Easy Synthesis of Doped Graphitic Carbon Nitride Nanosheets as New Material for Enhanced DNA Extraction from Vegetal Tissues Using a Simple and Fast Protocol
Abstract
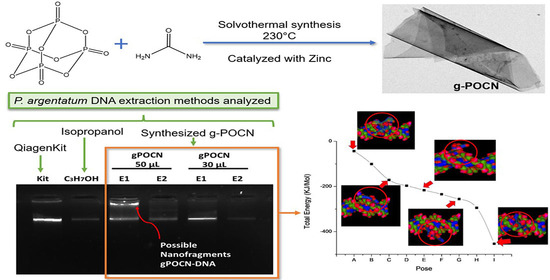
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Solvothermal Synthesis of Phosphorous-Doped g-POCN Catalyzed with Metallic Zinc
2.3. Nuclear Magnetic Resonance Spectroscopy (NMR)
2.4. Sample Preparation
2.5. Elemental Analysis
2.6. Powder X-ray Diffraction (PRXD)
2.7. Fourier Transform Infrared Spectroscopy (FTIR)
2.8. X-ray Photoelectron Spectroscopy (XPS)
2.9. Transmission Electron Microscopy (TEM)
2.10. Atomic Force Microscopy (AFM)
2.11. Protocol of DNA Extraction by Isopropanol
2.12. DNA Extraction with g-POCN
2.13. DNA Extraction Protocol with Qiagen Kit
2.14. DNA Amplification by PCR
2.15. Molecular Docking DNA/g-POCN
3. Results
3.1. PXRD Patterns
3.2. X-ray Photoelectron Spectroscopy
3.3. Fourier Transform Infrared Spectroscopy
3.4. Nuclear Magnetic Resonance
3.5. Elemental Analysis
3.6. TEM
3.7. AFM
3.8. DNA Extraction
3.9. DNA Extraction by Second Elution
3.10. Molecular Docking DNA/g-POCN
3.11. DNA Amplification by PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rono, N.; Kibet, J.K.; Martincigh, B.S.; Nyamori, V.O. A review of the current status of graphitic carbon nitride. Crit. Rev. Solid State Mater. Sci. 2020, 46, 189–217. [Google Scholar] [CrossRef]
- Alaghmandfard, A.; Ghandi, K. A Comprehensive Review of Graphitic Carbon Nitride (g-C3N4)–Metal Oxide-Based Nanocomposites: Potential for Photocatalysis and Sensing. Nanomaterials 2022, 12, 294. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.-H.; Ge, Q.; Jiang, N.; Cong, H.; Liu, M.; Zhang, Y.-Q. Graphitic carbon nitride (g-C3N4)-based photocatalytic materials for hydrogen evolution. Front. Chem. 2022, 10, 1369. [Google Scholar] [CrossRef] [PubMed]
- Goglio, G.; Denis, A.; Gaudin, E.; Labrugère, C.; Foy, D.; Largeteau, A. Solvothermal processes for nitride synthesis: Examples of Li 3GaN2 and graphitic C3N4 elaboration. Zeitschrift Naturforsch. Sect. B J. Chem. Sci. 2008, 63, 730–738. [Google Scholar] [CrossRef]
- Hu, C.C.; Wang, M.S.; Hung, W.Z. Influence of solvothermal synthesis on the photocatalytic degradation activity of carbon nitride under visible light irradiation. Chem. Eng. Sci. 2017, 167, 1–9. [Google Scholar] [CrossRef]
- Ong, W.J.; Tan, L.L.; Ng, Y.H.; Yong, S.T.; Chai, S.P. Graphitic Carbon Nitride (g-C3N4)-Based Photocatalysts for Artificial Photosynthesis and Environmental Remediation: Are We a Step Closer to Achieving Sustainability? Chem. Rev. 2016, 116, 7159–7329. [Google Scholar] [CrossRef]
- Darkwah, W.K.; Ao, Y. Mini Review on the Structure and Properties (Photocatalysis), and Preparation Techniques of Graphitic Carbon Nitride Nano-Based Particle, and Its Applications. Nanoscale Res. Lett. 2018, 13, 388. [Google Scholar] [CrossRef]
