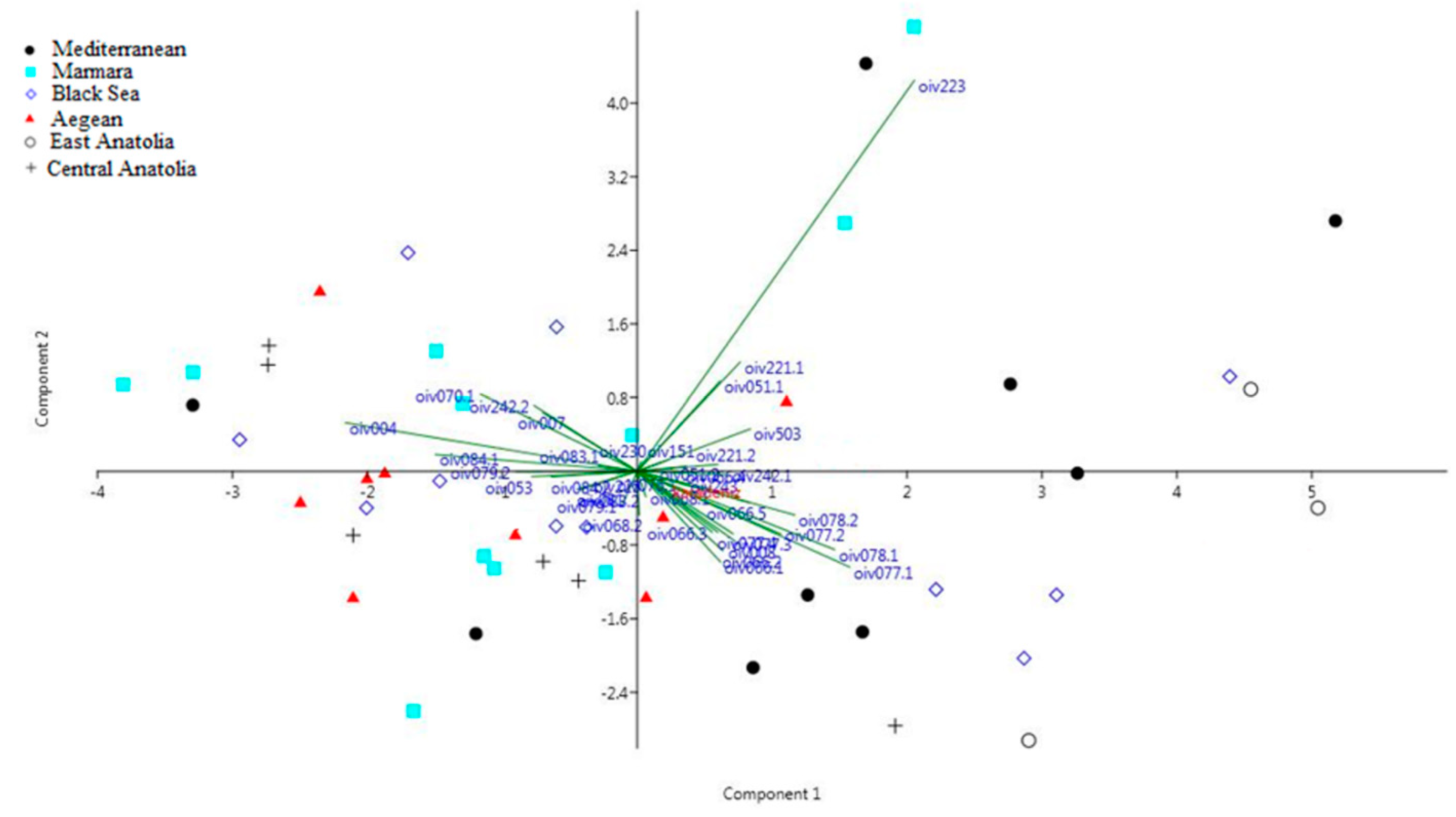
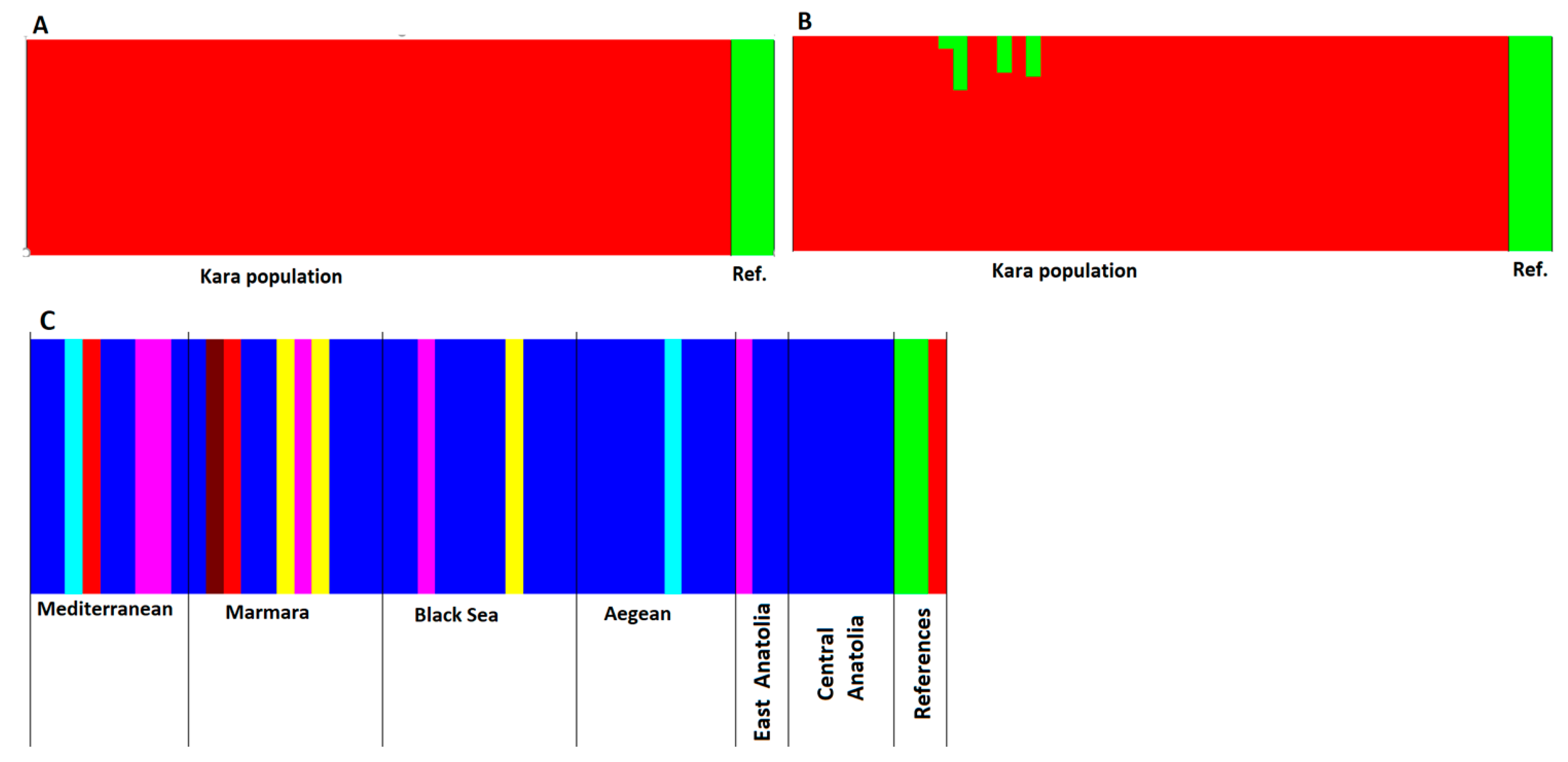
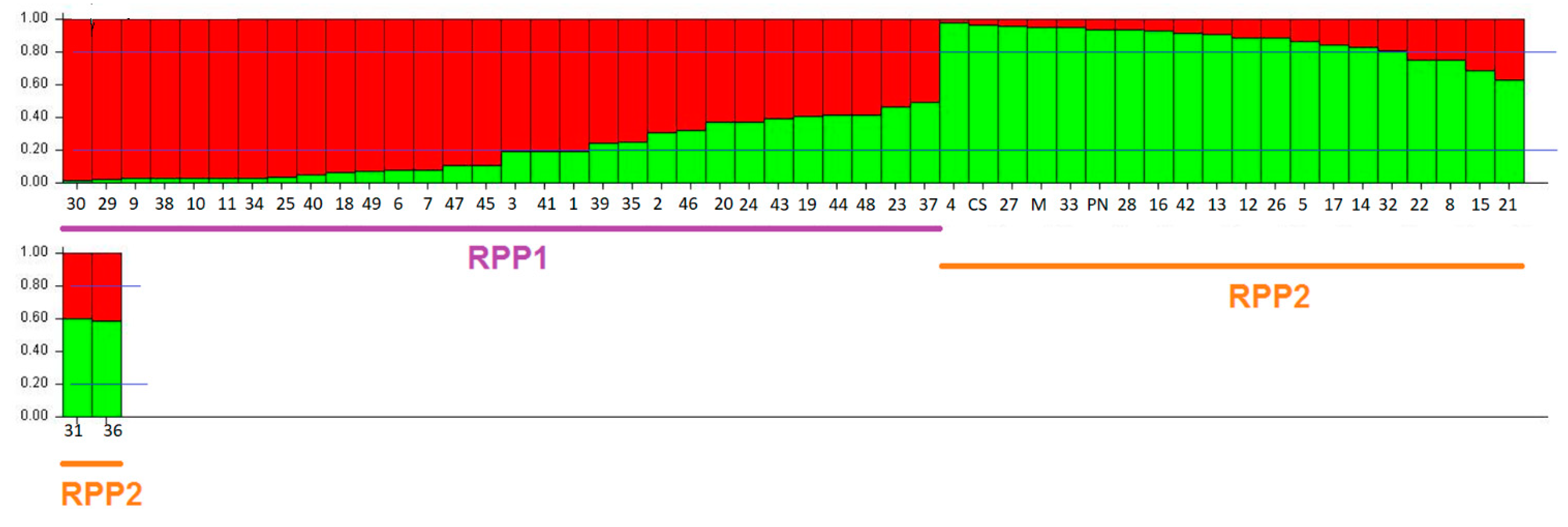
3.2.2. Genetic Structure Analysis among Kara Sub-Populations
3.2.3. Clonal Analysis
4. Discussion
4.1. OIV Data Analysis
4.2. SSR Analysis
4.3. Genetic Relations among the Kara Grape Cultivars
4.4. Genetic Structure Analysis among Kara Sub-Populations
4.5. Clonal Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ekhvaia, J.; Akhalkatsi, M. Morphological variation and relationships of Georgian populations of Vitis vinifera L. subsp. sylvestris (C.C. Gmel.). Hegi. Flora-Morphol. Distrib. Funct. Ecol. Plants 2010, 205, 608–617. [Google Scholar] [CrossRef]
- Naqinezhad, A.; Ramezani, E.; Djamali, M.; Schnitzler, A.; Arnold, C. Wild grapevine (Vitis vinifera subsp. sylvestris) in the Hyrcanian relict forests of northern Iran: An overview of current taxonomy, ecology and palaeorecords. J. For. Res. 2018, 29, 1757–1768. [Google Scholar] [CrossRef] [Green Version]
- da Silva Haas, I.C.; Toaldo, I.M.; de Gois, J.S.; Borges, D.L.; de Oliveira Petkowicz, C.L.; Bordignon-Luiz, M.T. Phytochemicals, monosaccharides and elemental composition of the non-pomace constituent of organic and conventional grape juices (Vitis labrusca L.): Effect of drying on the bioactive content. Plant Foods Hum. Nutr. 2016, 71, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Glevitzky, I.; Dumitrel, G.A.; Glevitzky, M.; Pasca, B.; Otrisal, P.; Bungau, S.; Cioca, G.; Pantis, C.; Popa, M. Statistical analysis of the relationship between antioxidant activity and the structure of flavonoid compounds. Rev. Chim. 2019, 70, 3103–3107. [Google Scholar] [CrossRef]
- Taymaz, E.R.; Uslu, M.E. Investigation and Determination of the Presence of Resveratrol in the Leaves of 99 Grapevine Varieties from the Western Anatolia Region in Turkey. JMPB 2022, 1, 1–8. [Google Scholar]
- Arroyo-García, R.; Ruiz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezoğlu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ergül, A.; Perez-Rivera, G.; Söylemezoğlu, G.; Kazan, K.; Arroyo-Garcia, R. Genetic diversity in Anatolian wild grapes (Vitis vinifera subsp. sylvestris) estimated by SSR markers. Plant Genet. Resour. 2011, 9, 375–383. [Google Scholar] [CrossRef]
- Samah, M.I.; Sahar, S.A.S.; Khaled, A.S.; Hoda, M.H.A. Phenolic compounds and antioxidant activity of white, red, black grape skin and white grape seeds. Life Sci. J. 2012, 9, 3464–3474. [Google Scholar]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Vezzulli, S.; Doligez, A.; Bellin, D. Molecular Mapping of Grapevine Genes. In The Grape Genome. Compendium of Plant Genomes; Cantu, D., Walker, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 103–136. [Google Scholar]
- Upadhyay, A.; Saboji, M.D.; Reddy, S.; Deokar, K.; Karibasappa, G.S. AFLP and SSR marker analysis of grape rootstocks in Indian grape germplasm. Sci. Hortic. 2007, 112, 176–183. [Google Scholar] [CrossRef]
