Assessment of Genetic Diversity among Pleurotus spp. Isolates from Jordan
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Pure Culture Preparation
2.2. Morphological Characterization
2.3. Dna Extraction
2.4. ITS and Nlsu Amplification and Sequence Analysis
2.5. Issr Analysis
3. Results
3.1. Morphological Characteristics of Jordanian Pleurotus Isolates
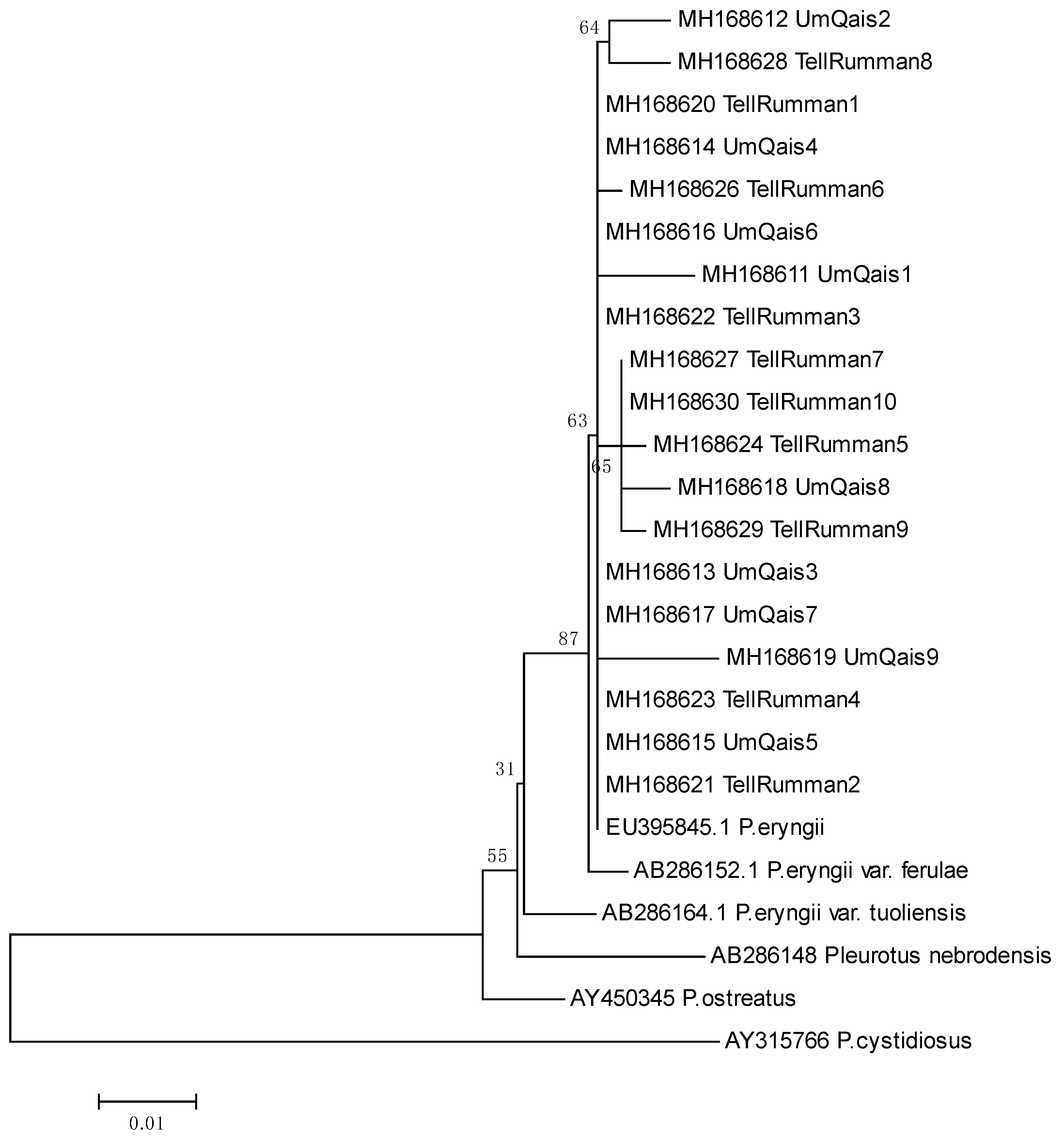
3.2. Molecular Identification Using ITS Analysis
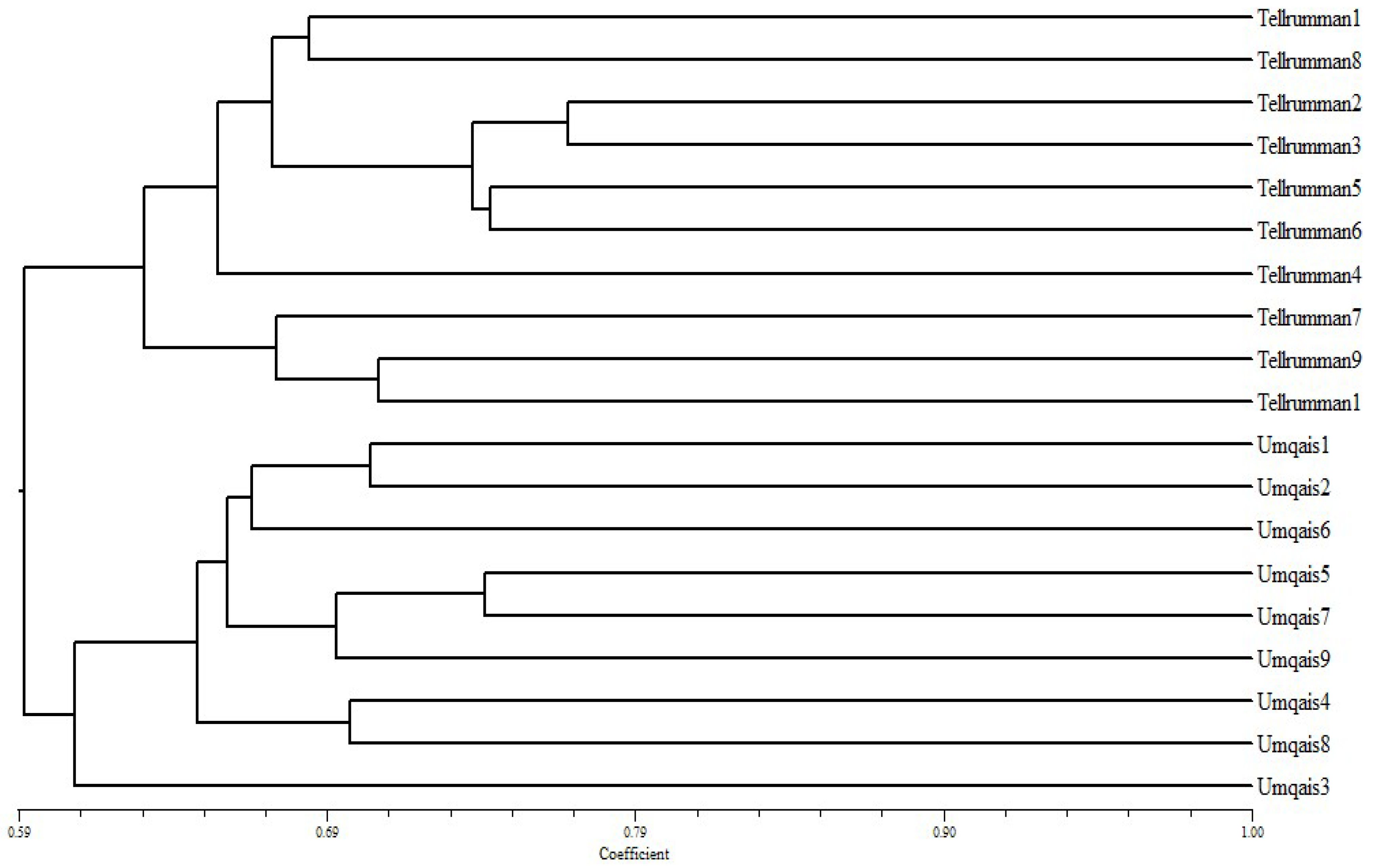
3.3. Genetic Diversity by Using ISSR
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| BLAST | Basic Local Alignment Search Tool |
| ISSR | Inter simple short repeat |
| ITS | Inter transcribed spacer |
| PDA | Potato dextrose Agar |
| BlASTn | nucleotide-nucleotide blast |
| NTSys | Numerical Taxonomy Multivariate Analysis System |
| PIC | Polymorphism Information Content |
| UPGMA | Unweighted Pair Group Method with Average |
References
- Alam, N.; Shim, M.J.; Lee, M.W.; Shin, P.G.; Yoo, Y.B.; Lee, T.S. Vegetative Growth and Phylogenetic Relationship of Commercially Cultivated Strains of Pleurotus eryngii based on ITS sequence and RAPD. MYCO 2009, 37, 258–266. [Google Scholar]
- Hassan, F.R.; Medany, G.M.; Abou Hussein, S.D. Cultivation of the King Oyster Mushroom (Pleurotus eryngii) in Egypt. Aust. J. Basic Appl. Sci. 2010, 4, 99–105. [Google Scholar]
- Rodriguez Estrada, A.E.; Royse, D.J. Yield, size and bacterial blotch resistance of Pleurotus eryngii grown on cotton seed hulls/oak sawdust supplemented with manganese, copper and whole ground soybean. Bioresour. Technol. 2007, 98, 1898–1906. [Google Scholar] [CrossRef] [PubMed]
- Behnamian, M.; Mohammadi, S.A.; Sonnenberg, A.; Goltapeh, E.M.; Alavi, A.; Hendrickx, P. Genetic diversity and population structure of Iranian wild Pleurotus eryngii species-complex strains revealed by URP-PCR markers. J. Food Agric. Environ. 2010, 8, 1203–1207. [Google Scholar]
