Considering the Genetic Architecture of Hypoplastic Left Heart Syndrome
Abstract
1. Evidence for Important Genetic Components in HLHS
1.1. Background and Motivation
1.2. Familiality
1.3. Heritability
1.4. Epidemiology
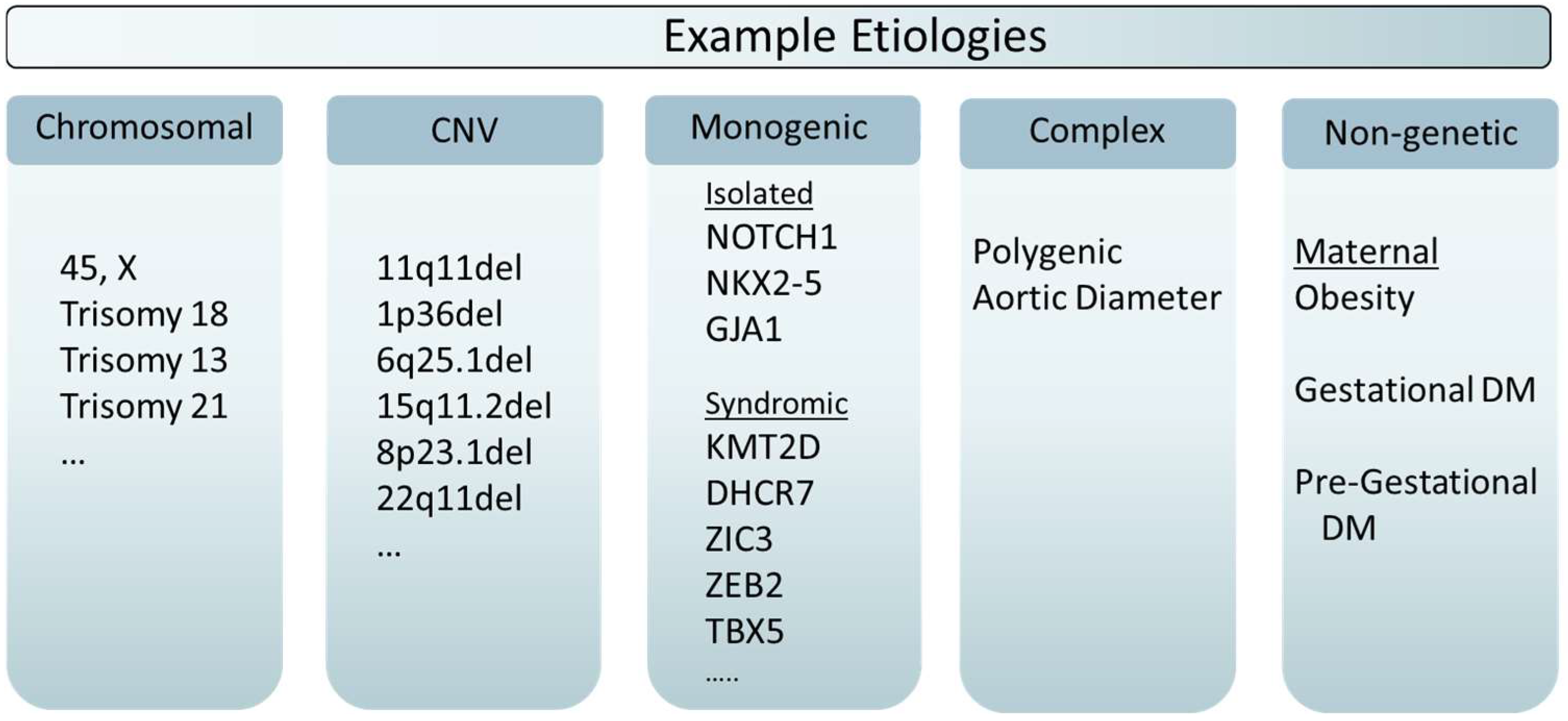
2. Monogenic and Chromosomal Inheritance
2.1. Chromosomal
2.2. De Novo Mutation
2.3. Single Nucleotide Variants and Other Variant Types
2.4. Autosomal Dominant
2.5. Mosaicism
2.6. Locus Heterogeneity
3. Complex Inheritance
3.1. Transmitted Deleterious DNA Variants
3.2. Common Variants
3.3. Non-Genetic Causes
4. Clinical Implications of Genetic Testing
4.1. Clinical Utility
4.2. Possible Harms and Disutility
4.3. Cost Effectiveness
4.4. Clinical Data Sharing
5. Next Steps in Research
5.1. Large Scale WGS
5.2. Phenotypic Heterogeneity and Integrative Studies
5.3. Exploratory Long Read WGS
5.4. Clinical Utility of Newborn WGS
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, J.Y.; Phan, K.; Ayer, J.; Celermajer, D.S.; Winlaw, D.S. Long term survival of hypoplastic left heart syndrome infants: Meta-analysis comparing outcomes from the modified Blalock-Taussig shunt and the right ventricle to pulmonary artery shunt. Int. J. Cardiol. 2018, 254, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.S.; Rausa, J.; Farias, J.S.; Villarreal, E.G.; Acosta, S.; Savorgnan, F.; Flores, S. Impact of Medical Interventions and Comorbidities on Norwood Admission for Patients with Hypoplastic Left Heart Syndrome. Pediatr. Cardiol. 2022, 43, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Dean, P.N.; Hillman, D.G.; McHugh, K.E.; Gutgesell, H.P. Inpatient costs and charges for surgical treatment of hypoplastic left heart syndrome. Pediatrics 2011, 128, e1181–e1186. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.L.; Song, A.Y.; Horak, R.; Friedlich, P.S.; Lakshmanan, A.; Pruetz, J.D.; Yieh, L.; Ram Kumar, S.; Williams, R.G. Impact of Confounding on Cost, Survival, and Length-of-Stay Outcomes for Neonates with Hypoplastic Left Heart Syndrome Undergoing Stage 1 Palliation Surgery. Pediatr. Cardiol. 2020, 41, 996–1011. [Google Scholar] [CrossRef]
- Epstein’s Inborn Errors of Development: The Molecular Basis of Clinical Disorders of Morphogenesis; Oxford University Press: Oxford, UK, 2016.
- Pierpont, M.E.; Brueckner, M.; Chung, W.K.; Garg, V.; Lacro, R.V.; McGuire, A.L.; Mital, S.; Priest, J.R.; Pu, W.T.; Roberts, A.; et al. Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association. Circulation 2018, 138, e653–e711. [Google Scholar] [CrossRef] [PubMed]
- Parker, L.E.; Landstrom, A.P. Genetic Etiology of Left-Sided Obstructive Heart Lesions: A Story in Development. J. Am. Heart Assoc. 2021, 10, e019006. [Google Scholar] [CrossRef] [PubMed]
- Øyen, N.; Poulsen, G.; Boyd, H.A.; Wohlfahrt, J.; Jensen, P.K.; Melbye, M. Recurrence of congenital heart defects in families. Circulation 2009, 120, 295–301. [Google Scholar] [CrossRef]