- Meneghetti, S.; Costacurta, A.; Morreale, G.; Calò, A. Study of intra-varietal genetic variability in grapevine cultivars by PCR-derived molecular markers and correlations with the geographic origins. Mol. Biotechnol. 2012, 50, 72–85. [Google Scholar] [CrossRef]
- Jahnke, G.; Májer, J.; Varga, P.; Szőke, B. Analysis of clones of pinots grown in Hungary by SSR markers. Sci. Hortic. 2011, 129, 32–37. [Google Scholar] [CrossRef]
- Huang, H.; Lu, J.; Ren, Z.; Hunter, W.; Dowd, S.E.; Dang, P. Mining and validating grape (Vitis L.) ESTs to develop EST-SSR markers for genotyping and mapping. Mol. Breed. 2011, 28, 241–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef] [Green Version]
- Kantartzi, S.K. (Ed.) Microsatellites Methods and Protocols, in Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1006. [Google Scholar]
- Tello, J.; Forneck, A. Use of DNA markers for grape phylloxera population and evolutionary genetics: From RAPDs to SSRs and beyond. Insects 2019, 10, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasco, R.; Zharkikh, A.; Troggio, M.; Cartwright, D.A.; Cestaro, A.; Pruss, D.; Pindo, M.; FitzGerald, L.M.; Vezzulli, S.; Reid, J.; et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2007, 2, e1326. [Google Scholar] [CrossRef] [Green Version]
- Stavrakaki, M.; Bouza, D.; Biniari, K. Differentiation of Greek grapevine cultivars (Vitis vinifera L.) based on the combination of ampelographic description and microsatellite markers. Genet. Resour. Crop. Evol. 2020, 67, 21–40. [Google Scholar] [CrossRef]
- Anonim. Descriptor for Grapevine (Vitis spp.); International Plant Genetic Resources Institute: Roma, Italy, 1997; p. 58. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Lefort, F.; Lally, M.; Thompson, D.; Douglas, G.C. Morphological traits microsatellite fingerprinting and genetic relatedness of a stand of elite oaks (Q. Robur L.) at Tuallynally, Ireland. Silvae Genet. 1998, 47, 257–262. [Google Scholar]
- Akçay, M.E.; Burak, M.; Kazan, K.; Yüksel, C.; Mutaf, F.; Bakir, M.; Ayanoğlu, H.; Ergül, A. Genetic analysis of Anatolian pear germplasm by simple sequence repeats. Ann. Appl. Biol. 2014, 164, 441–452. [Google Scholar] [CrossRef]
- Ergül, A.; Büyük, B.P.; Hazrati, N.; Yılmaz, F.; Kazan, K.; Arslan, N.; Özmen Yüksel, C.; Aydın Soydam, S.; Bakir, M.; Tan, N.; et al. Genetic characterization and population structure analysis of Anatolian figs (Ficus carica L.) by SSR markers. Folia Hortic. 2021, 33, 49–78. [Google Scholar] [CrossRef]
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar] [CrossRef]
- Sefc, K.M.; Regner, F.; Turetshek, E.; Glössl, J.; Steınkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Goto-Yamamoto, N.; Mouri, H.; Azumi, M.; Edwards, K.J. Development of grape microsatellite analysis including oriental cultivars. Am. J. Enol. Vitic. 2006, 57, 105–108. [Google Scholar] [CrossRef]