- Choi, U.K.; Lee, O.H.; Kim, Y.C. Effect of Calcinated Oyster Shell Powder on Growth, Yield, Spawn Run, and Primordial Formation of King Oyster Mushroom (Pleurotus eryngii). Molecules 2011, 16, 2313–2322. [Google Scholar] [CrossRef] [PubMed]
- Ola, I.O.; Unni, B.G.; Oloke, J.K.; Bordoloi, A.K. Morphological, ultrastructural and molecular variability studies of wild and mutant strains of edible Pleurotus species using growth yield, scanning electron microscopy and random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR). Afr. J. Micro Res. 2013, 7, 5057–5065. [Google Scholar] [CrossRef]
- Prasad, M.P.; Agarwal, K. DNA fingerprinting of commercial mushrooms by ISSR and SSR markers for genetic determination. Int. J. Pharma Bio Sci. 2013, 4, 1243–1249. [Google Scholar]
- Wang, S.; Yin, Y.; Liu, Y. Evaluation of Genetic diversity among Chinese Pleurotus eryngii cultivars by combined RAPD/ISSR marker. Curr. Microbiol. 2012, 65, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Boa, E.R. Wild Edible Fungi: A Global Overview of Their Use and Importance to People; Food and Agriculture Organization of the United Nations (FAO): Roma, Italy, 2004; p. 147. [Google Scholar]
- Urbanelli, S.; Della Rosa, V.; Punelli, F.; Porreta, D.; Reverberi, M.; Fabbri, A.A.; Fanelli, C. DNA-fingerprinting (AFLP and RFLP) for genotypic identification in species of the Pleurotus eryngii complex. Appl. Microbiol. Biotechnol. 2007, 74, 92–600. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Ghosh, K.; Acharya, K. Comparative study on the Indian cultivated Pleurotus species by RAPD fingerprinting. Nat. Sci. 2010, 8, 90–94. [Google Scholar]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coisacc, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Imtiaj, A.; Lee, T.S.; Ohga, S. Sequence variation of Pleurotus species collected from Eastern Asia. Micol. Apl. Int. 2011, 23, 1–10. [Google Scholar]