- Brodwall, K.; Greve, G.; Leirgul, E.; Tell, G.S.; Vollset, S.E.; Øyen, N. Recurrence of congenital heart defects among siblings-a nationwide study. Am. J. Med. Genet. A 2017, 173, 1575–1585. [Google Scholar] [CrossRef]
- Wessels, M.W.; Berger, R.M.; Frohn-Mulder, I.M.; Roos-Hesselink, J.W.; Hoogeboom, J.J.; Mancini, G.S.; Bartelings, M.M.; Krijger, R.; Wladimiroff, J.W.; Niermeijer, M.F.; et al. Autosomal dominant inheritance of left ventricular outflow tract obstruction. Am. J. Med. Genet. A 2005, 134A, 171–179. [Google Scholar] [CrossRef]
- Lewin, M.B.; McBride, K.L.; Pignatelli, R.; Fernbach, S.; Combes, A.; Menesses, A.; Lam, W.; Bezold, L.I.; Kaplan, N.; Towbin, J.A.; et al. Echocardiographic evaluation of asymptomatic parental and sibling cardiovascular anomalies associated with congenital left ventricular outflow tract lesions. Pediatrics 2004, 114, 691–696. [Google Scholar] [CrossRef]
- McBride, K.L.; Pignatelli, R.; Lewin, M.; Ho, T.; Fernbach, S.; Menesses, A.; Lam, W.; Leal, S.M.; Kaplan, N.; Schliekelman, P.; et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: Segregation, multiplex relative risk, and heritability. Am. J. Med. Genet. A 2005, 134A, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Hinton, R.B., Jr.; Martin, L.J.; Tabangin, M.E.; Mazwi, M.L.; Cripe, L.H.; Benson, D.W. Hypoplastic left heart syndrome is heritable. J. Am. Coll. Cardiol. 2007, 50, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.I. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr. Cardiol. 1995, 16, 155–165. [Google Scholar] [CrossRef] [PubMed]
- McBride, K.L.; Marengo, L.; Canfield, M.; Langlois, P.; Fixler, D.; Belmont, J.W. Epidemiology of noncomplex left ventricular outflow tract obstruction malformations (aortic valve stenosis, coarctation of the aorta, hypoplastic left heart syndrome) in Texas, 1999–2001. Birth Defects Res. A Clin. Mol. Teratol. 2005, 73, 555–561. [Google Scholar] [CrossRef]
- Canfield, M.A.; Mai, C.T.; Wang, Y.; O’Halloran, A.; Marengo, L.K.; Olney, R.S.; Borger, C.L.; Rutkowski, R.; Fornoff, J.; Irwin, N.; et al. The association between race/ethnicity and major birth defects in the United States, 1999–2007. Am. J. Public Health 2014, 104, e14–e23. [Google Scholar] [CrossRef]
- Eghtesady, P.; Brar, A.; Hall, M. Seasonality of hypoplastic left heart syndrome in the United States: A 10-year time-series analysis. J. Thorac. Cardiovasc. Surg. 2011, 141, 432–438. [Google Scholar] [CrossRef]
- Boyd, R.; McMullen, H.; Beqaj, H.; Kalfa, D. Environmental Exposures and Congenital Heart Disease. Pediatrics 2022, 149, e2021052151. [Google Scholar] [CrossRef]
- Zakaria, D.; Tang, X.; Bhakta, R.; ElHassan, N.O.; Prodhan, P. Chromosomal Abnormalities Affect the Surgical Outcome in Infants with Hypoplastic Left Heart Syndrome: A Large Cohort Analysis. Pediatr. Cardiol. 2018, 39, 11–18. [Google Scholar] [CrossRef]
