Non-Coding RNA Databases in Cardiovascular Research
Abstract
:1. Introduction
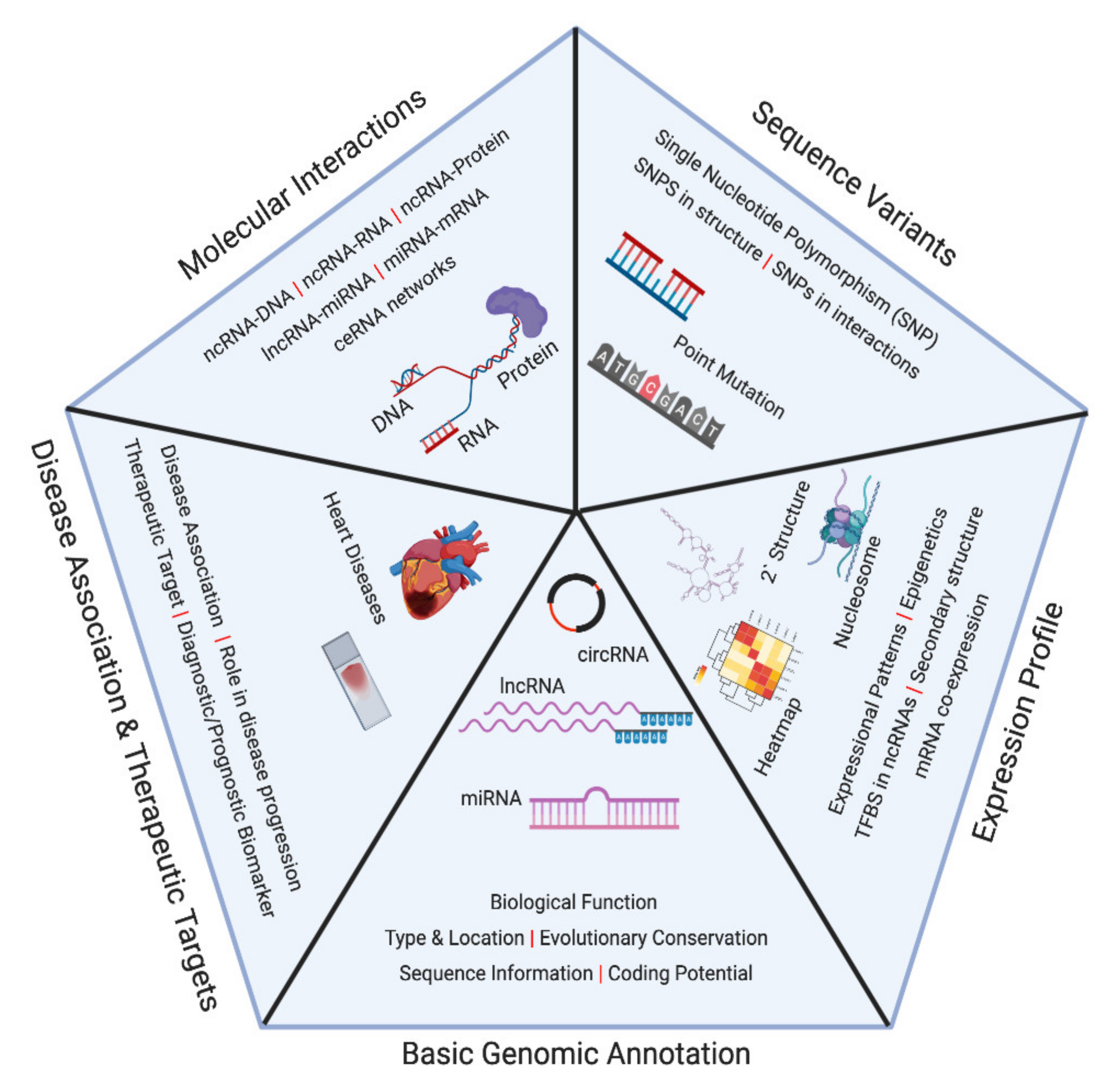
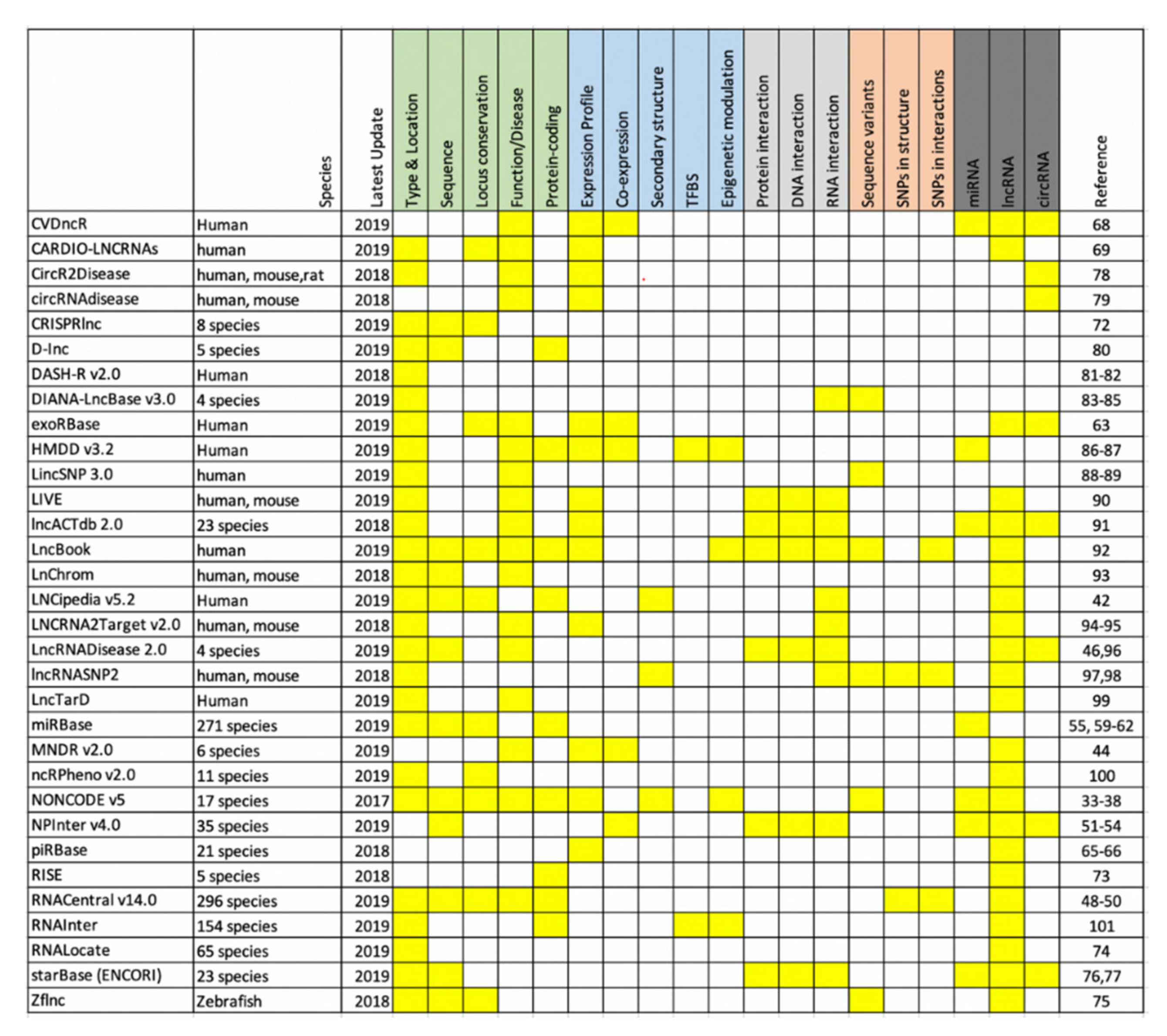
2. Noncoding RNA Databases
2.1. NONCODE V5
2.2. RNAcentral v14.0
2.3. NPInter v4.0
2.4. miRbase v22
2.5. exoRBase
2.6. piRBase
3. Noncoding RNA Databases in Cardiovascular Diseases
3.1. CVDncR
3.2. CARDIO-LNCRNAs
4. Other Resources
5. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AF | Atrial Fibrillation |
| AS | Atherosclerosis |
| asRNA | Antisense RNA |
| CAD | Coronary Artery Disease |
| cANRIL | Circular Antisense Noncoding RNA in the INK4 Locus |
| CCNL2 | Cyclin L2 |
| ceRNA | Competing endogenous RNA |
| CHD | Congenital Heart Disease |
| circRNA | Circular RNA |
| CLIP-Seq | Cross-linking Immunoprecipitation sequencing |
| ChiRP-Seq | Chromatin Isolation by RNA purification |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CSF | Cerebrospinal Fluid |
| CVD | Cardiovascular Diseases |
| eRNA | Enhancer RNA |
| GEO | Gene Expression Omnibus |
| GWAS | Genome Wide Association Studies |
| HF | Heart Failure |
| IHF | Ischemic Heart Failure |
| KLF4 | Kruppel Like Factor 4 |
| lncRNA | Long non-coding RNA |
| MeSH | Medical Subject Headings |
| MHRT | Myosin Heavy Chain Associated RNA Transcript |
| miRNA | MicroRNA |
| ncRNA | Non-coding RNA |
| OMIM | Online Mendelian Inheritance in Man |
| ORF | Open Reading Frame |
| piRNA | Piwi-interacting RNA |
| RNA | Ribo Nucleic Acid |
| rRNA | Ribosomal RNA |
| SLE | Systemic Lupus Erythematosus |
| SNP | Single Nucleotide Polymorphism |
| siRNA | Small interfering RNA |
| snRNA | Small nuclear RNA |
| snoRNA | Small nucleolar RNA |
| tRNA | Transfer RNA |
| WBC | White Blood Cells |
References
- World Health Organization. Cardiovascular Diseases (CVDs) Fact Sheet. Available online: http://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 7 November 2019).
- Wilkins, E.; Wilson, L.; Wickramasinghe, K.; Bhatnagar, P.; Leal, J.; Luengo-Fernandez, R.; Burrns, R.; Rayner, M.; Townsend, N. European Cardiovascular Disease Statistics 2017, 5th ed.; Løgstrup, S., Ed.; European Heart Network AISBL: Brussels, Belgium, 2017; pp. 1–192. [Google Scholar]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.M.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M. The Human Transcriptome: An Unfinished Story. Genes 2012, 3, 344–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalali, S.; Gandhi, S.; Scaria, V. Navigating the dynamic landscape of long noncoding RNA and protein-coding gene annotations in GENCODE. Hum. Genom. 2016, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zheng, X.; Zheng, Y.; Cao, R.; Zhang, M.; Sun, Y.; Wu, J. Construction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveals functional genes in heart failure. Mol. Med. Rep. 2018, 19, 994–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.; Gao, S.; Chen, Y.; Xu, B.; Yu, C.; Yue, M.; Tan, X. Integrative analysis of competing endogenous RNA networks reveals the functional lncRNAs in heart failure. J. Cell. Mol. Med. 2018, 22, 4818–4829. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Pedrotty, D.M.; Morley, M.P.; Cappola, T.P. Transcriptomic biomarkers of cardiovascular disease. Prog. Cardiovasc. Dis. 2012, 55, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.-Y.; Chandramohan, R.; Phan, J.H.; Mahle, W.T.; Gaynor, J.W.; Maher, K.O.; Wang, M.D. Cardiovascular transcriptomics and epigenomics using next-generation sequencing: Challenges, progress, and opportunities. Circ. Cardiovasc. Genet. 2014, 7, 701–710. [Google Scholar] [CrossRef] [Green Version]
- De Franciscis, S.; Metzinger, L.; Serra, R. The Discovery of Novel Genomic, Transcriptomic, and Proteomic Biomarkers in Cardiovascular and Peripheral Vascular Disease: The State of the Art. BioMed Res. Int. 2016, 2016, 7829174. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Luo, Y.; Liang, C.; Zhang, T.C. LncRNA-Mhrt regulates cardiac hypertrophy by modulating the miR-145a-5p/KLF4/myocardin axis. J. Mol. Cell. Cardiol. 2020, 139, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Xuan, L.; Sun, L.; Zhang, Y.; Huang, Y.; Hou, Y.; Li, Q.; Guo, Y.; Feng, B.; Cui, L.; Wang, X. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J. Cell. Mol. Med. 2017, 21, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-E.; Wu, Y.-J.; Zhang, S.-L. Circulating lncRNA MHRT predicts survival of patients with chronic heart failure. J. Geriatr. Cardiol. 2019, 16, 818–821. [Google Scholar] [PubMed]
- Lin, F.; Gong, X.; Yu, P.; Yue, A.; Meng, Q.; Zheng, L.; Chen, T.; Han, L.; Cao, H.; Cao, J. Distinct Circulating Expression Profiles of Long Noncoding RNAs in Heart Failure Patients With Ischemic and Nonischemic Dilated Cardiomyopathy. Front. Genet. 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier, S.; Henkens, M.; Heymans, S.; Robinson, E.L. Unlocking the Value of White Blood Cells for Heart Failure Diagnosis. J. Cardiovasc. Transl. Res. 2020, 1–10. [Google Scholar] [CrossRef]
- Bartekova, M.; Radosinska, J.; Jelemensky, M.; Dhalla, N.S. Role of cytokines and inflammation in heart function during health and disease. Heart Fail. Rev. 2018, 23, 733–758. [Google Scholar] [CrossRef]
- Mann, D.L. The emerging role of innate immunity in the heart and vascular system: For whom the cell tolls. Circ. Res. 2011, 108, 1133–1145. [Google Scholar] [CrossRef]
- Adamo, L.; Rocha-Resende, C.; Prabhu, S.D.; Mann, D.L. Reappraising the role of inflammation in heart failure. Nat. Rev. Cardiol. 2020, 17, 269–285. [Google Scholar] [CrossRef]
- Fritah, S.; Niclou, S.P.; Azuaje, F. Databases for lncRNAs: A comparative evaluation of emerging tools. RNA 2014, 20, 1655–1665. [Google Scholar] [CrossRef] [Green Version]