- Wang, A.; Wang, C.; Fu, L.; Wong-Ng, W.; Lan, Y. Recent Advances of Graphitic Carbon Nitride-Based Structures and Applications in Catalyst, Sensing, Imaging, and LEDs. Nano-Micro Lett. 2017, 9, 47. [Google Scholar] [CrossRef]
- Hashemi, E.; Akhavan, O.; Shamsara, M.; Valimehr, S.; Rahighi, R. DNA and RNA extractions from eukaryotic and prokaryotic cells by graphene nanoplatelets. RSC Adv. 2014, 4, 60720–60728. [Google Scholar] [CrossRef]
- Sreelakshmi, Y.; Gupta, S.; Bodanapu, R.; Chauhan, V.S.; Hanjabam, M.; Thomas, S.; Mohan, V.; Sharma, S.; Srinivasan, R.; Sharma, R. NEATTILL: A simplified procedure for nucleic acid extraction from arrayed tissue for TILLING and other high-throughput reverse genetic applications. Plant Methods 2010, 6, 3. [Google Scholar] [CrossRef]
- Ali, N.; Rampazzo, R.D.C.P.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res. Int. 2017, 2017, 9306564. [Google Scholar] [CrossRef] [Green Version]
- Meselson, M.; Stahl, F.W.; Vinograd, J. Equilibrium Sedimentation of Macromolecules in Density Gradients. Proc. Natl. Acad. Sci. USA 1957, 43, 581–588. [Google Scholar] [CrossRef] [Green Version]
- Bimboim, H.C.; Doly, J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef] [Green Version]
- Carpi, F.M.; Di Pietro, F.; Vincenzetti, S.; Mignini, F.; Napolioni, V. Human DNA extraction methods: Patents and applications. Recent Pat. DNA Gene Seq. 2011, 5, 1–7. [Google Scholar] [CrossRef]
- Saiyed, Z.M.; Ramchand, C.N. Extraction of Genomic DNA Using Magnetic Nanoparticles (Fe3O4) as a Solid-Phase Support. Am. J. Infect. Dis. 2007, 3, 225–229. [Google Scholar] [CrossRef] [Green Version]
- Dairawan, M.; Shetty, P.J. The Evolution of DNA Extraction Methods. Am. J. Biomed. Sci. Res. 2020, 8, 39–45. [Google Scholar] [CrossRef]
- Siegel, C.S.; Stevenson, F.O.; Zimmer, E.A. Evaluation and comparison of FTA card and CTAB DNA extraction methods for non-agricultural taxa. Appl. Plant Sci. 2017, 5, 1600109. [Google Scholar] [CrossRef]
- Pham, X.-H.; Baek, A.; Kim, T.H.; Lee, S.H.; Rho, W.-Y.; Chung, W.-J.; Kim, D.-E.; Jun, B.-H. Graphene Oxide Conjugated Magnetic Beads for RNA Extraction. Chem. Asian J. 2017, 12, 1883–1888. [Google Scholar] [CrossRef]
- Balaji, A.; Yang, S.; Wang, J.; Zhang, J. Graphene Oxide-Based Nanostructured DNA Sensor. Biosensors 2019, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Lubin, A.A.; Baker, B.R.; Plaxco, K.W.; Heeger, A.J. Single-step electronic detection of femtomolar DNA by target-induced strand displacement in an electrode-bound duplex. Proc. Natl. Acad. Sci. USA 2006, 103, 16677–16680. [Google Scholar] [CrossRef]
- Deshmukh, R.; Prusty, A.K.; Roy, U.; Bhand, S. A capacitive DNA sensor for sensitive detection of Escherichia coli O157:H7 in potable water based on the z3276 genetic marker: Fabrication and analytical performance. Analyst 2020, 145, 2267–2278. [Google Scholar] [CrossRef]