- Merdinoglu, D.; Butterlin, G.; Baur, C.; Balthazard, D.J. Comparison of RAPD, AFLP and SSR (microsatellite) markers for genetic diversity analysis in Vitis vinifera L. Acta Hortic. 2000, 528, 193–197. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Constantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreria-Montteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Maul, E.; Sudharma, K.N.; Kecke, S.; Marx, G.; Müller, C.; Audeguin, L.; Bosellı, M.; Boursiquot, J.M.; Bucchetti, B.; Cabello, F.; et al. The European Vitis Database (www.eu-vitis.de)—A technical innovation through an online uploading and interactive modification system. Vitis 2012, 51, 79–86. [Google Scholar]
- Yılmaz, F.; Shidfar, M.; Hazrati, N.; Kazan, K.; Yüksel Özmen, C.; Uysal, T.; Özer, C.; Yaşasın, A.S.; Söylemezoğlu, G.; Boz, Y.; et al. Genetic analysis of central Anatolian grapevine (Vitis vinifera L.) germplasm by simple sequence repeats. Tree Genet. Genomes 2020, 16, 55. [Google Scholar] [CrossRef]
- Wagner, H.W.; Sefc, K.M. Identity 1.0. Centre for Applied Genetics; University of Agricultural Science: Vienna, Austria, 1999. [Google Scholar]
- Bowcock, A.M.; Ruiz-Linares, A.; Tomfohrde, J.; Minch, E.; Kidd, J.R.; Cavalli-Sforza, L.L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 1994, 368, 455–457. [Google Scholar] [CrossRef]
- Minch, E.; Ruiz-Linares, A.; Goldstein, D.B.; Feldman, M.; Cavallisforza, L.L. Microsat (Version 1.4d): A Computer Program for Calculating Various Statistics on Microsatellite Allele Data; Stanford University: Stanford, CA, USA, 1995. [Google Scholar]
- Rohlf, F. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Version 2.1; Exeter Software: Setoukat, NY, USA, 2000. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Rohlf, F. NTSYS-pc: Numeric Taxonomic Systems; Version 2.20; Exeter Software, Applied Biostatistics Inc.: Setoukat, NY, USA, 2004. [Google Scholar]
- Belkhir, K.; Borsa, P.; Goudet, J.; Bonhomme, F. GENETIX, Logiciel sous Windows TM pour la Génétique des Populations; Laboratoire Génome et Populations, CNRS UPR 9060; Université de Montpellier II: Montpellier, France, 1998. [Google Scholar]
- Corender, J.; Marttinen, P.; Siren, J.; Tang, J. Enhanced Bayesian Modelling in BAPS software for learning genetic structure of populations. BMC Bioinform. 2008, 9, 539. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Ferreira, V.; Díaz-Hernández, M.B.; Carnide, V.; Pinto-Carnide, O.; Rodrigues, R.; Velázquez-Barrera, M.E.; Rios-Mesa, D.; Ascasíbar-Errasti, J.; et al. Genetic diversity and core collection of Malus × domestica in northwestern Spain, Portugal and the Canary Islands by SSRs. Sci. Hortic. 2018, 240, 49–56. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Gould, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.G. 6.5: Genetic analysis in Excel. Population genetic software for teaching and researchdan update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Meirmans, P.G.; van Tienderen, P.H. Genotype and Genodive: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
- Leão, P.C.D.S.; Cruz, C.D.; Motoike, S.Y. Genetic diversity of table grape based on morphoagronomic traits. Sci. Agric. 2011, 68, 42–49. [Google Scholar] [CrossRef]
- Khadivi-Khub, A.; Salimpour, A.; Rasouli, M. Analysis of grape germplasm from Iran based on fruit characteristics. Rev. Bras. Bot. 2014, 37, 105–113. [Google Scholar] [CrossRef]
