Analyses of Sporocarps, Morphotyped Ectomycorrhizae, Environmental ITS and LSU Sequences Identify Common Genera that Occur at a Periglacial Site
Abstract
:1. Introduction
2. Results
2.1. Sporocarp Data for Ectomycorrhizal Fungi
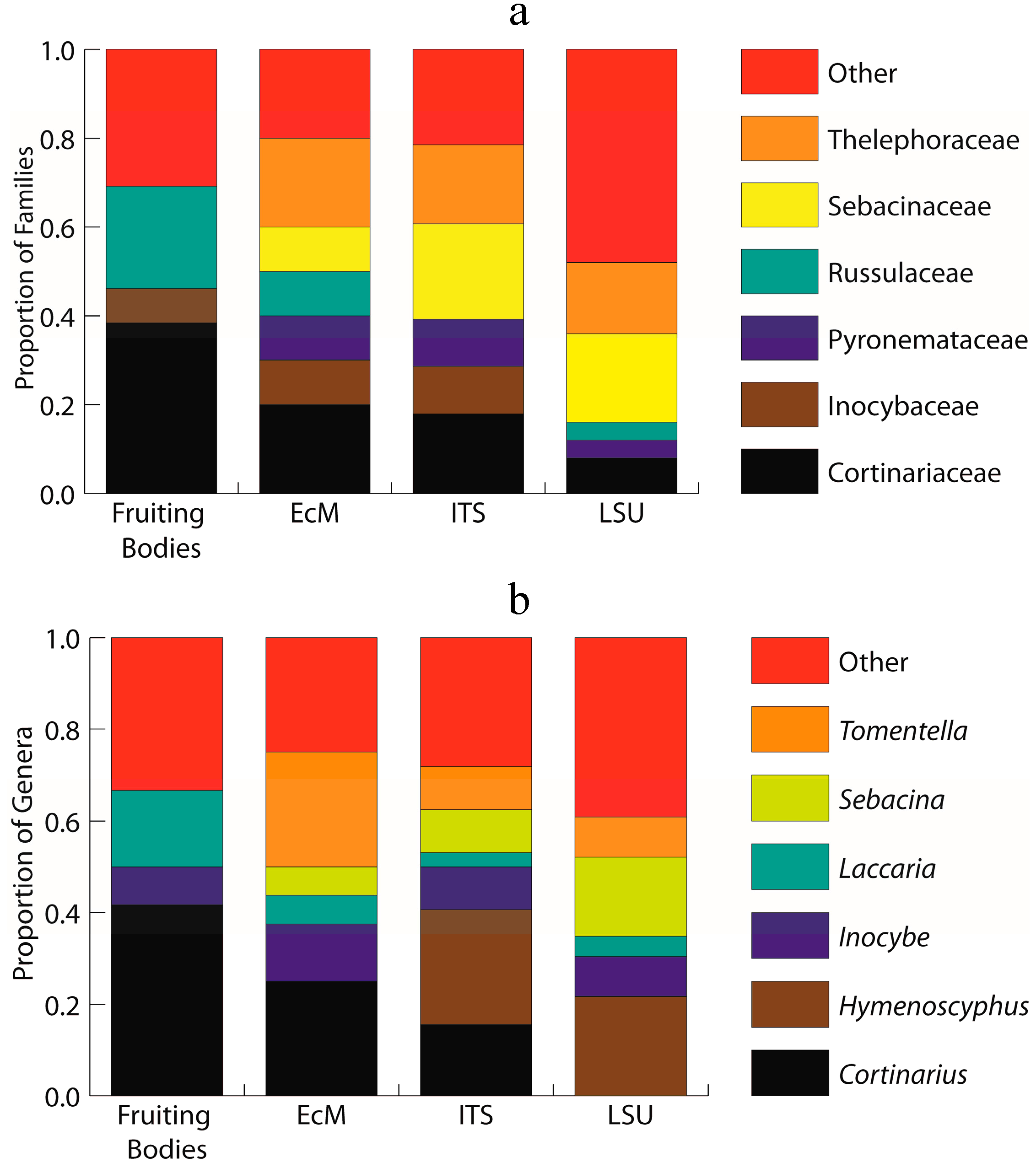
2.2. ITS Sequence Data for Morphotyped Ectomycorrhizae

2.3. Environmental ITS Amplicon Data
2.4. Environmental LSU Amplicon Data
3. Discussion
4. Materials and Methods
4.1. Study Site
4.2. Sporocarp Data
4.3. Morphotyped Ectomycorrhizae Sequence Data
4.4. Environmental ITS Amplicon Data
4.5. Environmental LSU Amplicon Data
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Hodge, S.M.; Trabant, D.C.; Krimmel, R.M.; Heinrichs, T.A.; March, R.S.; Josberger, E.G. Climate variations and changes in mass of three glaciers in western North America. J. Clim. 1998, 11, 2161–2179. [Google Scholar] [CrossRef]
- Dyurgerov, M.B.; Meier, M.F. Twentieth century climate change: Evidence from small glaciers. Proc. Natl. Acad. Sci. USA 2000, 97, 1406–1411. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Mirabella, A.; Fitze, P. Clay mineral formation in soils of two different chronosequences in the Swiss Alps. Geoderma 2001, 104, 145–175. [Google Scholar] [CrossRef]
- Matthews, J.A. The Ecology of Recently-Deglaciated Terrain: A Geoecological Approach to Glacier Forelands and Primary Succession; Cambridge University Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Jumpponen, A.; Väre, H.; Mattson, K.G.; Ohtonen, R.; Trappe, J.M. Characterization of ‘safe sites’ for pioneers in primary succession on recently deglaciated terrain. J. Ecol. 1999, 87, 98–105. [Google Scholar] [CrossRef]
- Jones, C.C.; del Moral, R. Dispersal and establishment both limit colonization during primary succession on a glacier foreland. Plant Ecol. 2009, 204, 217–230. [Google Scholar] [CrossRef]
- Cázares, E.; Trappe, J.M.; Jumpponen, A. Mycorrhiza-plant colonization patterns on a subalpine glacier forefront as a model system of primary succession. Mycorrhiza 2005, 15, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Blaalid, R.; Carlsen, T.; Kumar, S.; Halvorsen, R.; Ugland, K.I.; Fontana, G.; Kauserud, H. Changes in the root-associated fungal communities along a primary successional gradient analysed by 454 pyrosequencing. Mol. Ecol. 2012, 21, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Jumpponen, A.; Egerton-Warburton, L.M. Mycorrhizal fungi in successional environments—a community assembly model incorporating host plant, environmental and biotic filters. In The Fungal Community; Dighton, J., White, J., Oudemans, P., Eds.; CRC Press: New York, NY, USA, 2005; pp. 139–180. [Google Scholar]
- Diamond, J.M. Assembly of species communities. In Ecology and Evolution of Communities; Cody, M.L., Diamond, J.M., Eds.; Belknap Press, Harvard University: Cambridge, MA, USA, 1975; pp. 342–444. [Google Scholar]