- Avin, F.A.; Yee, S.B.; Tan, S.; Shahbazi, P.; Vikineswary, S. Molecular Divergence and Species Delimitation of the Cultivated Oyster Mushrooms: Integration of IGS1 and ITS. Sci. World J. 2014, 2014, 793414. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Estrada, A.E.; Jimenez-Gasco, M.M.; Royse, D.J. Pleurotus eryngii species complex: Sequence analysis and phylogeny based on partial EF1α and RPB2 genes. Fungal Biol. 2010, 114, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, A.; Janusz, G.; Koszerny, J.; Małek, W.; Rogalski, J. Genetic diversity of the edible mushroom Pleurotus sp. By amplified fragment length polymorphism. Curr. Microbiol. 2012, 65, 438–445. [Google Scholar] [PubMed]
- Yin, Y.; Liu, Y.; Wang, S.; Zhao, S.; Xu, F. Examining genetic relationships of Chinese Pleurotus ostreatus cultivars by combined RAPD and SRAP markers. Mycoscience 2014, 54, 221–225. [Google Scholar] [CrossRef]
- Phillips, R.; Mushrooms, A. Comprehensive Guide with over 1250 Detailed Photographs of Mushrooms and Other Fungi, 1st ed.; Macdonaki, C., Price, P., Eds.; Pan Macmillan: London, UK, 2006. [Google Scholar]
- Lincoff, G. The Audubon Society Field Guide to North American Mushrooms; Knopf: New York, NY, USA, 1981. [Google Scholar]
- Al-Momany, A.M. Nutritional Aspects of Two Widely Consumed Wild Edible Mushrooms in Jordan. Life Sci. 2013, 7, 653–657. [Google Scholar]
- Das, S.K.; Mandal, A.; Datta, A.K.; Gupta, S.; Paul, R.; Saha, A.; Sengupta, S.; Kumari, P. Nucleotide sequencing and identification of some wild mushrooms. Sci. World J. 2013, 2013, 403191. [Google Scholar] [CrossRef] [PubMed]