- Tello, J.; Torres-Pérez, R.; Grimplet, J.; Carbonell-Bejerano, P.; Martínez-Zapater, J.M.; Ibáñez, J. Polymorphisms and minihaplotypes in the VvNAC26 gene associate with berry size variation in grapevine. BMC Plant Biol. 2015, 15, 253. [Google Scholar] [CrossRef] [Green Version]
- Lamine, M.; Zemni, H.; Ziadi, S.; Chabaane, A.; Melki, I.; Mejri, S.; Zoghlami, N. Multivariate analysis and clustering reveal high morphological diversity in Tunisian autochthonous grapes (Vitis vinifera): Insights into characterization, conservation and commercialization. Oeno One 2014, 48, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Khalil, S.; Tello, J.; Hamed, F.; Forneck, A. A multivariate approach for the ampelographic discrimination of grapevine (Vitis vinifera) cultivars: Application to local Syrian genetic resources. Genet. Resour. Crop. Evol. 2017, 64, 1841–1851. [Google Scholar] [CrossRef] [Green Version]
- Sabir, A.; Tangolar, S.; Buyukalaca, S.; Kafkas, S. Ampelographic and molecular diversity among grapevine (Vitis spp.) cultivars. Czech J. Genet. 2009, 45, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Dilli, Y.; Ünal, A.; Kesgin, M.; İnan, M.S.; Söylemezoğlu, G. Comparison of ampelographic characteristics of some important grape varieties are grown in the Aegean Region, rootstock and clones. TURKJANS 2014, 1, 1546–1553. [Google Scholar]
- Knezović, Z.; Mandić, A.; Perić, N.; Beljo, J.; Žulj Mihaljević, M. Morphological and genetic characterization of vine grape cultivars of Herzegovina. CREBSS 2017, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhou, J.; Xu, X.; Perl, A.; Chen, S.; Ma, H. Adoption of table grape cultivars: An attribute preference study on Chinese grape growers. Sci. Hortic. 2017, 216, 66–75. [Google Scholar] [CrossRef]
- Doligez, A.; Bertrand, Y.; Farnos, M.; Grolier, M.; Romieu, C.; Esnault, F.; Dias, S.; Berger, G.; François, P.; Pons, T.; et al. New stable QTLs for berry weight do not colocalize with QTLs for seed traits in cultivated grapevine (Vitis vinifera L.). BMC Plant Biol. 2013, 13, 217. [Google Scholar] [CrossRef]
- Vouillamoz, J.F.; McGovern, P.E.; Ergul, A.; Söylemezoğlu, G.; Tevzadze, G.; Meredith, C.P.; Grando, M.S. Genetic characterization and relationships of traditional grape cultivars from Transcaucasia and Anatolia. Plant Genet. Resour. 2006, 4, 144–158. [Google Scholar] [CrossRef] [Green Version]
- Ibañez, J.; Andrés, M.T.; Molino, A.; Borrego, J. Genetic study of key Spanish grapevine varieties using microsatellite analysis. Am. J. Enol. Vitic. 2003, 54, 22–30. [Google Scholar] [CrossRef]
- Nebish, A.; ris Ochssner, I.; Maul, E.; Topfer, R.; Hausmann, L.; Hovhannisyan, A.; Devejyan, H.; Melyan, G.; Aroutiounian, R. Genetic identification and characterization of Armenian grapevine cultivars. In Proceedings of the BIO Web of Conferences, 40th World Congress of Vine and Wine, Sofia, Bulgaria, May 29–June 2 2017; Volume 9. [Google Scholar]
