C1QL1/CTRP14 Is Largely Dispensable for Atherosclerosis Formation in Apolipoprotein-E-Deficient Mice
Abstract
:1. Introduction
2. Material and Methods
2.1. Animals and Diets
2.2. Construction of the Adenoviral C1QL1 Vector and Infection of HEK293 Cells
2.3. Biochemical Analyses
2.4. Quantification of Atherosclerotic Lesions
2.5. Extraction of Total RNA and Construction of a cDNA Library
2.6. Analysis of Differentially Expressed Genes
2.7. Real-Time Polymerase Chain Reaction (PCR) Analysis
2.8. Statistical Analysis
3. Results
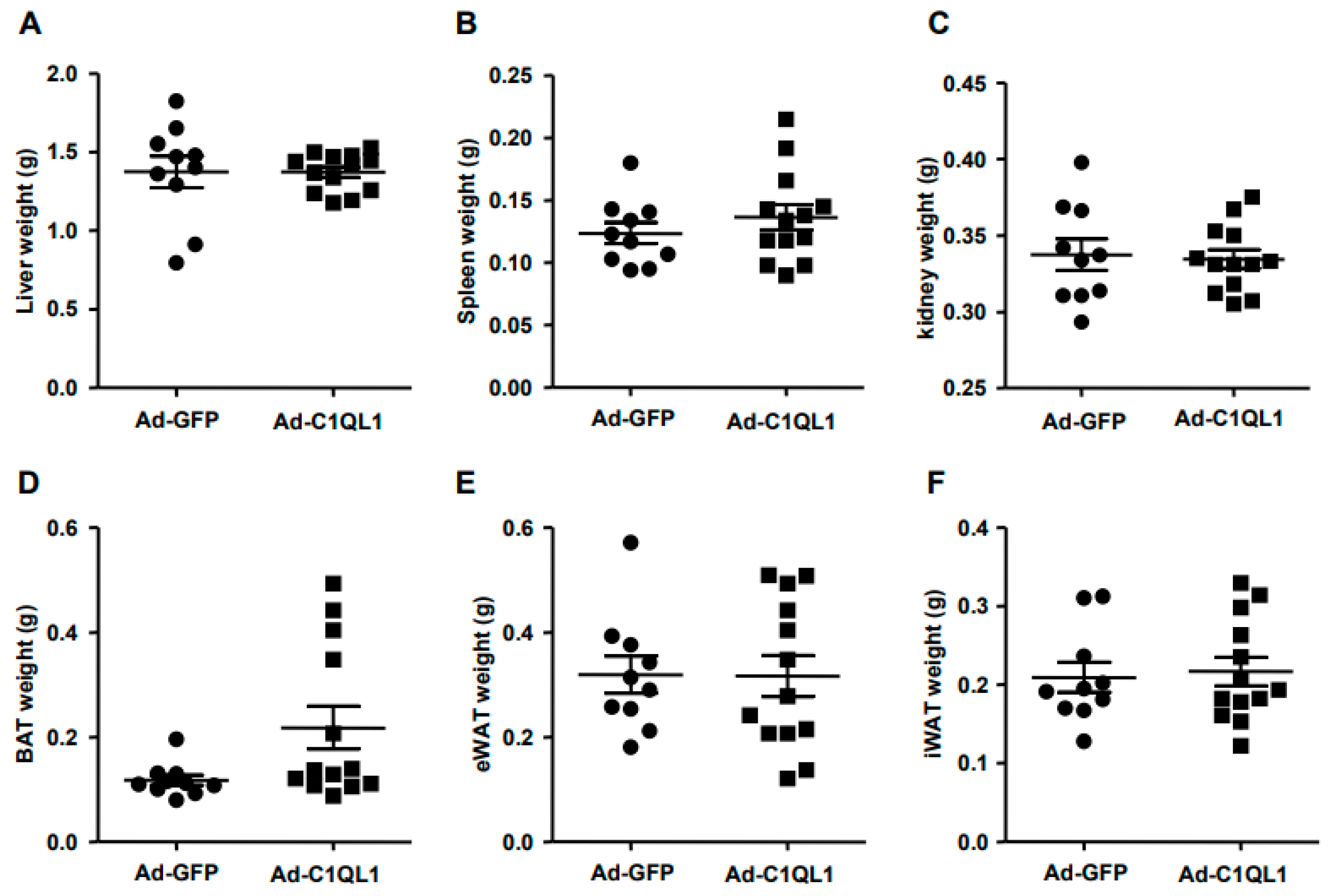
3.1. Overexpression of C1QL1 in ApoE KO Mice
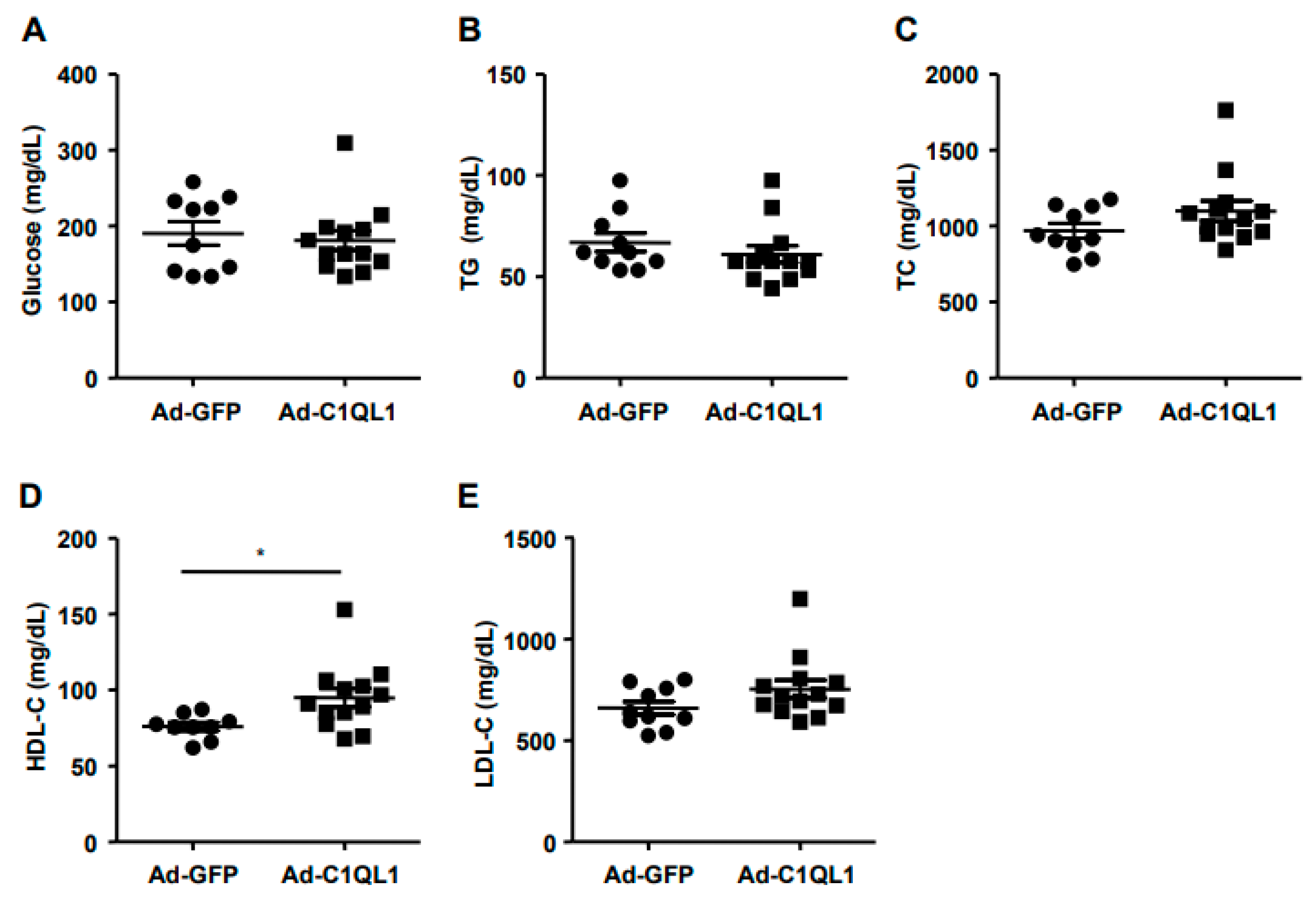
3.2. Overexpression of C1QL1 Elevated HDL-C in ApoE KO Mice
3.3. Increased Production of C1QL1 Did Not Affect the Formation of Atherosclerosis
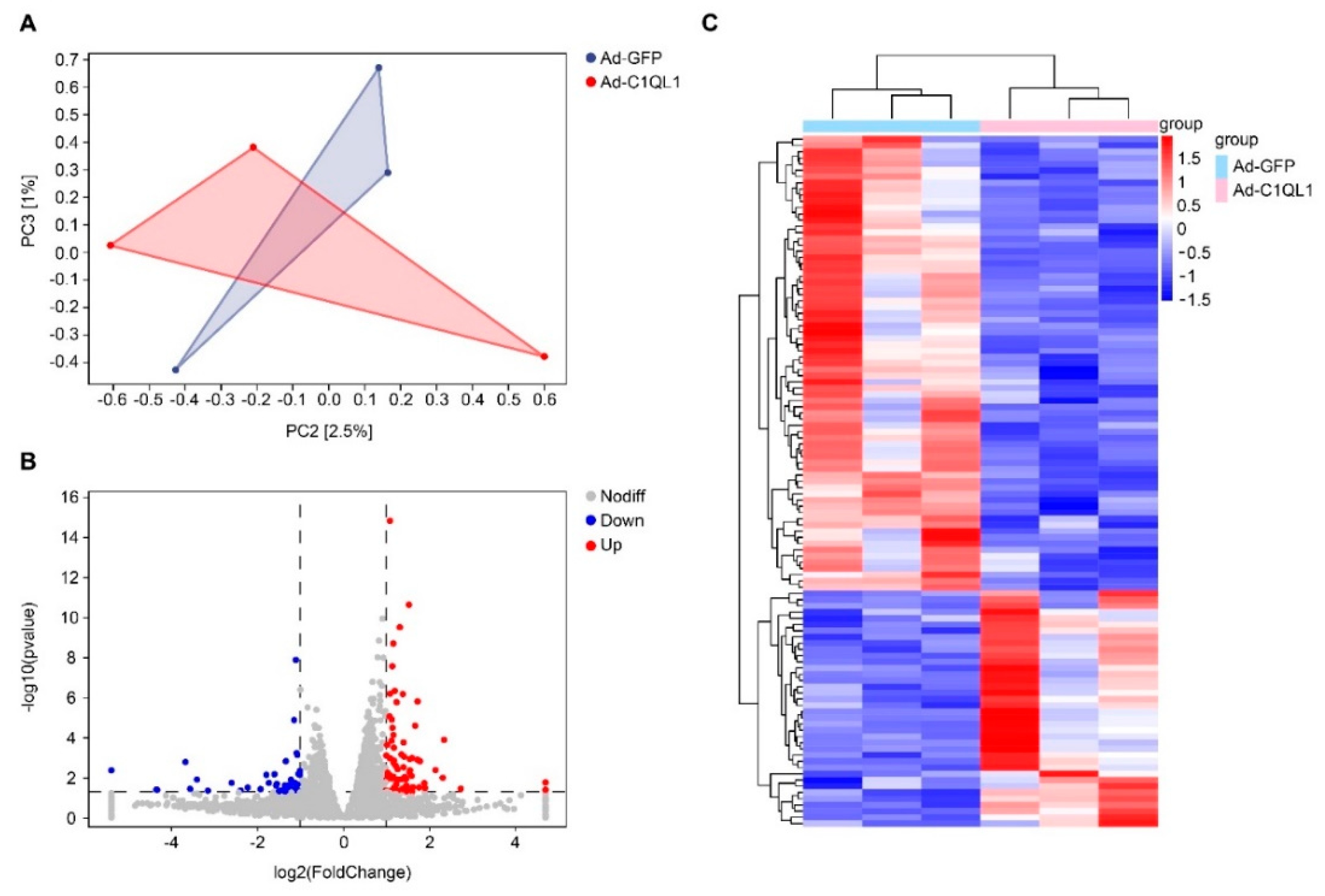
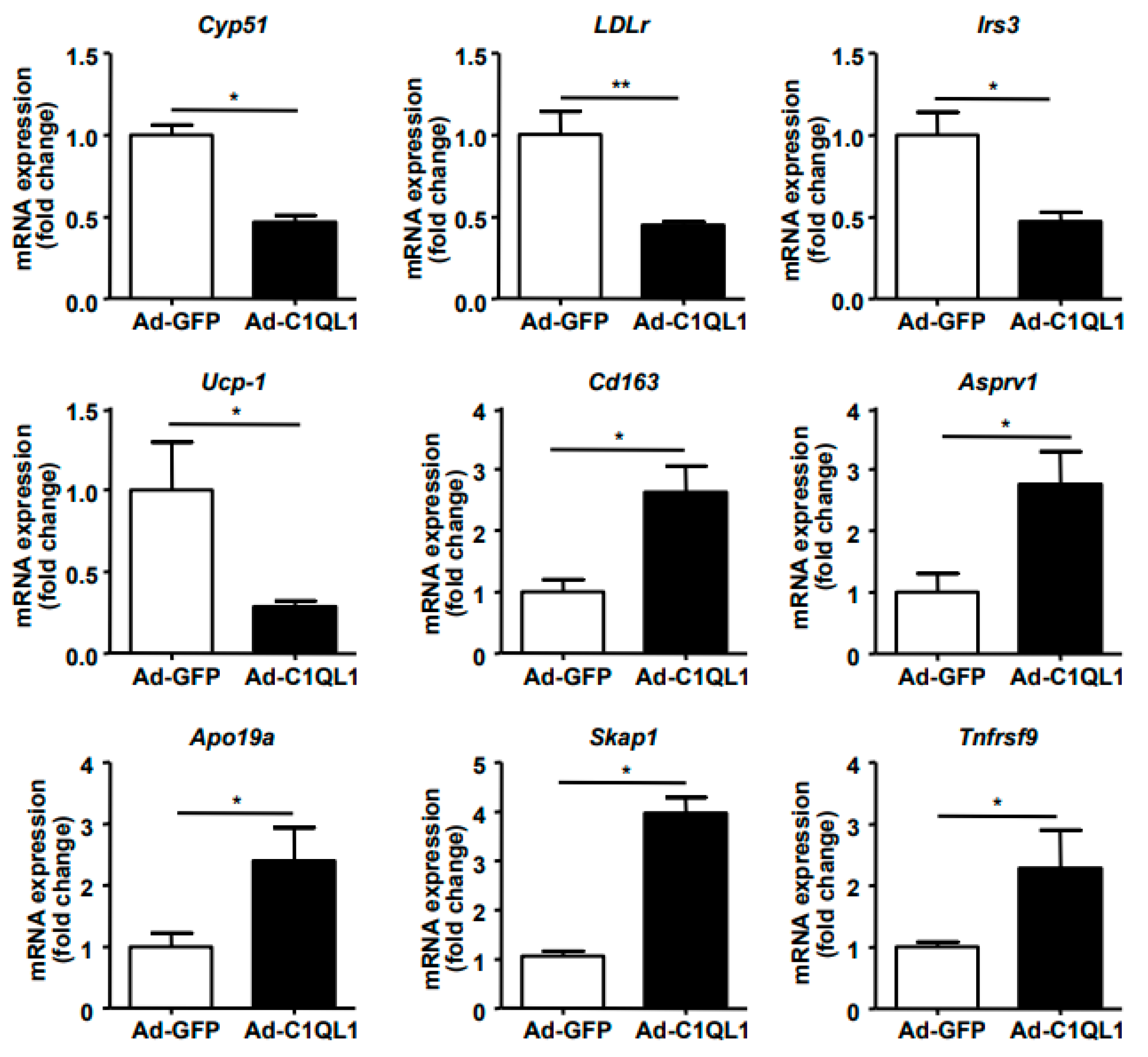
3.4. Gene Expression Profile in the Aorta
4. Discussion
Abbreviation
| Ad | adenovirus |
| C1QL1 | C1ql-like 1 |
| CRF | C1q-related factor |
| FDR | false discovery rate |
| KO | knockout |
| PCA | principal components analysis |
| GPCR | G protein-coupled bile acid receptor 1 |
| CTRPs | C1q/tumor-necrosis-factor-related proteins |
| CVD | cardiovascular diseases |
| Cyp51 | cytochrome P450 family 51 subfamily A member 1 |
| GFP | green fluorescent protein |
| H&E | hematoxylin–eosin |
| HDL-C | high-density lipoprotein cholesterol |
| LDLr | low-density lipoprotein receptor |
| LDL-C | low-density lipoprotein cholesterol |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| TC | total cholesterol |
| TG | triglyceride |