- Junior, N.M.; Asia, T.; Capelari, M.; Meirellus, L.D.P. Morphological and molecular identification of four Brazilian commercial isolates of Pleurotus spp. and cultivation on corncob. Braz. Arch. Biol. Technol. 2010, 53, 397–408. [Google Scholar] [CrossRef]
- De Gioia, T.; Sisto, D.; Rana, G.L.; Figliuolo, G. Genetic structure of the Pleurotus eryngii species-complex. Mycol. Res. 2005, 109, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, version 2.02; Exeter Publications: New York, NY, USA, 2000. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Kawai, G.; Babasaki, K.; Neda, H. Taxonomic position of a Chinese Pleurotus “Bai-Ling-Gu”: It belongs to Pleurotus eryngii (DC.: Fr.) Quél. and evolved independently in China. Mycoscience 2008, 49, 75–87. [Google Scholar] [CrossRef]
- Chengshu, Q.; Yan, W.; Li, P.; Deng, W.; Song, B.; Li, T. Evaluation of growth characterstics and genetic diversity of commercial and stored lines of Hypsizygus marmoreus. Int. J. Agric. Biol. 2013, 15, 479–485. [Google Scholar]
- Abdulmalk, H.W. Rapid Differentiation of Pleurotus ostreatus from Pleurotus sapidus using PCR Technique. Curr. Res. J. Biol. Sci. 2013, 5, 157–160. [Google Scholar]
- Liu, S.; Wu, X.; Liu, X.; Ke, B. Correlation between mating compatibility and the phylogenetic relationship of a rare edible mushroom, Pleurotus nebrodensis, with different Pleurotus species. Int. J. Agric. Biol. 2016, 18, 198–203. [Google Scholar] [CrossRef]
- Rodriguez Estrada, A.E.; Royse, D.J.; Jimenez-Gasco, M.M. Nucleotide sequence polymorphisms of the partial b-tubulin gene in two varieties of Pleurotus eryngii. Mushroom Sci. 2008, 17, 83–96. [Google Scholar]
- Ro, H.S.; Kim, S.S.; Ryu, J.S.; Jeon, C.O.; Lee, T.S.; Lee, H.S. Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics. Mycol. Res. 2007, 111, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Mang, S.; Figliuolo, G. Species delimitation in Pleurotus eryngii species-complex inferred from ITS and EF-1α gene sequences. Mycology 2010, 1, 269–280. [Google Scholar] [CrossRef]