- Riaz, S.; De Lorenzis, G.; Velasco, D.; Koehmstedt, A.; Maghradze, D.; Bobokashvili, Z.; Musayev, M.; Zdunic, G.; Laucou, V.; Walker, M.A.; et al. Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) cultivars around the Mediterranean basin and Central Asia. BMC Plant Biol. 2018, 18, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karataş, D.D.; Karatas, H.; Laucou, V.; Sarikamiş, G.; Riahi, L.; Bacilieri, R.; This, P. Genetic diversity of wild and cultivated grapevine accessions from southeast Turkey. Hereditas 2014, 151, 73–80. [Google Scholar] [CrossRef]
- Eyduran, P.S.; Ercisli, S.; Akin, M.; Eyduran, E. Genetic characterization of autochthonous grapevine cultivars from Eastern Turkey by simple sequence repeats (SSRs). Biotechnol. Biotechnol. Equip. 2015, 30, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture. Theor. Appl. Genet. 2010, 12, 1569–1585. [Google Scholar] [CrossRef]
- Boz, Y.; Bakır, M.; Çelikkol, B.P.; Kazan, K.; Yılmaz, F.; Çakır, B.; Aslantaş, Ş.; Söylemezoğlu, G.; Yaşasın, A.S.; Özer, C.; et al. Genetic characterization of grape (Vitis vinifera L.) germplasm from Southeast Anatolia by SSR markers. Vitis 2011, 50, 99–106. [Google Scholar]
- Shen, Z.; Zhang, K.; Ma, L.; Duan, J.; Yan, A. Analysis of the genetic relationships and diversity among 11 populations of Xanthoceras sorbifolia using phenotypic and microsatellite marker data. Electron. J. Biotechnol. 2017, 26, 33–39. [Google Scholar] [CrossRef]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, J.-M.; Ware, D.; et al. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3457–3458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ergül, A.; Kazan, K.; Aras, S.; Çevik, V.; Çelik, H.; Söylemezoğlu, G. AFLP analysis of genetic variation within the two economically important Anatolian grapevine (Vitis vinifera L.) varietal groups. Genome 2006, 49, 467–495. [Google Scholar] [CrossRef]
- Hou, L.; Cui, Y.; Li, X.; Chen, W.; Zhang, Z.; Pang, X.; Li, Y. Genetic Evaluation of Natural Populations of the Endangered Conifer Thuja koraiensis Using Microsatellite Markers by Restriction-Associated DNA Sequencing. Genes 2018, 9, 218. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, J.; Liu, L.; Zhang, L.; Wei, L.; Hu, D. Genetic diversity of grape germplasm as revealed by microsatellite (SSR) markers. Afr. J. Biotechnol. 2015, 14, 990–998. [Google Scholar]
- Laucou, V.; Launay, A.; Bacilieri, R.; Lacombe, T.; Adam-Blondon, A.F.; Beârard, A.; Chauveau, A.; de Andrés, M.T.; Hausmann, L.; Ibáñez, J.; et al. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS ONE 2018, 13, e0192540. [Google Scholar] [CrossRef] [Green Version]
- Latch, E.K.; Dharmarajan, G.; Glaubitz, J.C.; Rhodes, O.E. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv. Genet. 2006, 7, 295–302. [Google Scholar] [CrossRef]