- Lara, D.A.; Ethen, M.K.; Canfield, M.A.; Nembhard, W.N.; Morris, S.A. A population-based analysis of mortality in patients with Turner syndrome and hypoplastic left heart syndrome using the Texas Birth Defects Registry. Congenit. Heart Dis. 2017, 12, 105–112. [Google Scholar] [CrossRef]
- Bondy, C.; Bakalov, V.K.; Cheng, C.; Olivieri, L.; Rosing, D.R.; Arai, A.E. Bicuspid aortic valve and aortic coarctation are linked to deletion of the X chromosome short arm in Turner syndrome. J. Med. Genet. 2013, 50, 662–665. [Google Scholar] [CrossRef][Green Version]
- Grossfeld, P. ETS1 and HLHS: Implications for the Role of the Endocardium. J. Cardiovasc. Dev. Dis. 2022, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Hanchard, N.A.; Umana, L.A.; D’Alessandro, L.; Azamian, M.; Poopola, M.; Morris, S.A.; Fernbach, S.; Lalani, S.R.; Towbin, J.A.; Zender, G.A.; et al. Assessment of large copy number variants in patients with apparently isolated congenital left-sided cardiac lesions reveals clinically relevant genomic events. Am. J. Med. Genet. A 2017, 173, 2176–2188. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.A.; Hoffer, M.J.; van Velzen, C.L.; Plati, S.K.; Rijlaarsdam, M.E.; Clur, S.A.; Blom, N.A.; Pajkrt, E.; Bhola, S.L.; Knegt, A.C.; et al. Chromosomal abnormalities and copy number variations in fetal left-sided congenital heart defects. Prenat. Diagn. 2016, 36, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Warburton, D.; Ronemus, M.; Kline, J.; Jobanputra, V.; Williams, I.; Anyane-Yeboa, K.; Chung, W.; Yu, L.; Wong, N.; Awad, D.; et al. The contribution of de novo and rare inherited copy number changes to congenital heart disease in an unselected sample of children with conotruncal defects or hypoplastic left heart disease. Hum. Genet. 2014, 133, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Hitz, M.P.; Lemieux-Perreault, L.P.; Marshall, C.; Feroz-Zada, Y.; Davies, R.; Yang, S.W.; Lionel, A.C.; D’Amours, G.; Lemyre, E.; Cullum, R.; et al. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012, 8, e1002903. [Google Scholar] [CrossRef]
- Engwerda, A.; Leenders, E.; Frentz, B.; Terhal, P.A.; Löhner, K.; de Vries, B.B.A.; Dijkhuizen, T.; Vos, Y.J.; Rinne, T.; van den Berg, M.P.; et al. TAB2 deletions and variants cause a highly recognisable syndrome with mitral valve disease, cardiomyopathy, short stature and hypermobility. Eur. J. Hum. Genet. 2021, 29, 1669–1676. [Google Scholar] [CrossRef]
- Cheng, A.; Dinulos, M.B.P.; Neufeld-Kaiser, W.; Rosenfeld, J.; Kyriss, M.; Madan-Khetarpal, S.; Risheg, H.; Byers, P.H.; Liu, Y.J. 6q25.1 (TAB2) microdeletion syndrome: Congenital heart defects and cardiomyopathy. Am. J. Med. Genet. A 2017, 173, 1848–1857. [Google Scholar] [CrossRef]