- Ravash, R.; Shiran, B.; Alavi, A.; Bayat, F.; Rajaee, S.; Zervakis, G.I. Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol. Prog. 2010, 9, 181–194. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, W.; Dai, Y.H.; Fu, C.; Bian, Y.B. Using SSR markers to evaluate the genetic diversity of Lentinula edodes natural germplasm in China. World J. Microbiol. Biotechnol. 2010, 26, 527–536. [Google Scholar] [CrossRef]
- Du, P.; Cui, B.K.; Dai, Y.C. High genetic diversity in wild culinary medicinal wood ear mushroom, Auricularia polytricha (Mont) Sacc. in tropical China revealed by ISSR analysis. Int. J. Med. Mushrooms 2011, 13, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Huang, C.; Chen, Q.; Wu, X.; Qu, J. Genetic Variability and Population Structure of the Mushroom Pleurotus eryngii var. tuoliensis. PLoS ONE 2013, 8, e83253. [Google Scholar]
- Lewinsohn, D.; Nevo, E.; Wasser, S.P.; Hadar, Y.; Beharav, A. Genetic diversity in populations of Pleurotus eryngii complex in Israel. Mycol. Res. 2001, 8, 941–951. [Google Scholar] [CrossRef]
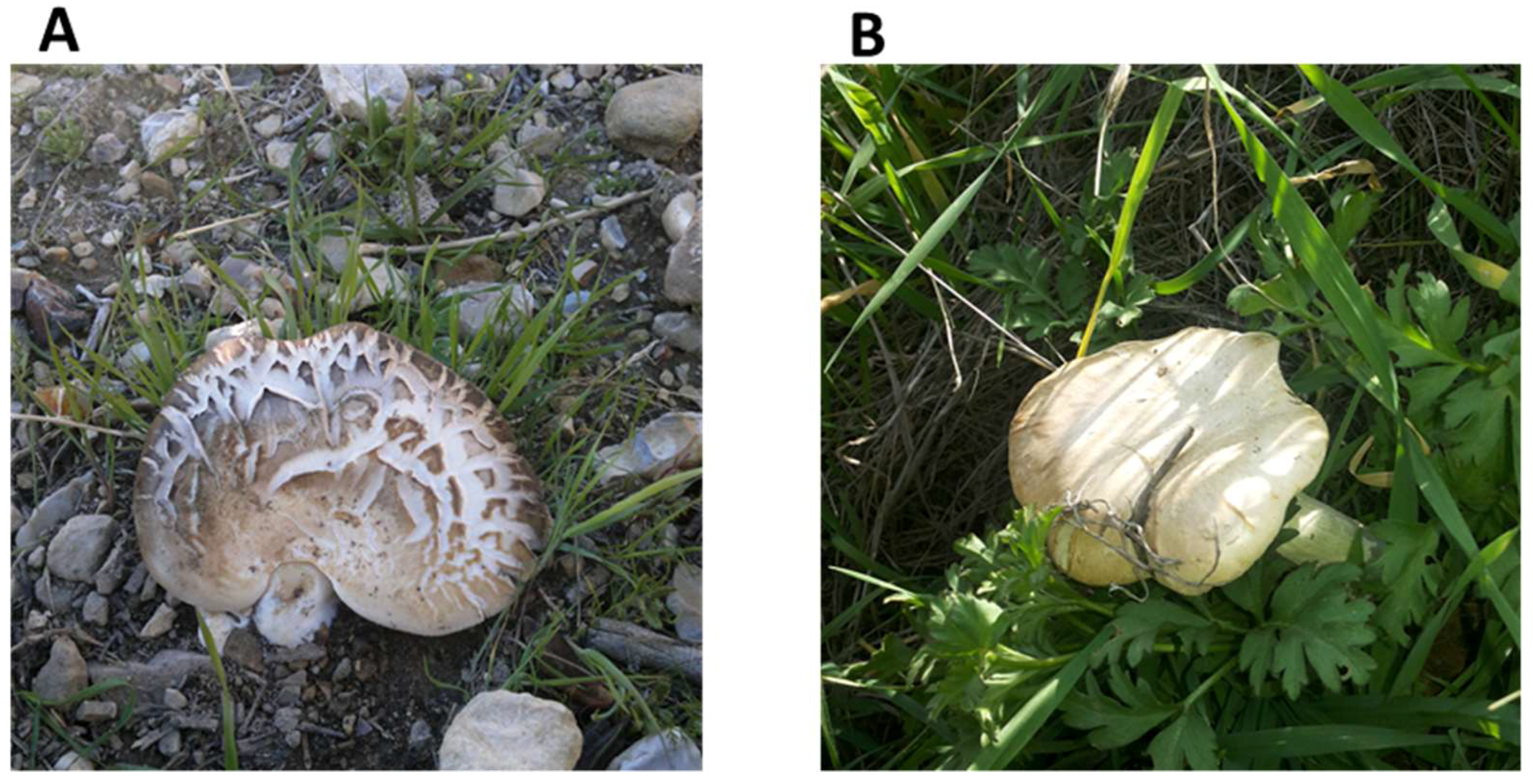

| Item | Description | Um-Qais | Tell ar-Rumman |
|---|---|---|---|
| Cap | Color | White, gray to beige | Flesh white to grey brown |
| Shape | Funnel shaped and irregular | Funnel shaped and irregular | |
| Diameter | 3–9 cm | 4–9 cm | |
| Stem | Color | Flesh white | White to brown |
| Shape | Eccentric thick, cylinder shape | Thick cylindrical | |
| Dimensions | Height:1–4 cm; Diameter: 1 cm | Height:1–4 cm; Diameter: 1 cm | |
| Spore | Color | White, creamy to yellow | White, creamy to yellow |
| Shape | Elliptic narrow; cylinder shape | Elliptic narrow, cylinder shape | |
| White | White | ||
| Measurements | Length: 5–7 µm; Width: 2–3 µm | Length: 5–10 µm; Width: 2–5 µm | |