- Yan, Y.; Cheng, Q.; Wang, G.; Li, C. Growth of polyaniline nanowhiskers on mesoporous carbon for supercapacitor application. J. Power Sources 2011, 196, 7835–7840. [Google Scholar] [CrossRef]
- Xiao, Y.; Sheng, Y.; Zhou, J.; Chen, M.; Wen, W.; Zhang, X.; Wang, S. A novel label-free strategy for pathogenic DNA detection based on metal ion binding-induced fluorescence quenching of graphitic carbon nitride nanosheets. Analyst 2017, 142, 2617–2623. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, F.; Liao, S.; Chen, M.; Zhu, Y.Q.; Liu, Q.; Chen, X. Single-stranded DNA modified protonated graphitic carbon nitride nanosheets: A versatile ratiometric fluorescence platform for multiplex detection of various targets. Talanta 2019, 197, 422–430. [Google Scholar] [CrossRef]
- Pal, A.; Levy, Y. Structure, stability and specificity of the binding of ssDNA and ssRNA with proteins. PLoS Comput. Biol. 2019, 15, e1006768. [Google Scholar] [CrossRef]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 16, 138–149. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, K.; Rawal, K. An In-Silico Approach for Analysing the interplay of Hepatitis B viral X protein with Human Adaptin protein. J. Res. Pharm. 2022, 26, 483–493. [Google Scholar] [CrossRef]
- Kharlamov, A.; Bondarenko, M.; Kharlamova, G.; Fomenko, V. Synthesis of reduced carbon nitride at the reduction by hydroquinone of water-soluble carbon nitride oxide (g-C3N4 ) O. J. Solid State Chem. 2016, 241, 115–120. [Google Scholar] [CrossRef]
- Putri, L.K.; Ng, B.J.; Er, C.C.; Ong, W.J.; Chang, W.S.; Mohamed, A.R.; Chai, S.P. Insights on the impact of doping levels in oxygen-doped gC3N4 and its effects on photocatalytic activity. Appl. Surf. Sci. 2020, 504, 144427. [Google Scholar] [CrossRef]
- Starukh, H.; Praus, P. Doping of Graphitic Carbon Nitride with Non-Metal Elements and Its Applications in Photocatalysis. Catalysts 2020, 10, 1119. [Google Scholar] [CrossRef]
- Zhu, Y.P.; Ren, T.Z.; Yuan, Z.Y. Mesoporous Phosphorus-Doped g-C3N4 Nanostructured Flowers with Superior Photocatalytic Hydrogen Evolution Performance. ACS Appl. Mater. Interfaces 2015, 7, 16850–16856. [Google Scholar] [CrossRef]
- Lu, Y.-C.; Chen, J.; Wang, A.-J.; Bao, N.; Feng, J.-J.; Wang, W.; Shao, L. Facile synthesis of oxygen and sulfur co-doped graphitic carbon nitride fluorescent quantum dots and their application for mercury(II ) detection and bioimaging. J. Mater. Chem. C 2015, 3, 73–78. [Google Scholar] [CrossRef]
- Mazov, I.; Krasnikov, D.; Stadnichenko, A.; Kuznetsov, V.; Romanenko, A.; Anikeeva, O.; Tkachev, E. Direct vapor-phase bromination of multiwall carbon nanotubes. J. Nanotechnol. 2012, 2012, 954084. [Google Scholar] [CrossRef] [Green Version]