| UCP-1 | uncoupling protein 1 |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdolmaleki, F.; Gheibi Hayat, S.M.; Bianconi, V.; Johnston, T.P.; Sahebkar, A. Atherosclerosis and immunity: A perspective. Trends Cardiovasc. Med. 2019, 29, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu-Ito, M.; Shibata, R.; Ohashi, K.; Uemura, Y.; Kanemura, N.; Kambara, T.; Enomoto, T.; Yuasa, D.; Matsuo, K.; Ito, M.; et al. Omentin attenuates atherosclerotic lesion formation in apolipoprotein E-deficient mice. Cardiovasc. Res. 2016, 110, 107–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, L.; Li, Z.; Li, Q.; Guan, H.; Zhao, S.; Liu, R.; Wang, R.; Zhang, J.; Jia, Y.; Fan, J.; et al. Mediator 1 Is Atherosclerosis Protective by Regulating Macrophage Polarization. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1470–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Cheng, X.W.; Hu, L.; Takeshita, K.; Hu, C.; Du, Q.; Li, X.; Zhu, E.; Huang, Z.; Yisireyili, M.; et al. Cathepsin S Activity Controls Injury-Related Vascular Repair in Mice via the TLR2-Mediated p38MAPK and PI3K-Akt/p-HDAC6 Signaling Pathway. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1549–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Bai, L.; Lin, Y.; Chen, Y.; Guan, H.; Zhu, N.; Li, Y.; Gao, S.; Sun, L.; Zhao, S.; et al. Combined use of probucol and cilostazol with atorvastatin attenuates atherosclerosis in moderately hypercholesterolemic rabbits. Lipids Health Dis. 2015, 14, 82. [Google Scholar] [CrossRef] [Green Version]
- Bérubé, N.G.; Swanson, X.H.; Bertram, M.J.; Kittle, J.D.; Didenko, V.; Baskin, D.S.; Smith, J.R.; Pereira-Smith, O.M. Cloning and characterization of CRF, a novel C1q-related factor, expressed in areas of the brain involved in motor function. Mol. Brain Res. 1999, 63, 233–240. [Google Scholar] [CrossRef]