- Thienpont, B.; Zhang, L.; Postma, A.V.; Breckpot, J.; Tranchevent, L.C.; Van Loo, P.; Møllgård, K.; Tommerup, N.; Bache, I.; Tümer, Z.; et al. Haploinsufficiency of TAB2 causes congenital heart defects in humans. Am. J. Hum. Genet. 2010, 86, 839–849. [Google Scholar] [CrossRef]
- Digilio, M.C.; Baban, A.; Marino, B.; Dallapiccola, B. Hypoplastic left heart syndrome in patients with Kabuki syndrome. Pediatr. Cardiol. 2010, 31, 1111–1113. [Google Scholar] [CrossRef]
- Musaad, W.; Lyons, A.; Allen, N.; Letshwiti, J. Mowat-Wilson syndrome presenting with Shone’s complex cardiac anomaly. BMJ Case Rep. 2022, 15, e246913. [Google Scholar] [CrossRef]
- Homsy, J.; Zaidi, S.; Shen, Y.; Ware, J.S.; Samocha, K.E.; Karczewski, K.J.; DePalma, S.R.; McKean, D.; Wakimoto, H.; Gorham, J.; et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015, 350, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.C.; Homsy, J.; Zaidi, S.; Lu, Q.; Morton, S.; DePalma, S.R.; Zeng, X.; Qi, H.; Chang, W.; Sierant, M.C.; et al. Contribution of rare inherited and de novo variants in 2871 congenital heart disease probands. Nat. Genet. 2017, 49, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Li, A.H.; Hanchard, N.A.; Furthner, D.; Fernbach, S.; Azamian, M.; Nicosia, A.; Rosenfeld, J.; Muzny, D.; D’Alessandro, L.C.A.; Morris, S.; et al. Whole exome sequencing in 342 congenital cardiac left sided lesion cases reveals extensive genetic heterogeneity and complex inheritance patterns. Genome Med. 2017, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Pasipoularides, A. The new era of whole-exome sequencing in congenital heart disease: Brand-new insights into rare pathogenic variants. J. Thorac. Dis. 2018, 10, S1923–S1929. [Google Scholar] [CrossRef]
- Boskovski, M.T.; Homsy, J.; Nathan, M.; Sleeper, L.A.; Morton, S.; Manheimer, K.B.; Tai, A.; Gorham, J.; Lewis, M.; Swartz, M.; et al. De Novo Damaging Variants, Clinical Phenotypes, and Post-Operative Outcomes in Congenital Heart Disease. Circ. Genom. Precis. Med. 2020, 13, e002836. [Google Scholar] [CrossRef]
- Gordon, D.M.; Cunningham, D.; Zender, G.; Lawrence, P.J.; Penaloza, J.S.; Lin, H.; Fitzgerald-Butt, S.M.; Myers, K.; Duong, T.; Corsmeier, D.J.; et al. Exome sequencing in multiplex families with left-sided cardiac defects has high yield for disease gene discovery. PLoS Genet. 2022, 18, e1010236. [Google Scholar] [CrossRef]
- Preuss, C.; Capredon, M.; Wünnemann, F.; Chetaille, P.; Prince, A.; Godard, B.; Leclerc, S.; Sobreira, N.; Ling, H.; Awadalla, P.; et al. Family Based Whole Exome Sequencing Reveals the Multifaceted Role of Notch Signaling in Congenital Heart Disease. PLoS Genet. 2016, 12, e1006335. [Google Scholar] [CrossRef]
- Durbin, M.D.; Cadar, A.G.; Williams, C.H.; Guo, Y.; Bichell, D.P.; Su, Y.R.; Hong, C.C. Hypoplastic Left Heart Syndrome Sequencing Reveals a Novel NOTCH1 Mutation in a Family with Single Ventricle Defects. Pediatr. Cardiol. 2017, 38, 1232–1240. [Google Scholar] [CrossRef]