- Pelsy, F. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity 2010, 104, 331–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aradhya, M.K.; Dangl, G.S.; Prins, B.H.; Boursiquot, J.-M.; Walker, M.A.; Meredith, C.P.; Simon, C.J. Genetic structure and differentiation in cultivated grape Vitis vinifera L. Genet. Res. 2003, 81, 179–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barazani, O.; Westberg, E.; Hanin, N.; Dag, A.; Kerem, Z.; Tugendhaft, Y.; Hmidat, M.; Hijawi, T.; Kadereit, J.W. A comparative analysis of genetic variation in rootstocks and scions of old olive trees–A window into the history of olive cultivation practices and past genetic variation. BMC Plant Biol. 2014, 14, 146. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.N.; Jiang, K.; Biswas, S.R.; Tong, X.; Wang, R.; Chen, X.Y. Clone configuration and spatial genetic structure of two Halophila ovalis populations with contrasting internode lengths. Front. Ecol. Evol. 2019, 7, 170. [Google Scholar] [CrossRef] [Green Version]
- Grigoriou, A.; Tsaniklidis, G.; Hagidimitriou, M.; Nikoloudakis, N. The Cypriot indigenous grapevine germplasm is a multi-clonal varietal mixture. Plants 2020, 9, 1034. [Google Scholar] [CrossRef] [PubMed]
- Zombardo, A.; Storchi, P.; Valentini, P.; Ciofini, A.; Migliaro, D.; Crespan, M. Recovery, molecular characterization, and ampelographic assessment of marginal grapevine germplasm from southern umbria (Central Italy). Plants 2021, 10, 1539. [Google Scholar] [CrossRef]
- Harris, A.M.; De Giorgio, M. An unbiased estimator of gene diversity with improved variance for samples containing related and inbred individuals of any ploidy. G3-Genes Genom Genet. 2017, 7, 671–691. [Google Scholar] [CrossRef] [Green Version]
| No | SSR Loci | n | He | Ho | PI |
|---|---|---|---|---|---|
| 1 | VVS1 | 7 | 0.551 | 0.500 | 0.315 |
| 2 | VVS2 | 12 | 0.843 | 0.885 | 0.075 |
| 3 | VVMD5 | 9 | 0.8036 | 0.769 | 0.111 |
| 4 | VVMD7 | 7 | 0.775 | 0.808 | 0.162 |
| 5 | VVMD21 | 7 | 0.743 | 0.692 | 0.190 |
| 6 | VVMD24 | 9 | 0.682 | 0.750 | 0.198 |
| 7 | VVMD27 | 9 | 0.823 | 0.769 | 0.099 |
| 8 | VVMD28 | 12 | 0.812 | 0.769 | 0.110 |
| 9 | VVMD31 | 9 | 0.714 | 0.692 | 0.196 |
| 10 | VrZAG21 | 8 | 0.709 | 0.750 | 0.185 |
| 11 | VrZAG47 | 11 | 0.831 | 0.731 | 0.090 |
| 12 | VrZAG62 | 10 | 0.778 | 0.904 | 0.140 |
| 13 | VrZAG64 | 8 | 0.802 | 0.846 | 0.122 |
| 14 | VrZAG79 | 9 | 0.815 | 0.846 | 0.100 |
| 15 | VrZAG83 | 6 | 0.660 | 0.673 | 0.277 |
| 16 | VrZAG112 | 9 | 0.621 | 0.615 | 0.219 |
| 17 | VMC2H4 | 13 | 0.871 | 0.962 | 0.056 |
| 18 | VMC2C3 | 7 | 0.696 | 0.712 | 0.240 |
| 19 | VVIH54 | 11 | 0.745 | 0.500 | 0.132 |
| 20 | VVIB01 | 4 | 0.526 | 0.558 | 0.557 |
| 21 | VVMD25 | 8 | 0.789 | 0.846 | 0.136 |
| 22 | VVMD32 | 11 | 0.820 | 0.577 | 0.094 |
| Total | 196 | 16.411 | 16.154 | 3.803 | |
| Average | 8.91 | 0.746 | 0.734 | 0.173 |
| No | Identical | Synonymous | Homonymous |
|---|---|---|---|
| (Cultivar Name/List no/Population (Region)-Location (Province)) | |||
| 1 | - | Patlak Kara/45/Central Anatolia-Sivas | Kara Üzüm/1/Mediterranean-Adana-Kara Üzüm/15/Black Sea-Gümüşhane-Kara Üzüm/16/Mediterranean-Hatay-Kara Üzüm/23/Aegean-İzmir-Kara Üzüm/25/Black Sea-Kastamonu-Kara Üzüm/26/Marmara-Kırklareli-Kara Üzüm/49/Black Sea-Zonguldak |
| Siyah Üzüm/47/Central Anatolia-Yozgat | |||