- Al-Gaashani, R.; Radiman, S.; Daud, A.R.; Tabet, N.; Al-Douri, Y. XPS and optical studies of different morphologies of ZnO nanostructures prepared by microwave methods. Ceram. Int. 2013, 39, 2283–2292. [Google Scholar] [CrossRef]
- Liu, J.; Li, W.; Duan, L.; Li, X.; Ji, L.; Geng, Z.; Huang, K.; Lu, L.; Zhou, L.; Liu, Z.; et al. A Graphene-like Oxygenated Carbon Nitride Material for Improved Cycle-Life Lithium/Sulfur Batteries. Nano Lett. 2015, 15, 5137–5142. [Google Scholar] [CrossRef]
- Rojas, J.V.; Toro-gonzalez, M.; Molina-higgins, M.C.; Castano, C.E. Facile radiolytic synthesis of ruthenium nanoparticles on graphene oxide and carbon nanotubes. Mater. Sci. Eng. B 2016, 205, 28–35. [Google Scholar] [CrossRef]
- She, X.; Liu, L.; Ji, H.; Mo, Z.; Li, Y.; Huang, L.; Du, D.; Xu, H.; Li, H. Template-free synthesis of 2D porous ultrathin nonmetal-doped g-C3N4 nanosheets with highly efficient photocatalytic H2 evolution from water under visible light. Appl. Catal. B Environ. 2016, 187, 144–153. [Google Scholar] [CrossRef]
- Boudou, J.P.; Paredes, J.I.; Cuesta, A.; Martínez-Alonso, A.; Tascón, J.M.D. Oxygen plasma modification of pitch-based isotropic carbon fibres. Carbon N. Y. 2003, 41, 41–56. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, L.; Liu, J.; Fan, X.; Wang, B.; Wang, M.; Ren, W.; Wang, J.; Li, M.; Shi, J.; et al. Brand new P-doped g-C3N4: Enhanced photocatalytic activity for H2 evolution and Rhodamine B degradation under visible light. J. Mater. Chem. A 2015, 3, 3862–3867. [Google Scholar] [CrossRef]
- Su, J.Y.; Geng, P.; Li, X.Y.; Zhao, Q.D.; Quan, X.; Chen, G.H. Novel phosphorus doped carbon nitride modified TiO2 nanotube arrays with improved photoelectrochemical performance. Nanoscale 2015, 7, 16282–16289. [Google Scholar] [CrossRef]
- Kumar, B.; Asadi, M.; Pisasale, D.; Sinha-ray, S.; Rosen, B.A.; Haasch, R.; Abiade, J.; Yarin, A.L.; Salehi-khojin, A. Renewable and metal-free carbon nanofibre catalysts for carbon dioxide reduction. Nat. Commun. 2013, 4, 2819. [Google Scholar] [CrossRef] [Green Version]
- Lan, D.H.; Wang, H.T.; Chen, L.; Au, C.T.; Yin, S.F. Phosphorous-modified bulk graphitic carbon nitride: Facile preparation and application as an acid-base bifunctional and efficient catalyst for CO2 cycloaddition with epoxides. Carbon N. Y. 2016, 100, 81–89. [Google Scholar] [CrossRef]
- Watson, V.J.; Delgado, C.N.; Logan, B.E. Improvement of activated carbons as oxygen reduction catalysts in neutral solutions by ammonia gas treatment and their performance in microbial fuel cells. POWER 2013, 242, 756–761. [Google Scholar] [CrossRef]
- Larkin, P.J. IR and Raman Spectroscopy—Principles and Spectral Interpretation; Elsevier: Oxford, UK, 2011; ISBN 9780123869845. [Google Scholar]
- WorldCat. Akademiiâ nauk SSSR.; Rossiĩskaiâ akademiiâ nauk. Zhurnal organicheskoĭ khimii; Izd-vo “Nauka”; 1965. Available online: https://www.worldcat.org/es/title/zhurnal-organicheskoi-khimii/oclc/2276562 (accessed on 10 November 2022).