- Qi, Y.; Xiong, W.; Yu, S.; Du, Z.; Qu, T.; He, L.; Wei, W.; Zhang, L.; Liu, K.; Li, Y.; et al. Deletion of C1ql1 Causes Hearing Loss and Abnormal Auditory Nerve Fibers in the Mouse Cochlea. Front. Cell Neurosci. 2021, 15, 713651. [Google Scholar] [CrossRef]
- Biswas, J.; Pijewski, R.S.; Makol, R.; Miramontes, T.G.; Thompson, B.L.; Kresic, L.C.; Burghard, A.L.; Oliver, D.L.; Martinelli, D.C. C1ql1 is expressed in adult outer hair cells of the cochlea in a tonotopic gradient. PLoS ONE 2021, 16, e0251412. [Google Scholar] [CrossRef]
- Liu, H.; Pecka, J.L.; Zhang, Q.; Soukup, G.A.; Beisel, K.W.; He, D.Z. Characterization of transcriptomes of cochlear inner and outer hair cells. J. Neurosci. 2014, 34, 11085–11095. [Google Scholar] [CrossRef] [Green Version]
- Sigoillot, S.M.; Iyer, K.; Binda, F.; Gonzalez-Calvo, I.; Talleur, M.; Vodjdani, G.; Isope, P.; Selimi, F. The Secreted Protein C1QL1 and Its Receptor BAI3 Control the Synaptic Connectivity of Excitatory Inputs Converging on Cerebellar Purkinje Cells. Cell Rep. 2015, 10, 820–832. [Google Scholar] [CrossRef]
- Gao, Y.J.; Chen, F.; Zhang, L.J. C1q-like 1 is frequently up-regulated in lung adenocarcinoma and contributes to the proliferation and invasion of tumor cells. J. Chemother. 2021, 33, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Salkowska, A.; Karas, K.; Karwaciak, I.; Walczak-Drzewiecka, A.; Krawczyk, M.; Sobalska-Kwapis, M.; Dastych, J.; Ratajewski, M. Identification of Novel Molecular Markers of Human Th17 Cells. Cells 2020, 9, 1611. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Yang, X.; Shi, T.; Zhang, Y.; Xiang, A.; Li, Y. CTRP9 Mitigates the Progression of Arteriovenous Shunt-Induced Pulmonary Artery Hypertension in Rats. Cardiovasc. Ther. 2021, 2021, 4971300. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, S.; Wang, Y.; Chen, Y.; Lin, Y.; Zhu, N.; Zheng, H.; Wu, M.; Cheng, D.; Li, Y.; et al. Urotensin II promotes atherosclerosis in cholesterol-fed rabbits. PLoS ONE 2014, 9, e95089. [Google Scholar] [CrossRef]
- Bodary, P.F.; Gu, S.; Shen, Y.; Hasty, A.H.; Buckler, J.M.; Eitzman, D.T. Recombinant leptin promotes atherosclerosis and thrombosis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2005, 25, e119–e122. [Google Scholar] [CrossRef] [Green Version]
- Guan, H.; Lin, Y.; Bai, L.; An, Y.; Shang, J.; Wang, Z.; Zhao, S.; Fan, J.; Liu, E. Dietary Cocoa Powder Improves Hyperlipidemia and Reduces Atherosclerosis in apoE Deficient Mice through the Inhibition of Hepatic Endoplasmic Reticulum Stress. Mediat. Inflamm. 2016, 2016, 1937572. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Bai, L.; Yu, Q.; Hu, F.; Wu, J.; Zhao, S.; Wang, R.; Wang, W.; Tao, Y.; Fan, J.; et al. iMarmot: An integrative platform for comparative and functional genomics of marmots. BMC Genom. 2020, 21, 266. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Y.; Bai, L.; Zhao, S.; Wang, R.; Liu, B.; Zhang, Y.; Fan, J.; Liu, E. Transcriptomic analysis of the liver of cholesterol-fed rabbits reveals altered hepatic lipid metabolism and inflammatory response. Sci. Rep. 2018, 8, 6437. [Google Scholar] [CrossRef]
- Go, G.W.; Mani, A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J. Biol. Med. 2012, 85, 19–28. [Google Scholar] [PubMed]