- Gardes, M.; Bruns, T.D. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above- and below-ground views. Can. J. Bot. 1996, 74, 1572–1583. [Google Scholar] [CrossRef]
- Horton, T.R.; Bruns, T.D. The molecular revolution in ectomycorrhizal ecology: Peeking into the black-box. Mol. Ecol. 2001, 10, 1855–1871. [Google Scholar] [CrossRef] [PubMed]
- Jumpponen, A.; Jones, K.L.; Mattox, J.D.; Yeage, C. Massively parallel 454-sequencing of fungal communities in Quercus spp. ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol. Ecol. 2010, 19 (Suppl. 1), 41–53. [Google Scholar] [CrossRef] [PubMed]
- Lothamer, K.; Brown, S.P.; Mattox, J.D.; Jumpponen, A. Comparison of root-associated communities of native and non-native ectomycorrhizal hosts in an urban landscape. Mycorrhiza 2014, 24, 267–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coince, A.; Caël, O.; Bach, C.; Lengelle, J.; Cruaud, C.; Gavory, F.; Morin, E.; Murat, C.; Marcais, B.; Buée, M. Below-ground fine-scale distribution and soil versus fine root detection of fungal and soil oomycete communities in a French beech forest. Fungal. Ecol. 2013, 6, 223–235. [Google Scholar] [CrossRef]
- Oliver, A.K.; Callaham, M.A.; Jumpponen, A. Soil fungal communities respond compositionally to recurring frequent prescribed burning in a managed southeastern US forest ecosystem. For. Ecol. Manag. 2015, 345, 1–9. [Google Scholar] [CrossRef]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Res. 2011, 11, 759–769. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Carlsen, T.; Kjøller, R.; Köljalg, U.; Pennanen, T.; Rosendahl, S.; Stenlid, J.; Kauserud, H. Fungal community analyses by high-throughput sequencing of amplified markers—a user’s guide. New Phytol. 2013, 199, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Öpik, M.; Metsis, M.; Daniell, T.J.; Zobel, M.; Moora, M. Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol. 2009, 184, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Pellissier, L.; Niculita-Hirzel, H.; Dubuis, A.; Pagni, M.; Guex, N.; Ndiribe, C.; Salamin, N.; Xenarios, I.; Goudet, J.; Sanders, I.R.; Guisan, A. Soil fungal communities of grasslands are environmentally structured at a regional scale in the Alps. Mol. Ecol. 2014, 23, 4274–4290. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Nilsson, R.H.; Abarenkov, K.; Jairus, T.; Saar, I.; Bahram, M.; Bechem, E.; Chuyong, G.; Köljalg, U. 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 2010, 188, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Morgado, L.N.; Semenova, T.A.; Welker, J.M.; Walker, M.D.; Smets, E.; Geml, J. Summer temperature increase has distinct effects on the ectomycorrhizal fungal communities of moist tussock and dry tundra in Arctic Alaska. Glob. Chang. Biol. 2014, 21, 952–972. [Google Scholar]