- Helle, E.; Córdova-Palomera, A.; Ojala, T.; Saha, P.; Potiny, P.; Gustafsson, S.; Ingelsson, E.; Bamshad, M.; Nickerson, D.; Chong, J.X.; et al. Loss of function, missense, and intronic variants in NOTCH1 confer different risks for left ventricular outflow tract obstructive heart defects in two European cohorts. Genet. Epidemiol. 2019, 43, 215–226. [Google Scholar] [CrossRef]
- Montano, C.; Britton, J.F.; Harris, J.R.; Kerkhof, J.; Barnes, B.T.; Lee, J.A.; Sadikovic, B.; Sobreira, N.; Fahrner, J.A. Genome-wide DNA methylation profiling confirms a case of low-level mosaic Kabuki syndrome 1. Am. J. Med. Genet. A 2022, 188, 2217–2225. [Google Scholar] [CrossRef]
- Theis, J.L.; Olson, T.M. Whole Genome Sequencing in Hypoplastic Left Heart Syndrome. J. Cardiovasc. Dev. Dis. 2022, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yagi, H.; Saeed, S.; Bais, A.S.; Gabriel, G.C.; Chen, Z.; Peterson, K.A.; Li, Y.; Schwartz, M.C.; Reynolds, W.T.; et al. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017, 49, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Sanders, S.J.; Liu, L.; De Rubeis, S.; Lim, E.T.; Sutcliffe, J.S.; Schellenberg, G.D.; Gibbs, R.A.; Daly, M.J.; Buxbaum, J.D.; et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013, 9, e1003671. [Google Scholar] [CrossRef]
- Povysil, G.; Petrovski, S.; Hostyk, J.; Aggarwal, V.; Allen, A.S.; Goldstein, D.B. Rare-variant collapsing analyses for complex traits: Guidelines and applications. Nat. Rev. Genet. 2019, 20, 747–759. [Google Scholar] [CrossRef]
- Theis, J.L.; Niaz, T.; Sundsbak, R.S.; Fogarty, Z.C.; Bamlet, W.R.; Hagler, D.J.; Olson, T.M. CELSR1 Risk Alleles in Familial Bicuspid Aortic Valve and Hypoplastic Left Heart Syndrome. Circ. Genom. Precis. Med. 2022, 15, e003523. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.E.; Agopian, A.J.; Bhalla, A.; Glessner, J.T.; Kim, C.E.; Swartz, M.D.; Hakonarson, H.; Goldmuntz, E. Genome-wide association study of maternal and inherited effects on left-sided cardiac malformations. Hum. Mol. Genet. 2015, 24, 265–273. [Google Scholar] [CrossRef]
- Hanchard, N.A.; Swaminathan, S.; Bucasas, K.; Furthner, D.; Fernbach, S.; Azamian, M.S.; Wang, X.; Lewin, M.; Towbin, J.A.; D’Alessandro, L.C.; et al. A genome-wide association study of congenital cardiovascular left-sided lesions shows association with a locus on chromosome 20. Hum. Mol. Genet. 2016, 25, 2331–2341. [Google Scholar] [CrossRef]
- Agopian, A.J.; Goldmuntz, E.; Hakonarson, H.; Sewda, A.; Taylor, D.; Mitchell, L.E. Genome-Wide Association Studies and Meta-Analyses for Congenital Heart Defects. Circ. Cardiovasc. Genet. 2017, 10, e001449. [Google Scholar] [CrossRef]