- Bajaj, A.; Susztak, K.; Damrauer, S.M. APOL1 and Cardiovascular Disease: A Story in Evolution. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1587–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libby, P. The changing landscape of atherosclerosis. Nature 2021, 592, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Xian, W.; Cao, J.; Yuan, X.; Wang, G.; Jin, Q.; Zhang, H.; Zhou, G.; You, L. Deficiency of Intellectual Disability-Related Gene Brpf1 Attenuated Hippocampal Excitatory Synaptic Transmission and Impaired Spatial Learning and Memory Ability. Front. Cell Dev. Biol. 2021, 9, 711792. [Google Scholar] [CrossRef] [PubMed]
- Sarver, D.C.; Xu, C.; Aja, S.; Wong, G.W. CTRP14 inactivation alters physical activity and food intake response to fasting and refeeding. Am. J. Physiol.-Endocrinol. Metab. 2022, 322, E480–E493. [Google Scholar] [CrossRef] [PubMed]
- Casula, M.; Colpani, O.; Xie, S.; Catapano, A.L.; Baragetti, A. HDL in Atherosclerotic Cardiovascular Disease: In Search of a Role. Cells 2021, 10, 1869. [Google Scholar] [CrossRef] [PubMed]
- The Emerging Risk Factors Collaboration. Major lipids, apolipoproteins, and risk of vascular disease. JAMA 2009, 302, 1993–2000. [Google Scholar] [CrossRef] [Green Version]
- Marz, W.; Kleber, M.E.; Scharnagl, H.; Speer, T.; Zewinger, S.; Ritsch, A.; Parhofer, K.G.; von Eckardstein, A.; Landmesser, U.; Laufs, U. HDL cholesterol: Reappraisal of its clinical relevance. Clin. Res. Cardiol. Off. J. Ger. Card. Soc. 2017, 106, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Franczyk, B.; Rysz, J.; Lawinski, J.; Rysz-Gorzynska, M.; Gluba-Brzozka, A. Is a High HDL-Cholesterol Level Always Beneficial? Biomedicines 2021, 9, 1083. [Google Scholar] [CrossRef]
- Piko, P.; Fiatal, S.; Werissa, N.A.; Bekele, B.B.; Racz, G.; Kosa, Z.; Sandor, J.; Adany, R. The Effect of Haplotypes in the CETP and LIPC Genes on the Triglycerides to HDL-C Ratio and Its Components in the Roma and Hungarian General Populations. Genes 2020, 11, 56. [Google Scholar] [CrossRef]
