Getting to the Heart of Left–Right Asymmetry: Contributions from the Zebrafish Model
Abstract
1. Introduction
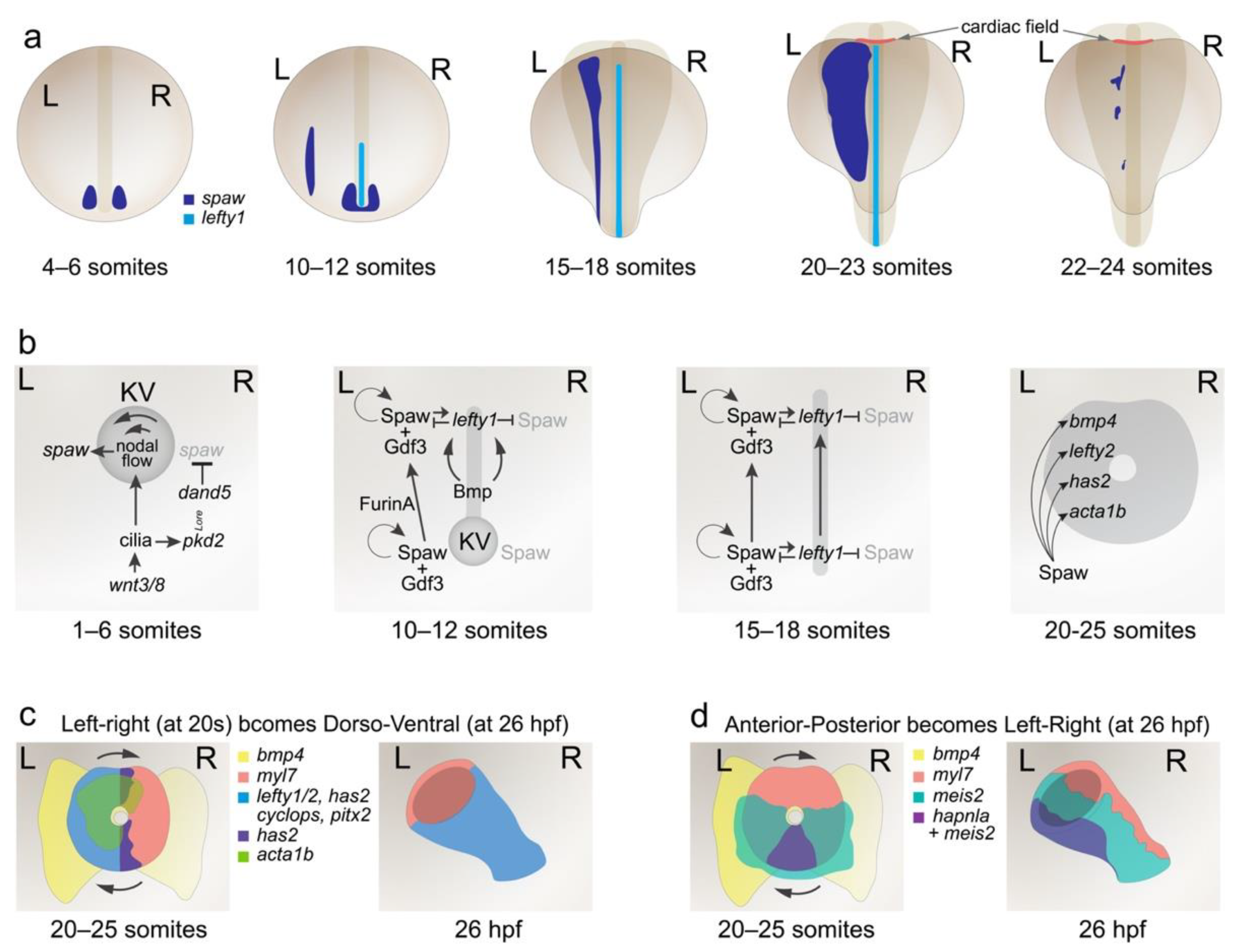
2. Establishment of the Left–Right Axis around the Kupffer’s Vesicle
3. Propagation of Left–Right Signalling from Posterior to Anterior LPM
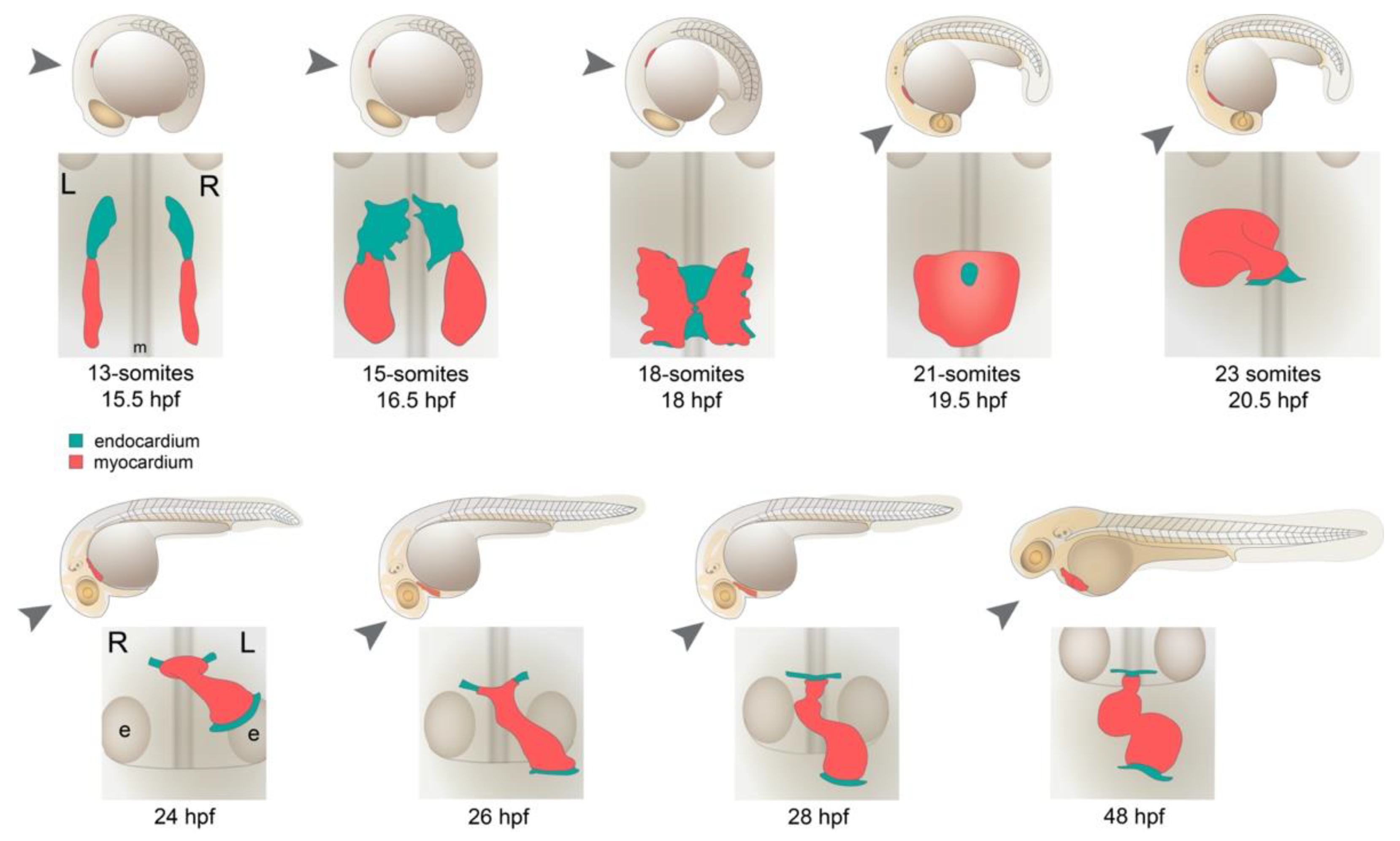
4. Differentiation of Cardiac Progenitors and Cardiac Fusion
5. Formation of the Cardiac Tube
6. Looping Morphogenesis
7. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sempou, E.; Khokha, M.K. Genes and mechanisms of heterotaxy: Patients drive the search. Curr. Opin. Genet. Dev. 2019, 56, 34–40. [Google Scholar] [CrossRef]
- Ramsdell, A.F.; Bernanke, J.M.; Johnson, J.; Trusk, T.C. Left-right lineage analysis of AV cushion tissue in normal and laterality defective Xenopus hearts. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2005, 287, 1176–1182. [Google Scholar] [CrossRef]
- Degenhardt, K.; Rychik, J. Fetal Situs, Isomerism, Heterotaxy Syndrome: Diagnostic Evaluation and Implication for Postnatal Management. Curr. Treat. Options Cardiovasc. Med. 2016, 18, 77. [Google Scholar] [CrossRef] [PubMed]
- Pierpont, M.E.; Brueckner, M.; Chung, W.K.; Garg, V.; Lacro, R.V.; McGuire, A.L.; Mital, S.; Priest, J.R.; Pu, W.; Roberts, A.; et al. Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association. Circulation 2018, 138, e653–e711. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, G.C.; Lo, C.W. Left–right patterning in congenital heart disease beyond heterotaxy. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.E.; Krikov, S.; Riehle-Colarusso, T.; Frías, J.L.; Belmont, J.; Anderka, M.; Geva, T.; Getz, K.; Botto, L.D.; the National Birth Defects Prevention Study. Laterality defects in the national birth defects prevention study (1998–2007): Birth prevalence and descriptive epidemiology. Am. J. Med. Genet. Part A 2014, 164, 2581–2591. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, J.; Verhoeven, M.C.; Abdelilah-Seyfried, S. Shaping the zebrafish heart: From left–right axis specification to epithelial tissue morphogenesis. Dev. Biol. 2009, 330, 213–220. [Google Scholar] [CrossRef]
- Desgrange, A.; Le Garrec, J.-F.; Meilhac, S.M. Left-right asymmetry in heart development and disease: Forming the right loop. Development 2018, 145, dev162776. [Google Scholar] [CrossRef]
- Essner, J.J.; Vogan, K.J.; Wagner, M.K.; Tabin, C.J.; Yost, H.J.; Brueckner, M. Conserved function for embryonic nodal cilia. Nature 2002, 418, 37–38. [Google Scholar] [CrossRef]
- Amack, J.D.; Yost, H. The T Box Transcription Factor No Tail in Ciliated Cells Controls Zebrafish Left-Right Asymmetry. Curr. Biol. 2004, 14, 685–690. [Google Scholar] [CrossRef]