- Jalali, S.; Kapoor, S.; Sivadas, A.; Bhartiya, D.; Scaria, V. Computational approaches towards understanding human long non-coding RNA biology. Bioinformatics 2015, 31, 2241–2251. [Google Scholar] [CrossRef] [Green Version]
- Iwakiri, J.; Hamada, M.; Asai, K. Bioinformatics tools for lncRNA research. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2016, 1859, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Rühle, F.; Stoll, M. Long non-coding RNA Databases in Cardiovascular Research. Genom. Proteom. Bioinform. 2016, 14, 191–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Arora, P. Long Noncoding Mhrt RNA. Circ. Cardiovasc. Genet. 2015, 8, 213–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Pan, M.; Zhou, J.; Yin, F. Overexpression of long non-coding RNA ANRIL promotes post-ischaemic angiogenesis and improves cardiac functions by targeting Akt. J. Cell. Mol. Med. 2020, 24, 6860–6868. [Google Scholar] [CrossRef]
- Ucar, A.; Gupta, S.K.; Fiedler, J.; Erikci, E.; Kardasinski, M.; Bátkai, S.; Dangwal, S.; Kumarswamy, R.; Bang, C.; Holzmann, A. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012, 3, 1078. [Google Scholar] [CrossRef]
- Foinquinos, A.; Batkai, S.; Genschel, C.; Viereck, J.; Rump, S.; Gyöngyösi, M.; Traxler, D.; Riesenhuber, M.; Spannbauer, A.; Lukovic, D.; et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat. Commun. 2020, 11, 633. [Google Scholar] [CrossRef]
- Cao, H.; Li, D.; Lu, H.; Sun, J.; Li, H. Uncovering potential lncRNAs and nearby mRNAs in systemic lupus erythematosus from the Gene Expression Omnibus dataset. Epigenomics 2019, 11, 1795–1809. [Google Scholar] [CrossRef]
- Hiraki, Y.; Fujita, H.; Yamamori, S.; Ohashi, H.; Eguchi, M.; Harada, N.; Mizuguchi, T.; Matsumoto, N. Mild craniosynostosis with 1p36.3 trisomy and 1p36.3 deletion syndrome caused by familial translocation t(Y;1). Am. J. Med. Genet. Part A 2006, 140, 1773–1777. [Google Scholar] [CrossRef]
- Gajecka, M.; Yu, W.; Ballif, B.C.; Glotzbach, C.D.; Bailey, K.A.; Shaw, C.A.; Kashork, C.D.; Heilstedt, H.A.; Ansel, D.A.; Theisen, A. Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur. J. Hum. Genet. 2004, 13, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Bai, B.; Skogerbø, G.; Cai, L.; Deng, W.; Zhang, Y.; Bu, N.; Zhao, Y.; Chen, R. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2004, 33, D112–D115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.; Liu, C.; Skogerbø, G.; Zhao, H.; Wang, J.; Liu, T.; Bai, B.; Zhao, Y.; Chen, R. NONCODE v2.0: Decoding the non-coding. Nucleic Acids Res. 2007, 36, D170–D172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, D.; Yu, K.; Sun, S.; Xie, C.; Skogerbø, G.; Miao, R.; Xiao, H.; Liao, Q.; Luo, H.; Zhao, G. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2011, 40, D210–D215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Yuan, J.; Li, H.; Li, M.; Zhao, G.; Bu, D.; Zhu, W.; Wu, W.; Chen, R.; Zhao, Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2013, 42, D98–D103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2015, 44, D203–D208. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2017, 46, D308–D314. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2020, 48, D84–D86. [Google Scholar] [CrossRef] [Green Version]
- Yates, A.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J. Ensembl 2020. Nucleic Acids Res. 2019, 48, D682–D688. [Google Scholar] [CrossRef]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.V.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2014, 43, D168–D173. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2018, 47, D135–D139. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Qu, R.; Huang, Y.; Shi, B.; Liu, M.; Li, Y.; Lu, Z.J. RNAex: An RNA secondary structure prediction server enhanced by high-throughput structure-probing data. Nucleic Acids Res. 2016, 44, W294–W301. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Zhang, L.; Huang, Y.; Yi, Y.; Tan, P.; Zhao, Y.; Hu, Y.; Xu, L.-Y.; Li, E.; Wang, D. MNDR v2.0: An updated resource of ncRNA–disease associations in mammals. Nucleic Acids Res. 2017, 46, D371–D374. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, P.; Wang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Li, X.; Fang, Y.; Shen, W.; Xu, Y. Lnc2Cancer v2.0: Updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2018, 47, D1028–D1033. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, N. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2018, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [Green Version]
- The RNAcentral Consortium; Sweeney, B.A.; Petrov, A.I.; Burkov, B.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Gorodkin, J.; Seemann, E.S.; et al. RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2018, 47, D221–D229. [Google Scholar] [CrossRef] [Green Version]
- The RNAcentral Consortium; Petrov, A.I.; Kay, S.J.E..; Gibson, R.C.; Kulesha, E.; Staines, D.M.; Bruford, E.A.; Wright, M.W.; Burge, S.; Finn, R.D.; et al. RNAcentral: An international database of ncRNA sequences. Nucleic Acids Res. 2014, 43, D123–D129. [Google Scholar] [CrossRef] [Green Version]
- The RNAcentral Consortium; Petrov, A.I.; Kay, S.J.E.; Kalvari, I.; Howe, K.L.; Gray, K.; Bruford, E.A.; Kersey, P.J.; Cochrane, G.; Finn, R.D.; et al. RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2016, 45, D128–D134. [Google Scholar] [CrossRef]
- Wu, T.P. NPInter: The noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006, 34, D150–D152. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Wu, W.; Xie, C.; Zhao, G.; Zhao, Y.; Chen, R. NPInter v2.0: An updated database of ncRNA interactions. Nucleic Acids Res. 2013, 42, D104–D108. [Google Scholar] [CrossRef]