| Gills | Color | Greyish | Whitish honey |
| Shape | Long extended internally to stem | Long extended internally to stem | |
| Attachment | Descending with stem | Descending with stem | |
| Flesh | Color and smell | White with taste and smell pleasant | White with taste and smell pleasant |
| Isolates | Best nr Database Similarity Hit Using nLSU | Best nr Database Similarity Hit Using ITS | |||||
|---|---|---|---|---|---|---|---|
| No | Code * | Taxon | ID% | GenBank Accession No. | Taxon | ID% | GenBank Accession No. |
| 1 | Q1 | P. eryngii | 98 | KY963032.1 | P. eryngii var. ferulae | 99 | KY962437.1 |
| 2 | Q2 | P. eryngii | 99 | AB777519.1 | P. eryngii | 97 | KY962448.1 |
| 3 | Q3 | P. eryngii | 99 | AY450347.1 | P. eryngii | 99 | KX977448.1.1 |
| 4 | Q4 | P. eryngii | 100 | AB777519.1 | P. eryngii | 99 | MG282489.1 |
| 5 | Q5 | P. eryngii | 99 | AY450347.1 | P. eryngii var. ferulae | 99 | MG282459.1 |
| 6 | Q6 | P. eryngii | 99 | AY450347.1 | P. eryngii | 97 | KX977448.1 |
| 7 | Q7 | P. eryngii | 99 | AB777519.1 | P. eryngii | 99 | MG282489.1 |
| 8 | Q8 | P. eryngii | 99 | KY963084.1 | P. eryngii | 99 | KX977448.1 |
| 9 | Q9 | P. eryngii | 99 | KY963084.1 | P. eryngii | 98 | JX429941.1 |
| 10 | R1 | P. eryngii | 99 | AB777519.1 | P. eryngii | 100 | MG282489.1 |
| 11 | R2 | P. eryngii | 99 | AB777519.1 | P. eryngii | 99 | MG282489.1 |
| 12 | R3 | P. eryngii | 99 | MG282549.1 | P. eryngii | 100 | MG282489.1 |
| 13 | R4 | P. eryngii | 99 | MG282549.1 | P. eryngii | 99 | KX836359.1 |
| 14 | R5 | P. eryngii | 99 | MG282549.1 | P. eryngii | 98 | MG282489.1 |
| 15 | R6 | P. eryngii | 99 | MG282549.1 | P. eryngii | 99 | MG282489.1 |
| 16 | R7 | P. eryngii | 98 | MG282549.1 | P. eryngii | 99 | MG282489.1 |
| 17 | R8 | P. eryngii | 98 | MG282549.1 | P. eryngii | 99 | MG282489.1 |