- Essner, J.J.; Amack, J.D.; Nyholm, M.K.; Harris, E.B.; Yost, H.J. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 2005, 132, 1247–1260. [Google Scholar] [CrossRef]
- Kramer-Zucker, A.G.; Olale, F.; Haycraft, C.J.; Yoder, B.K.; Schier, A.F.; Drummond, I.A. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development 2005, 132, 1907–1921. [Google Scholar] [CrossRef] [PubMed]
- Roux, W. Foundations of Experimental Embryology; Oppenheimer, J.M., Willier, B.H., Eds.; Hafner Press: New York, NY, USA, 1964. [Google Scholar]
- Vandenberg, L.N.; Levin, M. A unified model for left–right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev. Biol. 2013, 379, 1–15. [Google Scholar] [CrossRef]
- Long, S.; Ahmad, N.; Rebagliati, M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 2003, 130, 2303–2316. [Google Scholar] [CrossRef] [PubMed]
- Bisgrove, B.W.; Su, Y.-C.; Yost, H.J. Maternal Gdf3 is an obligatory cofactor in Nodal signaling for embryonic axis formation in zebrafish. ELife 2017, 6, e28534. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Snarr, B.S.; Emrazian, A.; Yost, H.J. Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer’s vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 2005, 287, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, M.; Overman, J.P.; Tessadori, F.; Noël, E.; Bakkers, J.; Hertog, J.D. Noonan and LEOPARD syndrome Shp2 variants induce heart displacement defects in zebrafish. Development 2014, 141, 1961–1970. [Google Scholar] [CrossRef] [PubMed]
- Schneider, I.; Schneider, P.N.; Derry, S.W.; Lin, S.; Barton, L.J.; Westfall, T.; Slusarski, D.C. Zebrafish Nkd1 promotes Dvl degradation and is required for left–right patterning. Dev. Biol. 2010, 348, 22–33. [Google Scholar] [CrossRef]
- Hashimoto, H.; Rebagliati, M.; Ahmad, N.; Muraoka, O.; Kurokawa, T.; Hibi, M.; Suzuki, T. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 2004, 131, 1741–1753. [Google Scholar] [CrossRef]
- Montague, T.G.; Gagnon, J.A.; Schier, A.F. Conserved regulation of Nodal-mediated left-right patterning in zebrafish and mouse. Development 2018, 145, dev.171090. [Google Scholar] [CrossRef]
- Schottenfeld, J.; Sullivan-Brown, J.; Burdine, R.D. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development 2007, 134, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Zhao, L.; Brueckner, M.; Sun, Z. Intraciliary Calcium Oscillations Initiate Vertebrate Left-Right Asymmetry. Curr. Biol. 2015, 25, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Xu, X. Distinct functions of Wnt/beta-catenin signaling in KV development and cardiac asymmetry. Development 2009, 136, 207–217. [Google Scholar] [CrossRef]