- Hao, Y.; Wu, W.; Li, H.; Yuan, J.; Luo, J.; Zhao, Y.; Chen, R. NPInter v3.0: An upgraded database of noncoding RNA-associated interactions. Database 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Teng, X.; Chen, X.; Xue, H.; Tang, Y.; Zhang, P.; Kang, Q.; Hao, Y.; Chen, R.; Zhao, Y.; He, S. NPInter v4.0: An integrated database of ncRNA interactions. Nucleic Acids Res. 2020, 48, D160–D165. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- The UniProt Consortium; UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; Van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010, 39, D152–D157. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2017, 46, D106–D112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Si, X.; Skogerbø, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database 2014, 2014, bau110. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, P.; Lu, Y.; Li, Y.; Zheng, Y.; Kan, Y.; Chen, R.; He, S. piRBase: A comprehensive database of piRNA sequences. Nucleic Acids Res. 2018, 47, D175–D180. [Google Scholar] [CrossRef] [Green Version]
- Lakshmi, S.S.; Agrawal, S. piRNABank: A web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2007, 36, D173–D177. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Lin, Y.; Liu, X.; Zhan, C.; He, H.; Shi, M.; Jiang, Z.; Shen, B. Phenotype–genotype network construction and characterization: A case study of cardiovascular diseases and associated non-coding RNAs. Database 2020, 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda-Silva, D.; Wüst, R.C.I.; Conceição, G.; Gonçalves-Rodrigues, P.; Gonçalves, N.; Gonçalves, A.; Kuster, D.W.D.; Leite-Moreira, A.F.; Van Der Velden, J.; Beleza, J.M.D.S.; et al. Disturbed cardiac mitochondrial and cytosolic calcium handling in a metabolic risk-related rat model of heart failure with preserved ejection fraction. Acta Physiol. 2019, 228, e13378. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Kolesnikov, N.; Hastings, E.; Keays, M.; Melnichuk, O.; Tang, Y.; Williams, E.; Dylag, M.; Kurbatova, N.; Brandizi, M.; Burdett, T.; et al. ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 2014, 43, D1113–D1116. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, G.; Li, J.; Zhang, X.; Huang, S.; Xiang, S.; Hu, X.; Liu, C. CRISPRlnc: A manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 2018, 47, D63–D68. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Shao, D.; Xu, K.; Lu, Z.; Lu, Z.J.; Yang, Y.T.; Zhang, Q.C. RISE: A database of RNA interactome from sequencing experiments. Nucleic Acids Res. 2017, 46, D194–D201. [Google Scholar] [CrossRef]
- Zhang, T.; Tan, P.; Wang, L.; Jin, N.; Li, Y.; Zhang, L.; Yang, H.; Hu, Z.; Zhang, L.; Hu, C.; et al. RNALocate: A resource for RNA subcellular localizations. Nucleic Acids Res. 2016, 45, D135–D138. [Google Scholar] [CrossRef]