- Agopian, A.J.; Hoang, T.T.; Goldmuntz, E.; Hakonarson, H.; Musfee, F.I.; Mitchell, L.E. X-chromosome association studies of congenital heart defects. Am. J. Med. Genet. A 2020, 182, 250–254. [Google Scholar] [CrossRef]
- Rashkin, S.R.; Cleves, M.; Shaw, G.M.; Nembhard, W.N.; Nestoridi, E.; Jenkins, M.M.; Romitti, P.A.; Lou, X.Y.; Browne, M.L.; Mitchell, L.E.; et al. A genome-wide association study of obstructive heart defects among participants in the National Birth Defects Prevention Study. Am. J. Med. Genet. A 2022, 188, 2303–2314. [Google Scholar] [CrossRef]
- Cordova-Palomera, A.; Priest, J.R. Association between the 4p16 genomic locus and different types of congenital heart disease: Results from adult survivors in the UK Biobank. Sci. Rep. 2019, 9, 16515. [Google Scholar] [CrossRef] [PubMed]
- Tcheandjieu, C.; Zanetti, D.; Yu, M.; Priest, J.R. Inherited Extremes of Aortic Diameter Confer Risk for a Specific Class of Congenital Heart Disease. Circ. Genom. Precis. Med. 2020, 13, e003170. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.J.; Sun, X.X.; Zhang, L.; Hong, Q. Association between maternal body mass index and congenital heart defects in offspring: A systematic review. Am. J. Obstet. Gynecol. 2014, 211, 91–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, T.; Chen, L.; Wang, L.; Wang, T.; Zhao, L.; Ye, Z.; Zhang, S.; Luo, L.; Zheng, Z.; et al. Risk of congenital heart defects in offspring exposed to maternal diabetes mellitus: An updated systematic review and meta-analysis. Arch. Gynecol. Obstet. 2019, 300, 1491–1506. [Google Scholar] [CrossRef] [PubMed]
- Tinker, S.C.; Gilboa, S.M.; Moore, C.A.; Waller, D.K.; Simeone, R.M.; Kim, S.Y.; Jamieson, D.J.; Botto, L.D.; Fisher, S.C.; Reefhuis, J.; et al. Modification of the association between diabetes and birth defects by obesity, National Birth Defects Prevention Study, 1997–2011. Birth Defects Res. 2021, 113, 1084–1097. [Google Scholar] [CrossRef]
- Hoang, T.T.; Marengo, L.K.; Mitchell, L.E.; Canfield, M.A.; Agopian, A.J. Original Findings and Updated Meta-Analysis for the Association between Maternal Diabetes and Risk for Congenital Heart Disease Phenotypes. Am. J. Epidemiol. 2017, 186, 118–128. [Google Scholar] [CrossRef]
- Nie, X.; Liu, X.; Wang, C.; Wu, Z.; Sun, Z.; Su, J.; Yan, R.; Peng, Y.; Yang, Y.; Wang, C.; et al. Assessment of evidence on reported non-genetic risk factors of congenital heart defects: The updated umbrella review. BMC Pregnancy Childbirth 2022, 22, 371. [Google Scholar] [CrossRef]
- Dimmock, D.; Caylor, S.; Waldman, B.; Benson, W.; Ashburner, C.; Carmichael, J.L.; Carroll, J.; Cham, E.; Chowdhury, S.; Cleary, J.; et al. Project Baby Bear: Rapid precision care incorporating rWGS in 5 California children’s hospitals demonstrates improved clinical outcomes and reduced costs of care. Am. J. Hum. Genet. 2021, 108, 1231–1238. [Google Scholar] [CrossRef]