| 2 | - | Deli Kara/3/Marmara-Balıkesir | Siyah Üzüm/12/East Anatolia-Diyarbakır-Siyah Üzüm/17/Mediterranean-Hatay-Siyah Üzüm/29/Central Anatolia-Konya-Siyah Üzüm/31/East Anatolia-Malatya-Siyah Üzüm/32/East Anatolia-Mardin-Siyah Üzüm/42/Black Sea-Samsun-Siyah Üzüm/43/Black Sea-Sinop-Siyah Üzüm/44/Black Sea-Sinop/-Siyah Üzüm/47/Central Anatolia-Yozgat |
| Yerli Kara/41/Marmara-Sakarya | |||
| 3 | - | Eski Kara/11/Aegean-Denizli | Ekşi Kara/10/Aegean-Denizli-Ekşi Kara/48/Central Anatolia-Yozgat |
| Yerli Kara/34/Aegean-Muğla | |||
| 4 | - | Siyah Üzüm/12/East Anatolia-Diyarbakır | Yerli Kara/34/Aegean-Muğla-Yerli Kara/41/Marmara-Sakarya |
| Kara Üzüm/26/Marmara-Kırklareli | |||
| 5 | - | - | Katı Kara/36/Aegean-Muğla-Katı Kara/38/Black Sea-Ordu-Katı Kara/39/Black Sea-Ordu |
| Sub-Population | Number of Cultivars (n) | Heterozygosity | Polymorphic Locus | Mean of Alleles/Locus | |||
|---|---|---|---|---|---|---|---|
| Hexp | Hobs | p (0.95) | p (0.99) | ||||
| Mediterranean | 9 | 0.700 | 0.732 | 1.0000 | 1.0000 | 5.27 | |
| Std. error | 0.086 | 0.170 | |||||
| Marmara | 11 | 0.752 | 0.743 | 1.0000 | 1.0000 | 6.72 | |
| Std. error | 0.084 | 0.155 | |||||
| Black Sea | 11 | 0.700 | 0.714 | 1.0000 | 1.0000 | 5.95 | |
| Std. error | 0.148 | 0.178 | |||||
| Aegean | 9 | 0.651 | 0.717 | 1.0000 | 1.0000 | 4.54 | |
| Std. error | 0.118 | 0.226 | |||||
| East Anatolia | 3 | 0.596 | 0.727 | 1.0000 | 1.0000 | 3.31 | |
| Std. error | 0.170 | 0.284 | |||||
| Central Anatolia | 6 | 0.613 | 0.765 | 1.0000 | 1.0000 | 3.72 | |
| Std. error | 0.125 | 0.255 | |||||
| References | 3 | 0.611 | 0.772 | 1.0000 | 1.0000 | 3.50 | |
| Std. error | 0.151 | 0.238 | |||||
| Sub-Populations | Mediterranean | Marmara | Black Sea | Aegean | East Anatolia | Central Anatolia | Ref. |
|---|---|---|---|---|---|---|---|
| Mediterranean | - | ||||||
| Marmara | 0.149 | - | |||||
| Black Sea | 0.170 | 0.147 | - | ||||
| Aegean | 0.248 | 0.180 | 0.195 | - | |||
| East Anatolia | 0.300 | 0.311 | 0.352 | 0.478 | - | ||
| Central Anatolia | 0.229 | 0.232 | 0.157 | 0.231 | 0.470 | - | |
| Ref. | 0.632 | 0.519 | 0.699 | 0.641 | 0.815 | 0.757 | - |
| Sub-Population (Fst/Nm) | Mediterranean | Marmara | Black Sea | Aegean | East Anatolia | Central Anatolia | Ref. |
|---|---|---|---|---|---|---|---|
| Mediterranean | - | ||||||
| Marmara | 0.00027/İnf. | - | |||||
| Black Sea | 0.01284/21.03 | 0.00500/84.93 | - | ||||
| Aegean | 0.04291 ***/5.47 | 0.02003/12.82 | 0.03029 */7.96 | - | |||
| East Anatolia | 0.01655/15.66 | 0.01518/25.11 | 0.03902 */6.50 | 0.08739 ***/2.48 | - | ||
| Central Anatolia | 0.03113/6.97 | 0.03068/7.80 | 0.00898/22.23 | 0.04072 */5.08 | 0.08424 */2.28 | - | |
| Ref. | 0.09815 ***/2.29 | 0.06173 */4.08 | 0.11655 ***/1.91 | 0.12209 */1.71 | 0.12159/1.68 | 0.14787 */1.28 | - |
| RPP | Number of Genotypes | qI > 0.8 | qI < 0.8 | The Most Representative Populations and the Number of Individuals |
|---|---|---|---|---|
| RPP1 | 30 | 18 (60%) | 12 (40%) | Mediterranean (5), Marmara (5), Black Sea (7), Aegean (7), Central Anatolia (6) |
| RPP2 | 22 | 16 (73%) | 6 (27%) | Mediterranean (4), Marmara (6), Black Sea (4), Aegean (2), East Anatolia (3), References (3) |