- Hu, X.; Chen, W.; Li, J.; Huang, S.; Xu, X.; Zhang, X.; Xiang, S.; Liu, C. ZFLNC: A comprehensive and well-annotated database for zebrafish lncRNA. Database 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.-H.; Li, J.-H.; Shao, P.; Zhou, H.; Chen, Y.-Q.; Qu, L.-H. starBase: A database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2010, 39, D202–D209. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Lei, X.; Fang, Z.; Jiang, Q.; Wu, F.-X. CircR2Disease: A manually curated database for experimentally supported circular RNAs associated with various diseases. Database 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Wang, K.; Wu, F.; Wang, W.; Zhang, K.; Hu, H.; Liu, Y.; Jiang, T. circRNA disease: A manually curated database of experimentally supported circRNA-disease associations. Cell. Death Dis. 2018, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Qu, Y.; Yang, Q.; Ma, X.; Meng, Q.; Xu, J.; Liu, X.; Wang, S. D-lnc: A comprehensive database and analytical platform to dissect the modification of drugs on lncRNA expression. RNA Biol. 2019, 16, 1586–1591. [Google Scholar] [CrossRef]
- Kuksa, P.P.; Amlie-Wolf, A.; Katanić, Ž.; Valladares, O.; Wang, L.-S.; Leung, Y.Y. DASHR 2.0: Integrated database of human small non-coding RNA genes and mature products. Bioinformatics 2019, 35, 1033–1039. [Google Scholar] [CrossRef] [Green Version]
- Leung, Y.Y.; Kuksa, P.P.; Amlie-Wolf, A.; Valladares, O.; Ungar, L.H.; Kannan, S.; Gregory, B.D.; Wang, L.-S. DASHR: Database of small human noncoding RNAs. Nucleic Acids Res. 2015, 44, D216–D222. [Google Scholar] [CrossRef] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2019, 48, D101–D110. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Vlachos, I.; Karagkouni, D.; Georgakilas, G.; Kanellos, I.; Vergoulis, T.; Zagganas, K.; Tsanakas, P.; Floros, E.; Dalamagas, T.; et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2015, 44, D231–D238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Reczko, M.; Maragkakis, M.; Dalamagas, T.M.; Hatzigeorgiou, A.G. DIANA-LncBase: Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2012, 41, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA–disease associations. Nucleic Acids Res. 2018, 47, D1013–D1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2013, 42, D1070–D1074. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Yue, M.; Wang, P.; Liu, Y.; Zhi, H.; Zhang, Y.; Zhang, J.; Gao, Y.; Guo, M.; Zhou, D. LincSNP 2.0: An updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2016, 45, D74–D78. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Zhao, Z.; Ye, J.; Wang, P.; Zhi, H.; Li, R.; Wang, T.; Li, X. LincSNP: A database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinform. 2014, 15, 152. [Google Scholar] [CrossRef] [Green Version]