- Caron, A.; Xu, X.; Lin, X. Wnt/β-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer’s vesicle. Development 2012, 139, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Mine, N.; Mochida, K.; Sakai, Y.; Saijoh, Y.; Meno, C.; Hamada, H. Nodal signaling induces the midline barrier by activating Nodal expression in the lateral plate. Development 2003, 130, 1795–1804. [Google Scholar] [CrossRef]
- Tessadori, F.; Noël, E.S.; Rens, E.G.; Magliozzi, R.; Gogh, I.J.E.-V.; Guardavaccaro, D.; Merks, R.M.; Bakkers, J. Nodal Signaling Range Is Regulated by Proprotein Convertase-Mediated Maturation. Dev. Cell 2015, 32, 631–639. [Google Scholar] [CrossRef]
- Peterson, A.G.; Wang, X.; Yost, H.J. Dvr1 transfers left-right asymmetric signals from Kupffer’s vesicle to lateral plate mesoderm in zebrafish. Dev. Biol. 2013, 382, 198–208. [Google Scholar] [CrossRef]
- Tanaka, C.; Sakuma, R.; Nakamura, T.; Hamada, H.; Saijoh, Y. Long-range action of Nodal requires interaction with GDF1. Genes Dev. 2007, 21, 3272–3282. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Van Eeden, F.J.; Warren, K.S.; Chin, A.; Nüsslein-Volhard, C.; Haffter, P.; Fishman, M.C. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 1997, 124, 4373–4382. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Essner, J.J.; Yost, H.J. Regulation of midline development by antagonism of lefty and nodal signaling. Development 1999, 126, 3253–3262. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.W.; Lord, N.D.; Gagnon, J.A.; Pauli, A.; Zimmerman, S.; Aksel, D.C.; Reyon, D.; Tsai, S.Q.; Joung, J.K.; Schier, A.F. Nodal patterning without Lefty inhibitory feedback is functional but fragile. ELife 2017, 6, e28785. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yost, H.J. Initiation and propagation of posterior to anterior (PA) waves in zebrafish left-right development. Dev. Dyn. 2008, 237, 3640–3647. [Google Scholar] [CrossRef]
- Branford, W.W.; Yost, H. Lefty-Dependent Inhibition of Nodal- and Wnt-Responsive Organizer Gene Expression Is Essential for Normal Gastrulation. Curr. Biol. 2002, 12, 2136–2141. [Google Scholar] [CrossRef]
- Lenhart, K.F.; Lin, S.-Y.; Titus, T.A.; Postlethwait, J.H.; Burdine, R.D. Two additional midline barriers function with midline lefty1 expression to maintain asymmetric Nodal signaling during left-right axis specification in zebrafish. Development 2011, 138, 4405–4410. [Google Scholar] [CrossRef]
- Smith, K.A.; Noël, E.; Thurlings, I.; Rehmann, H.; Chocron, S.; Bakkers, J. Bmp and Nodal Independently Regulate lefty1 Expression to Maintain Unilateral Nodal Activity during Left-Right Axis Specification in Zebrafish. PLoS Genet 2011, 7, e1002289. [Google Scholar] [CrossRef]