| 18 | R9 | P. eryngii | 99 | MG282549.1 | P. eryngii | 99 | MG282489.1 |
| 19 | R10 | P. eryngii | 99 | MG282549.1 | P. eryngii | 99 | MG282489.1 |
| Primer | Sequences | Total Bands | Polymorphic Bands | PIC (%) |
|---|---|---|---|---|
| UBC807 | (AG)8T | 14 | 10 | 71.4 |
| UBC808 | (AG)8C | 11 | 9 | 81.8 |
| UBC809 | (AG)8G | 15 | 1 | 6.66 |
| UBC810 | (GA)8T | 12 | 9 | 75 |
| UBC811 | (GA)8C | 0 | 0 | 0 |
| UBC812 | (GA)8A | 11 | 10 | 90.9 |
| UBC816 | (CA)8T | 11 | 8 | 72.7 |
| UBC823 | (TC)8C | 16 | 13 | 81.2 |
| UBC825 | (AC)8T | 12 | 11 | 91.6 |
| UBC826 | (AC)8C | 12 | 8 | 66.6 |
| UBC827 | (AC)8G | 13 | 12 | 92.3 |
| UBC864 | (ATG)5 | 9 | 7 | 77.7 |
| UBC866 | (CTC)6 | 8 | 5 | 62.5 |
| UBC868 | (GAA)6 | 9 | 8 | 88.8 |
| UBC873 | (GACA)4 | 7 | 4 | 57.1 |
| UBC874 | (CCCT)4 | 9 | 8 | 88.8 |
| UBC876 | (GATA)2(GACA)2 | 12 | 11 | 91.66 |
| UBC880 | (GGAGA)3 | 15 | 11 | 73.3 |
| Total | 196 | 141 | ||
| Mean | 10.89 | 7.83 | 70.56 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aref Hasan, H.; Mohamad Almomany, A.; Hasan, S.; Al-Abdallat, A.M. Assessment of Genetic Diversity among Pleurotus spp. Isolates from Jordan. J. Fungi 2018, 4, 52. https://doi.org/10.3390/jof4020052
Aref Hasan H, Mohamad Almomany A, Hasan S, Al-Abdallat AM. Assessment of Genetic Diversity among Pleurotus spp. Isolates from Jordan. Journal of Fungi. 2018; 4(2):52. https://doi.org/10.3390/jof4020052
Chicago/Turabian StyleAref Hasan, Hanan, Ahmad Mohamad Almomany, Shireen Hasan, and Ayed M. Al-Abdallat. 2018. "Assessment of Genetic Diversity among Pleurotus spp. Isolates from Jordan" Journal of Fungi 4, no. 2: 52. https://doi.org/10.3390/jof4020052
APA StyleAref Hasan, H., Mohamad Almomany, A., Hasan, S., & Al-Abdallat, A. M. (2018). Assessment of Genetic Diversity among Pleurotus spp. Isolates from Jordan. Journal of Fungi, 4(2), 52. https://doi.org/10.3390/jof4020052