| Overall | 52 | 34 (65%) | 18 (35%) | All populations |
| Sub-Population (Sub-Population Number) | MLG | Na | Ne | Ho | uHe | PAS (Locus No: Alleles (bp)) |
|---|---|---|---|---|---|---|
| Central Anatolia (6) | 2 | 3.72 | 2.83 | 0.76 | 0.66 | VVMD32:146 |
| East Anatolia (3) | 1 | 3.31 | 2.75 | 0.72 | 0.71 | VVS1:181, VVMD28:281, VVMD31:201, ZAG83:203 |
| Mediterranean (9) | 2 | 5.27 | 3.62 | 0.73 | 0.74 | VVS2:141, VVMD7:230, VVMD24:221, ZAG62:190, ZAG112:229, VVMD5:271 |
| Marmara (11) | 3 | 6.72 | 4.44 | 0.74 | 0.78 | VVS1:193, VVMD5:231, VVMD7:258, VVMD24:201, VVMD28:219, ZAG21:195, ZAG47:183, ZAG62:210, ZAG83:183, ZAG112:247, VMC2h4:196, VVIh54:142 and154, VVIb01:300, VVMD25:237 and247, VVMD32:258 |
| Black Sea (11) | 2 | 5.95 | 3.99 | 0.71 | 0.73 | VVMD5:219,VVMD28:245, ZAG21:211, ZAG47:169, ZAG62:208, ZAG64: 144, VMC2h4:208 and210; VMC2c3:187, VVMD32:262 |
| Aegean (9) | 2 | 4.54 | 3.14 | 0.71 | 0.68 | VVMD21:250, VVIb01:308 |
| Total | 12 | 24.97 | 20.77 | 4.37 | 4.3 | - |
| Sub-Population (Sub-Population Number) | Number of Genotypes (Accessions) (gen)/Clonality | Effective Number of Genotypes (Accessions) (eff) | Genotypic Diversity (div) | Evenness (eve) | Shannon–Wiener (shw) |
|---|---|---|---|---|---|
| Central Anatolia (6) | 5/1 | 4 | 0.933 | 0.900 | 0.677 |
| East Anatolia (3) | 3/0 | 3 | 1 | 1 | 0.477 |
| Mediterranean (9) | 9/0 | 9 | 1 | 1 | 0.954 |
| Marmara (11) | 10/1 | 9 | 0.981 | 0.930 | 0.986 |
| Black Sea (11) | 11/0 | 11 | 1 | 1 | 1.041 |
| Aegean (9) | 8/1 | 7 | 0.972 | 0.920 | 0.887 |
| Matched Number | Accession Name (List No-Region, Province) | Matches at T = 2 (List No-Region, Province) |
|---|---|---|
| 1 | Kokulu Kara (5-Marmara, Bilecik) | Siyah Üzüm (17-Mediterranean, Hatay) |
| 2 | Yerli Kara (34-Aegean, Muğla) | Eski Kara (11-Aegean, Denizli) |
| 3 | Patlak Kara (45-Central Anatolia, Sivas) | Kara Üzüm (26-Marmara, Kırklareli) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arslan, N.; Yılmaz Baydu, F.; Hazrati, N.; Yüksel Özmen, C.; Ergönül, O.; Uysal, T.; Yaşasın, A.S.; Özer, C.; Boz, Y.; Kuleyin, Y.S.; et al. Genetic Diversity and Population Structure Analysis of Anatolian Kara Grapevine (Vitis vinifera L.) Germplasm Using Simple Sequence Repeats. Horticulturae 2023, 9, 743. https://doi.org/10.3390/horticulturae9070743
Arslan N, Yılmaz Baydu F, Hazrati N, Yüksel Özmen C, Ergönül O, Uysal T, Yaşasın AS, Özer C, Boz Y, Kuleyin YS, et al. Genetic Diversity and Population Structure Analysis of Anatolian Kara Grapevine (Vitis vinifera L.) Germplasm Using Simple Sequence Repeats. Horticulturae. 2023; 9(7):743. https://doi.org/10.3390/horticulturae9070743
Chicago/Turabian StyleArslan, Nur, Funda Yılmaz Baydu, Nahid Hazrati, Canan Yüksel Özmen, Onur Ergönül, Tamer Uysal, Ahmet Semih Yaşasın, Cengiz Özer, Yılmaz Boz, Yusuf Serhat Kuleyin, and et al. 2023. "Genetic Diversity and Population Structure Analysis of Anatolian Kara Grapevine (Vitis vinifera L.) Germplasm Using Simple Sequence Repeats" Horticulturae 9, no. 7: 743. https://doi.org/10.3390/horticulturae9070743