- Brown, S.P.; Veach, A.M.; Grond, K.; Lickteig, S.K.; Lothamer, K.; Oliver, A.K.; Rigdon-Huss, A.R.; Jumpponen, A. Scraping the bottom of the barrel: Are rare high throughput sequences artifacts? Fungal. Ecol. 2015, 13, 221–225. [Google Scholar] [CrossRef]
- Brown, S.P.; Huss-Rigdon, A.; Jumpponen, A. Analyses of ITS and LSU gene regions provide congruent results on fungal community responses. Fungal. Ecol. 2014, 9, 65–68. [Google Scholar] [CrossRef]
- Jumpponen, A.; Brown, S.P.; Trappe, J.M.; Cázares, E.; Strömmer, R. Twenty years of research on Lyman Glacier forefront—lessons learned and questions yet unanswered. Fun. Ecol. 2012, 5, 430–442. [Google Scholar] [CrossRef]
- Jumpponen, A.; Trappe, J.M.; Cázares, E. Occurrence of ectomycorrhizal fungi on a receding glacier forefront. Mycorrhiza 2002, 12, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Trowbridge, J.; Jumpponen, A. Fungal colonization of shrub willow roots at the forefront of a receding glacier. Mycorrhiza 2004, 14, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.P.; Jumpponen, A. Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Mol. Ecol. 2014, 23, 481–497. [Google Scholar] [CrossRef]
- Cázares, E.; Trappe, J.M. Spore dispersal of ectomycorrhizal fungi on a glacier forefront by mammal mycophagy. Mycologia 1994, 86, 507–510. [Google Scholar] [CrossRef]
- Cázares, E.; Trappe, J.M. Alpine and subalpine fungi of the Cascade Mountains. I. Hymenogaster glacialis sp. nov. Mycotaxon 1990, 38, 245–249. [Google Scholar]
- Trappe, J.M.; Claridge, A.W. Hypogeous fungi: Evolution of reproductive and dispersal strategies through interactions with animals and mycorrhizal plants. In The Fungal Community—Its Organization and Role in the Ecosystem; Dighton, J., White, J.F., Oudemans, P., Eds.; Taylor & Francis: Boca Raton, FL, USA, 2005; pp. 613–623. [Google Scholar]
- Trappe, J.M. Lessons from alpine fungi. Mycologia 1988, 80, 1–10. [Google Scholar] [CrossRef]
- Gardes, M.; Dahlberg, A. Mycorrhizal diversity in arctic and alpine tundra: An open question. New Phytol. 1996, 133, 147–157. [Google Scholar] [CrossRef]
- Trappe, J.M.; Jumpponen, A.; Oregon State University, Corvallis, OR, US. Mycorrhizal fruiting bodies on the forefront of Exit Glacier, Alaska. Unpublished work. 1996. [Google Scholar]
- Cripps, C.L.; Horak, E. Checklist and ecology of the Agaricales, Russulales and Boletales in the alpine zone of the Rocky Moutains (Colorado, Montana, Wyoming) at 3000–4000 m.a.s.l. Sommerfeltia 2008, 31, 101–123. [Google Scholar] [CrossRef]
- Ohenoja, E.J.; Bauras, J.; Ohenoja, M. The Inocybe species found in the Canadian Arctic and West Siberian Sub-Arctic, with ecological notes. In Arctic and Alpine Mycology 5; Mukhin, V.A., Knudsen, H., Eds.; Yekaterina Publishers: Yekaterinburg, Russia, 1998; pp. 106–122. [Google Scholar]