- Kingsmore, S.F.; Cole, F.S. The Role of Genome Sequencing in Neonatal Intensive Care Units. Annu. Rev. Genom. Hum. Genet. 2022, 23, 427–448. [Google Scholar] [CrossRef]
- Manickam, K.; McClain, M.R.; Demmer, L.A.; Biswas, S.; Kearney, H.M.; Malinowski, J.; Massingham, L.J.; Miller, D.; Yu, T.W.; Hisama, F.M. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 2029–2037. [Google Scholar] [CrossRef]
- Bejjani, A.T.; Wary, N.; Gu, M. Hypoplastic left heart syndrome (HLHS): Molecular pathogenesis and emerging drug targets for cardiac repair and regeneration. Expert Opin. Ther. Targets 2021, 25, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Ethical and policy issues in genetic testing and screening of children. Pediatrics 2013, 131, 620–622. [CrossRef] [PubMed]
- Botkin, J.R.; Belmont, J.W.; Berg, J.S.; Berkman, B.E.; Bombard, Y.; Holm, I.A.; Levy, H.P.; Ormond, K.E.; Saal, H.M.; Spinner, N.B.; et al. Points to Consider: Ethical, Legal, and Psychosocial Implications of Genetic Testing in Children and Adolescents. Am. J. Hum. Genet. 2015, 97, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Austin-Tse, C.A.; Jobanputra, V.; Perry, D.L.; Bick, D.; Taft, R.J.; Venner, E.; Gibbs, R.A.; Young, T.; Barnett, S.; Belmont, J.W.; et al. Best practices for the interpretation and reporting of clinical whole genome sequencing. NPJ Genom. Med. 2022, 7, 27. [Google Scholar] [CrossRef]
- Marshall, C.R.; Chowdhury, S.; Taft, R.J.; Lebo, M.S.; Buchan, J.G.; Harrison, S.M.; Rowsey, R.; Klee, E.W.; Liu, P.; Worthey, E.A.; et al. Best practices for the analytical validation of clinical whole-genome sequencing intended for the diagnosis of germline disease. NPJ Genom. Med. 2020, 5, 47. [Google Scholar] [CrossRef]
- Incerti, D.; Xu, X.M.; Chou, J.W.; Gonzaludo, N.; Belmont, J.W.; Schroeder, B.E. Cost-effectiveness of genome sequencing for diagnosing patients with undiagnosed rare genetic diseases. Genet. Med. 2022, 24, 109–118. [Google Scholar] [CrossRef]
- Lavelle, T.A.; Feng, X.; Keisler, M.; Cohen, J.T.; Neumann, P.J.; Prichard, D.; Schroeder, B.E.; Salyakina, D.; Espinal, P.S.; Weidner, S.B.; et al. Cost-effectiveness of exome and genome sequencing for children with rare and undiagnosed conditions. Genet. Med. 2022, 24, 1349–1361. [Google Scholar] [CrossRef]
- Nurchis, M.C.; Riccardi, M.T.; Radio, F.C.; Chillemi, G.; Bertini, E.S.; Tartaglia, M.; Cicchetti, A.; Dallapiccola, B.; Damiani, G. Incremental net benefit of whole genome sequencing for newborns and children with suspected genetic disorders: Systematic review and meta-analysis of cost-effectiveness evidence. Health Policy 2022, 126, 337–345. [Google Scholar] [CrossRef]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef]
- ClinVar. 2022. Available online: https://www.ncbi.nlm.nih.gov/clinvar/?gr=0&term=hypoplastic+left+heart (accessed on 12 September 2022).