- Morton, R.E.; Mihna, D.; Liu, Y. The lipid substrate preference of CETP controls the biochemical properties of HDL in fat/cholesterol-fed hamsters. J. Lipid Res. 2021, 62, 100027. [Google Scholar] [CrossRef] [PubMed]
- Chowaniec, Z.; Skoczynska, A. Plasma lipid transfer proteins: The role of PLTP and CETP in atherogenesis. Adv. Clin. Exp. Med. 2018, 27, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and Reverse Cholesterol Transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Garruti, G.; Liu, M.; Portincasa, P.; Wang, D.Q. Cholesterol and Lipoprotein Metabolism and Atherosclerosis: Recent Advances In reverse Cholesterol Transport. Ann. Hepatol. 2017, 16 (Suppl. S1), s27–s42. [Google Scholar] [CrossRef]
- Ruiz-Ramie, J.J.; Barber, J.L.; Sarzynski, M.A. Effects of exercise on HDL functionality. Curr. Opin. Lipidol. 2019, 30, 16–23. [Google Scholar] [CrossRef]
- Zakiev, E.; Feng, M.; Sukhorukov, V.; Kontush, A. HDL-Targeting Therapeutics: Past, Present and Future. Curr. Pharm. Des. 2017, 23, 1207–1215. [Google Scholar] [CrossRef]
- Ressl, S.; Vu, B.K.; Vivona, S.; Martinelli, D.C.; Sudhof, T.C.; Brunger, A.T. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure 2015, 23, 688–699. [Google Scholar] [CrossRef] [Green Version]
- Bolliger, M.F.; Martinelli, D.C.; Sudhof, T.C. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 2534–2539. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.-F.; Li, G.-D.; Sun, X.; Li, X.-X.; Gao, Y.; Gao, C.; Cao, K.-X.; Yang, G.-W.; Yu, M.-W.; Wang, X.-M. Identification of FAM107A as a potential biomarker and therapeutic target for prostate carcinoma. Am. J. Transl. Res. 2021, 13, 10163–10177. [Google Scholar]