- Nara, K.; Nakaya, H.; Hogetsu, T. Ectomycorrhizal sporocarp succession and production during early primary succession on Mount Fuji. New Phytol. 2003, 158, 193–206. [Google Scholar] [CrossRef]
- Voříšková, J.; Bradcová, V.; Cajthaml, T.; Baldrian, P. Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol. 2013, 201, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Vrålstad, T.; Schumacher, T.; Taylor, A.F.S. Mycorrhizal synthesis between fungal strains of Hymenoscyphus ericae aggregate and potential ectomycorrhizal and ericoid hosts. New Phytol. 2002, 153, 143–152. [Google Scholar] [CrossRef]
- Agnelli, A.; Ascher, J.; Corti, G.; Ceccherini, M.T.; Pietramellara, G.; Nannipieri, P. Purification and isotopic signatures (delta C-13, delta N-15, Delta C-14) of soil extracellular DNA. Biol. Fertil. Soils 2007, 44, 353–361. [Google Scholar] [CrossRef]
- Anderson, I.C.; Parkin, P.I. Detection of active soil fungi by RT-PCR amplification of precursor rRNA molecules. J. Microbiol. Methods 2007, 68, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Girvan, M.S.; Bullimore, J.; Ball, A.S.; Pretty, J.N.; Osborn, A.M. Responses of active bacterial and fungal communities in soils under winter wheat to different fertilizer and pesticide regimens. Appl. Environ. Microbiol. 2004, 70, 2692–2701. [Google Scholar] [CrossRef] [PubMed]
- Jumpponen, A. Analysis of ribosomal RNA indicates seasonal community dynamics in Andropogon gerardii roots. Mycorrhiza 2011, 21, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Jumpponen, A. Analysis of rhizosphere fungal communities using rRNA and rDNA. In Symbiotic Fungi, Soil Biology 18; Varma, A., Kharkwal, A.C., Eds.; Springer-Verlag: Berlin, Germany, 2009; pp. 29–40. [Google Scholar]
- Reeves, F.B.; Wagner, D.; Moorman, T.; Kiel, J. Role of endomycorrhizae in revegetation practices in the semi-arid west. 1. Comparison of incidence of mycorrhizae in severely disturbed vs. natural environments. Am. J. Bot. 1979, 66, 6–13. [Google Scholar] [CrossRef]
- Allen, E.B.; Allen, M.F. Natural re-establishment of vesicular-arbuscular mycorrhizae following stripmine reclamation in Wyoming. J. Appl. Ecol. 1980, 17, 139–147. [Google Scholar] [CrossRef]
- Allen, M.F.; Crisafulli, C.M.; Morris, S.J.; Egerton-Warburton, L.M.; MacMahon, J.A.; Trappe, J.M. Mycorrhizae and Mount St. Helens: Story of a symbiosis. In Ecological Responses to the 1980 Eruption of Mount St. Helens; Dale, V.H., Swanson, F.J., Crisafulli, C.M., Eds.; Springer Verlag: New York, NY, USA, 2005; pp. 221–231. [Google Scholar]
- Allen, M.F.; Macmahon, J.A.; Andersen, D.C. Reestablishment of Endogonaceae on Mount St. Helens—survival of residuals. Mycologia 1984, 76, 1031–1038. [Google Scholar] [CrossRef]
- Bertness, M.D.; Callaway, R. Positive interactions in communities. TREE 1994, 9, 191–193. [Google Scholar] [PubMed]