- Crucean, A.; Alqahtani, A.; Barron, D.J.; Brawn, W.J.; Richardson, R.V.; O’Sullivan, J.; Anderson, R.H.; Henderson, D.J.; Chaudhry, B. Re-evaluation of hypoplastic left heart syndrome from a developmental and morphological perspective. Orphanet J. Rare Dis. 2017, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.C.; Kadow, Z.A.; Long, H.; Morikawa, Y.; Martin, T.J.; Birks, E.J.; Campbell, K.S.; Nerbonne, J.; Lavine, K.; Wadhwa, L.; et al. Integrated multi-omic characterization of congenital heart disease. Nature 2022, 608, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Suzuki, H.; Miya, F.; Takenouchi, T.; Kosaki, K. Deciphering complex rearrangements at the breakpoint of an apparently balanced reciprocal translocation t(4:18)(q31;q11.2)dn and at a cryptic deletion: Further evidence of TLL1 as a causative gene for atrial septal defect. Am. J. Med. Genet. A 2022, 188, 2472–2478. [Google Scholar] [CrossRef]
- Noyes, M.D.; Harvey, W.T.; Porubsky, D.; Sulovari, A.; Li, R.; Rose, N.R.; Audano, P.A.; Munson, K.M.; Lewis, A.P.; Hoekzema, K.; et al. Familial long-read sequencing increases yield of de novo mutations. Am. J. Hum. Genet. 2022, 109, 631–646. [Google Scholar] [CrossRef] [PubMed]
- OMIM® An Online Catalog of Human Genes and Genetic Disorders. 2022. Available online: https://omim.org/ (accessed on 9 August 2022).
- Human Phenotype Ontology. 2022. Available online: https://monarchinitiative.org/phenotype/HP:0004383 (accessed on 9 August 2022).
- Monarch Initiative. 2022. Available online: https://hpo.jax.org/app/browse/term/HP:0004383 (accessed on 9 August 2022).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belmont, J.W. Considering the Genetic Architecture of Hypoplastic Left Heart Syndrome. J. Cardiovasc. Dev. Dis. 2022, 9, 315. https://doi.org/10.3390/jcdd9100315
Belmont JW. Considering the Genetic Architecture of Hypoplastic Left Heart Syndrome. Journal of Cardiovascular Development and Disease. 2022; 9(10):315. https://doi.org/10.3390/jcdd9100315
Chicago/Turabian StyleBelmont, John W. 2022. "Considering the Genetic Architecture of Hypoplastic Left Heart Syndrome" Journal of Cardiovascular Development and Disease 9, no. 10: 315. https://doi.org/10.3390/jcdd9100315
APA StyleBelmont, J. W. (2022). Considering the Genetic Architecture of Hypoplastic Left Heart Syndrome. Journal of Cardiovascular Development and Disease, 9(10), 315. https://doi.org/10.3390/jcdd9100315