- Finkel’shtein, A.I.; Boitsov, E.N. The Molecular Structure of 1,3,5-Triazine and Its Derivatives. Russ. Chem. Rev. 1962, 31, 712–720. [Google Scholar] [CrossRef]
- Mostafaei, A.; Zolriasatein, A. Synthesis and characterization of conducting polyaniline nanocomposites containing ZnO nanorods. Prog. Nat. Sci. Mater. Int. 2012, 22, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.S.; Jorge, A.B.; Suter, T.M.; Sella, A.; Corà, F.; McMillan, P.F. Carbon nitrides: Synthesis and characterization of a new class of functional materials. Phys. Chem. Chem. Phys. 2017, 19, 15613–15638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alwin, E.; Kočí, K.; Wojcieszak, R.; Zieliński, M.; Edelmannová, M.; Pietrowski, M. Influence of High Temperature Synthesis on the Structure of Graphitic Carbon Nitride and Its Hydrogen Generation Ability. Materials 2020, 13, 2756. [Google Scholar] [CrossRef]
- Seifer, G.B. Cyanuric acid and Cyanurates. Russ. J. Coord. Chem. Khimiya 2002, 28, 301–324. [Google Scholar] [CrossRef]
- Cranwell, P.B.; Harwood, L.M.; Moody, C.J. Experimental Organic Chemistry/Philippa; John Wiley & Sons Ltd: West Sussex, UK, 2017; ISBN 9781118683804. [Google Scholar]
- Scheer, H.; Katz, J.J. Nuclear Magnetic Resonance Spectroscopy of Porphyrins and Metalloporphyrins; Elsevier: Amsterdam, The Netherlands, 1975; ISBN 0-444-41375-8. [Google Scholar]
- Silverstein, R.M.; Robert, M.; Webster, F.X.; Kiemle, D.J.; Bryce, D.L.; David, L. Spectrometric Identification of Organic Compounds; John Wiley & Sons: Hoboken, NJ, USA, 2005; ISBN 0470616377. [Google Scholar]
- Mohan, J. Organic Analytical Chemistry: Theory and Practice; Alpha Science International Ltd: Oxford, UK, 2003; ISBN 9781842651193. [Google Scholar]
- Zhang, Y.; Mori, T.; Ye, J.; Antonietti, M. Phosphorus-Doped Carbon Nitride Solid Enhanced Electrical Conductivity. J. Am. Chem. Soc. 2010, 132, 6294–6295. [Google Scholar] [CrossRef]
- Mason , J.; Verkade, J.G.; Quin, L.D. (Eds.) Phosphorus-31 NMR spectroscopy in stereochemical analysis: Organic compounds and metal complexes. VCH, Deerfield Beach, FL, 1987, DM 258. Magn. Reson. Chem. 1988, 26, 178. [Google Scholar] [CrossRef]
- Schwarzer, A.; Saplinova, T.; Kroke, E. Tri-s-triazines (s-heptazines)-From a “mystery molecule” to industrially relevant carbon nitride materials. Coord. Chem. Rev. 2013, 257, 2032–2062. [Google Scholar] [CrossRef]
- Song, M.; Yu, L.; Wu, Y. Simple Synthesis and Enhanced Performance of Graphene Oxide-Gold Composites. J. Nanomater. 2012, 2012, 37. [Google Scholar] [CrossRef] [Green Version]
- Xue, R.; Xin, X.; Wang, L.; Shen, J.; Ji, F.; Li, W.; Jia, C.; Xu, G. A systematic study of the effect of molecular weights of polyvinyl alcohol on polyvinyl alcohol–graphene oxide composite hydrogels. Phys. Chem. Chem. Phys. 2015, 17, 5431–5440. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Gao, D.; Wang, B.; Zhao, R.; Guan, M.; Zheng, L.; Zhou, X.; Chai, Z.; Feng, W. Graphene oxide as an anaerobic membrane scaffold for the enhancement of B. adolescentis proliferation and antagonistic effects against pathogens E. coli and S. aureus. Nanotechnology 2014, 25, 165101. [Google Scholar] [CrossRef]
- Son, M.; Lim, H.; Hong, M.; Choi, H.C. Direct growth of graphene pad on exfoliated hexagonal boron nitride surface. Nanoscale 2011, 3, 3089. [Google Scholar] [CrossRef] [Green Version]
- Štengl, V.; Henych, J.; Slušná, M.; Ecorchard, P. Ultrasound exfoliation of inorganic analogues of graphene. Nanoscale Res. Lett. 2014, 9, 167. [Google Scholar] [CrossRef]