- Jumpponen, A.; Mattson, K.; Trappe, J.M.; Ohtonen, R. Effects of established willows on primary succession on Lyman Glacier forefront: Evidence for simultaneous canopy inhibition and soil facilitation. Arct. Alp. Res. 1998, 30, 31–39. [Google Scholar] [CrossRef]
- Post, A.; Richardson, D.W.; Rossellot, F.F. Inventory of Glaciers in the North Cascades, Washington; U.S. Department of the Interior Geological Survey: Washington, DC, USA, 1971; Professional Paper 705-A.
- Castellano, M.A.; Trappe, J.M.; Maser, Z.; Maser, C. Key to Spores of the Genera of Hypogeous Fungi of North Temperate Forests with Special Reference to Animal Mycophagy; Mad River Press: Eureka, CA, USA, 1989; p. 186. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for higher fungi and basidiomycetes: Application to identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS-RFLP matching for the identification of fungi. In Methods in Molecular Biology, Volume 50: Species Diagnostics Protocols: PCR and other Nucleic Acid Methods; Clapp, J.P., Ed.; Humanan Press Inc.: Totowa, NJ, USA, 1996; pp. 177–186. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification for fungi—Recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfland, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Amend, A.S.; Seifert, K.A.; Samson, R.; Bruns, T.D. Indoor fungal composition is geographically pattered and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. USA 2010, 107, 13748–13753. [Google Scholar] [CrossRef] [PubMed]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [PubMed]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J.; et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Porras-Alfaro, A.; Kuske, C.R.; Eichorst, A.S.; Xie, G. Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes. Appl. Environ. Microbiol. 2012, 78, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Lcensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jumpponen, A.; Brown, S.P.; Trappe, J.M.; Cázares, E.; Strömmer, R. Analyses of Sporocarps, Morphotyped Ectomycorrhizae, Environmental ITS and LSU Sequences Identify Common Genera that Occur at a Periglacial Site. J. Fungi 2015, 1, 76-93. https://doi.org/10.3390/jof1010076
Jumpponen A, Brown SP, Trappe JM, Cázares E, Strömmer R. Analyses of Sporocarps, Morphotyped Ectomycorrhizae, Environmental ITS and LSU Sequences Identify Common Genera that Occur at a Periglacial Site. Journal of Fungi. 2015; 1(1):76-93. https://doi.org/10.3390/jof1010076
Chicago/Turabian StyleJumpponen, Ari, Shawn P. Brown, James M. Trappe, Efrén Cázares, and Rauni Strömmer. 2015. "Analyses of Sporocarps, Morphotyped Ectomycorrhizae, Environmental ITS and LSU Sequences Identify Common Genera that Occur at a Periglacial Site" Journal of Fungi 1, no. 1: 76-93. https://doi.org/10.3390/jof1010076