- Eto, N.; Miyagishi, M.; Inagi, R.; Fujita, T.; Nangaku, M. Mitogen-activated protein 3 kinase 6 mediates angiogenic and tumorigenic effects via vascular endothelial growth factor expression. Am. J. Pathol. 2009, 174, 1553–1563. [Google Scholar] [CrossRef] [Green Version]
- Melincovici, C.S.; Bosca, A.B.; Susman, S.; Marginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular endothelial growth factor (VEGF)—Key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. Rev. Roum. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar]
- Liu, F.; Tan, A.; Yang, R.; Xue, Y.; Zhang, M.; Chen, L.; Xiao, L.; Yang, X.; Yu, Y. C1ql1/Ctrp14 and C1ql4/Ctrp11 promote angiogenesis of endothelial cells through activation of ERK1/2 signal pathway. Mol. Cell Biochem. 2017, 424, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein-Coupled Receptor Activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, H.; Shi, T.; Liu, M.; Wang, X.; Guo, F. C1QL1/CTRP14 Is Largely Dispensable for Atherosclerosis Formation in Apolipoprotein-E-Deficient Mice. J. Cardiovasc. Dev. Dis. 2022, 9, 341. https://doi.org/10.3390/jcdd9100341
Guan H, Shi T, Liu M, Wang X, Guo F. C1QL1/CTRP14 Is Largely Dispensable for Atherosclerosis Formation in Apolipoprotein-E-Deficient Mice. Journal of Cardiovascular Development and Disease. 2022; 9(10):341. https://doi.org/10.3390/jcdd9100341
Chicago/Turabian StyleGuan, Hua, Tao Shi, Miaomiao Liu, Xue Wang, and Fengwei Guo. 2022. "C1QL1/CTRP14 Is Largely Dispensable for Atherosclerosis Formation in Apolipoprotein-E-Deficient Mice" Journal of Cardiovascular Development and Disease 9, no. 10: 341. https://doi.org/10.3390/jcdd9100341