- Becker, L.; Steglich, M.; Fuchs, S.; Werner, G.; Nübel, U. Comparison of six commercial kits to extract bacterial chromosome and plasmid DNA for MiSeq sequencing. Sci. Rep. 2016, 6, 28063. [Google Scholar] [CrossRef] [Green Version]
- Sebastianelli, A.; Sen, T.; Bruce, I.J. Extraction of DNA from soil using nanoparticles by magnetic bioseparation. Lett. Appl. Microbiol. 2008, 46, 488–491. [Google Scholar] [CrossRef]
- Qiao, Z.; Liu, H.; Noh, G.S.; Koo, B.; Zou, Q.; Yun, K.; Jang, Y.O.; Kim, S.H.; Shin, Y. A Simple and Rapid Fungal DNA Isolation Assay Based on ZnO Nanoparticles for the Diagnosis of Invasive Aspergillosis. Micromachines 2020, 11, 515. [Google Scholar] [CrossRef]
- Sajali, N.; Wong, S.C.; Hanapi, U.K.; Abu Bakar Jamaluddin, S.; Tasrip, N.A.; Mohd Desa, M.N. The Challenges of DNA Extraction in Different Assorted Food Matrices: A Review. J. Food Sci. 2018, 83, 2409–2414. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Q.; Lei, J.; Ju, H. Electrochemiluminescent DNA sensing using carbon nitride nanosheets as emitter for loading of hemin labeled single-stranded DNA. Biosens. Bioelectron. 2015, 73, 7–12. [Google Scholar] [CrossRef]
- Muraru, S.; Samoila, C.G.; Slusanschi, E.I.; Burns, J.S.; Ionita, M. Molecular Dynamics Simulations of DNA Adsorption on Graphene Oxide and Reduced Graphene Oxide-PEG-NH2 in the Presence of Mg2+ and Cl− ions. Coatings 2020, 10, 289. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.M.; Liu, J.W.; Adkins, G.B.; Shen, W.; Trinh, M.P.; Duan, L.Y.; Jiang, J.H.; Zhong, W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017, 89, 12327–12333. [Google Scholar] [CrossRef] [PubMed]










| Catalyzed | |
|---|---|
| Element | Content |
| Carbon | 15.44% |
| Nitrogen | 27.93% |
| Oxygen | 39.38% |
| Hydrogen | 3.065% |
| Phosphorus | 14.17% |
| Total Concentration gDNA (ng/uL) | OD (260 nm)/OD (280 nm) | |||
|---|---|---|---|---|
| Extraction Method | Elution 1 | Elution 2 | Elution 1 | Elution 2 |
| g-POCN 30 μL | 316.35 | 142.15 | 1.88 | 1.9 |
| g-POCN 50 μL | 365.58 | 86.13 | 1.85 | 1.75 |
| Isopropanol | 296.24 | - | 0.7 | - |
| Qiagen Kit | 97.39 | - | 1.79 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cartagena, M.E.; Bernal-Martínez, J.; Banda-Villanueva, A.; Lechuga-Islas, V.D.; Córdova, T.; Magaña, I.; Torres-Lubián, J.R.; Fernández-Tavizón, S.; Romero-García, J.; Rodríguez-Hernández, A.M.; et al. Easy Synthesis of Doped Graphitic Carbon Nitride Nanosheets as New Material for Enhanced DNA Extraction from Vegetal Tissues Using a Simple and Fast Protocol. C 2022, 8, 68. https://doi.org/10.3390/c8040068
Martínez-Cartagena ME, Bernal-Martínez J, Banda-Villanueva A, Lechuga-Islas VD, Córdova T, Magaña I, Torres-Lubián JR, Fernández-Tavizón S, Romero-García J, Rodríguez-Hernández AM, et al. Easy Synthesis of Doped Graphitic Carbon Nitride Nanosheets as New Material for Enhanced DNA Extraction from Vegetal Tissues Using a Simple and Fast Protocol. C. 2022; 8(4):68. https://doi.org/10.3390/c8040068
Chicago/Turabian StyleMartínez-Cartagena, Manuel Eduardo, Juan Bernal-Martínez, Arnulfo Banda-Villanueva, Víctor D. Lechuga-Islas, Teresa Córdova, Ilse Magaña, José Román Torres-Lubián, Salvador Fernández-Tavizón, Jorge Romero-García, Ana Margarita Rodríguez-Hernández, and et al. 2022. "Easy Synthesis of Doped Graphitic Carbon Nitride Nanosheets as New Material for Enhanced DNA Extraction from Vegetal Tissues Using a Simple and Fast Protocol" C 8, no. 4: 68. https://doi.org/10.3390/c8040068