- An, G.; Sun, J.; Ren, C.; Ouyang, Z.; Zhu, L.-Y.; Bo, X.; Peng, S.; Shu, W. LIVE: A manually curated encyclopedia of experimentally validated interactions of lncRNAs. Database 2019, 2019. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Li, X.; Gao, Y.; Guo, Q.; Wang, Y.; Fang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Shen, W.; et al. LncACTdb 2.0: An updated database of experimentally supported ceRNA interactions curated from low- and high-throughput experiments. Nucleic Acids Res. 2018, 47, D121–D127. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Cao, J.; Liu, L.; Du, Q.; Li, Z.; Zou, D.; Bajic, V.B.; Zhang, Z. LncBook: A curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2018, 47, D128–D134. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Zhang, G.; Shi, A.; Hu, J.; Li, F.; Zhang, X.; Zhang, Y.; Huang, J.; Xiao, Y.; Li, X.; et al. LnChrom: A resource of experimentally validated lncRNA–chromatin interactions in human and mouse. Database 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2018, 47, D140–D144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Q.; Wang, J.; Wu, X.; Ma, R.; Zhang, T.; Jin, S.; Han, Z.; Tan, R.; Peng, J.; Liu, G.; et al. LncRNA2Target: A database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2014, 43, D193–D196. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012, 41, D983–D986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.-R.; Liu, W.; Zhang, Q.; Guo, A.-Y. lncRNASNP2: An updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2017, 46, D276–D280. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Liu, W.; Zhang, J.; Miao, X.; Guo, A.-Y. lncRNASNP: A database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2014, 43, D181–D186. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Zhang, Y.; Xie, A.; Yu, L.; Zhang, C.; Lei, J.; Xu, H.; Leng, Z.; Li, T.; et al. LncTarD: A manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 2019, 48, D118–D126. [Google Scholar] [CrossRef]
- Zhang, W.; Yao, G.; Wang, J.; Yang, M.; Wang, J.; Zhang, H.; Li, W. ncRPheno: A comprehensive database platform for identification and validation of disease related noncoding RNAs. RNA Biol. 2020, 17, 943–955. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2019, 48, D189–D197. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balamurali, D.; Stoll, M. Non-Coding RNA Databases in Cardiovascular Research. Non-Coding RNA 2020, 6, 35. https://doi.org/10.3390/ncrna6030035
Balamurali D, Stoll M. Non-Coding RNA Databases in Cardiovascular Research. Non-Coding RNA. 2020; 6(3):35. https://doi.org/10.3390/ncrna6030035
Chicago/Turabian StyleBalamurali, Deepak, and Monika Stoll. 2020. "Non-Coding RNA Databases in Cardiovascular Research" Non-Coding RNA 6, no. 3: 35. https://doi.org/10.3390/ncrna6030035