- Chocron, S.; Verhoeven, M.C.; Rentzsch, F.; Hammerschmidt, M.; Bakkers, J. Zebrafish Bmp4 regulates left–right asymmetry at two distinct developmental time points. Dev. Biol. 2007, 305, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Tessadori, F.; De Bakker, D.E.M.; Barske, L.; Nelson, N.; Algra, H.A.; Willekers, S.; Nichols, J.T.; Crump, J.G.; Bakkers, J. Zebrafish prrx1a mutants have normal hearts. Nature 2020, 585, E14–E16. [Google Scholar] [CrossRef] [PubMed]
- Castroviejo, N.; Ocaña, O.H.; Rago, L.; Coskun, H.; Arcas, A.; Galcerán, J.; Nieto, M.A. Reply to: Zebrafish prrx1a mutants have normal hearts. Nature 2020, 585, E17–E19. [Google Scholar] [CrossRef] [PubMed]
- Schoenebeck, J.; Keegan, B.R.; Yelon, D. Vessel and Blood Specification Override Cardiac Potential in Anterior Mesoderm. Dev. Cell 2007, 13, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Haack, T.; Abdelilah-Seyfried, S. The force within: Endocardial development, mechanotransduction and signalling during cardiac morphogenesis. Development 2016, 143, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Schilling, T.F.; Concordet, J.-P.; Ingham, P.W. Regulation of Left–Right Asymmetries in the Zebrafish by Shh and BMP4. Dev. Biol. 1999, 210, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Yelon, D.; Horne, S.A.; Stainier, D. Restricted Expression of Cardiac Myosin Genes Reveals Regulated Aspects of Heart Tube Assembly in Zebrafish. Dev. Biol. 1999, 214, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, J.; Bakkers, J.; Schulte-Merker, S. Early endocardial morphogenesis requires Scl/Tal1. PLoS Genet 2007, 3, e140. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.M.; Anderson, E.A.B.; Ho, R.K. Anterior lateral plate mesoderm gives rise to multiple tissues and requires tbx5a function in left-right asymmetry, migration dynamics, and cell specification of late-addition cardiac cells. Dev. Biol. 2021, 472, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.A.; Chocron, S.; von der Hardt, S.; de Pater, E.; Soufan, A.; Bussmann, J.; Schulte-Merker, S.; Hammerschmidt, M.; Bakkers, J. Rotation and Asymmetric Development of the Zebrafish Heart Requires Directed Migration of Cardiac Progenitor Cells. Dev. Cell 2008, 14, 287–297. [Google Scholar] [CrossRef]
- Lenhart, K.F.; Holtzman, N.G.; Williams, J.R.; Burdine, R.D. Integration of Nodal and BMP Signals in the Heart Requires FoxH1 to Create Left–Right Differences in Cell Migration Rates That Direct Cardiac Asymmetry. PLoS Genet 2013, 9, e1003109. [Google Scholar] [CrossRef]
- Veerkamp, J.; Rudolph, F.; Cseresnyes, Z.; Priller, F.; Otten, C.; Renz, M.; Schaefer, L.; Abdelilah-Seyfried, S. Unilateral Dampening of Bmp Activity by Nodal Generates Cardiac Left-Right Asymmetry. Dev. Cell 2013, 24, 660–667. [Google Scholar] [CrossRef]
- Noël, E.; Verhoeven, M.; Lagendijk, A.K.; Tessadori, F.; Smith, K.; Choorapoikayil, S.; Hertog, J.D.; Bakkers, J. A Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat. Commun. 2013, 4, 2754. [Google Scholar] [CrossRef]
- Rohr, S.; Otten, C.; Abdelilah-Seyfried, S. Asymmetric Involution of the Myocardial Field Drives Heart Tube Formation in Zebrafish. Circ. Res. 2008, 102, e12–e19. [Google Scholar] [CrossRef]
- Baker, K.; Holtzman, N.G.; Burdine, R.D. Direct and indirect roles for Nodal signaling in two axis conversions during asymmetric morphogenesis of the zebrafish heart. Proc. Natl. Acad. Sci. USA 2008, 105, 13924–13929. [Google Scholar] [CrossRef]
- de Campos-Baptista, M.I.; Holtzman, N.G.; Yelon, D.; Schier, A.F. Nodal signaling promotes the speed and directional movement of cardiomyocytes in zebrafish. Dev. Dyn. 2008, 237, 3624–3633. [Google Scholar] [CrossRef]
- Guerra, A.; Germano, R.F.V.; Stone, O.; Arnaout, R.; Guenther, S.; Ahuja, S.; Uribe, V.; Vanhollebeke, B.; Stainier, D.Y.R.; Reischauer, S. Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish. Elife 2018, 7, e32833. [Google Scholar] [CrossRef]
- Derrick, C.J.; Sánchez-Posada, J.; Hussein, F.; Tessadori, F.; Pollitt, E.J.G.; Savage, A.M.; Wilkinson, R.N.; Chico, T.J.; van Eeden, F.J.; Bakkers, J.; et al. Asymmetric Hapln1a drives regionalised cardiac ECM expansion and promotes heart morphogenesis in zebrafish development. Cardiovasc. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Auman, H.J.; Coleman, H.; Riley, H.E.; Olale, F.; Tsai, H.-J.; Yelon, D. Functional Modulation of Cardiac Form through Regionally Confined Cell Shape Changes. PLoS Biol. 2007, 5, e53. [Google Scholar] [CrossRef] [PubMed]
- Merks, A.M.; Swinarski, M.; Meyer, A.M.; Müller, N.V.; Özcan, I.; Donat, S.; Burger, A.; Gilbert, S.; Mosimann, C.; Abdelilah-Seyfried, S.; et al. Planar cell polarity signalling coordinates heart tube remodelling through tissue-scale polarisation of actomyosin activity. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Singleman, C.; Holtzman, N.G. Analysis of postembryonic heart development and maturation in the zebrafish, Danio rerio. Dev. Dyn. 2012, 241, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Scherf, N.; Meyer, A.M.; Panáková, D.; Kohl, P.; Huisken, J. Cell-accurate optical mapping across the entire developing heart. ELife 2017, 6, e28307. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, V.A.; Heise, M.; Moghtadaei, M.; Bornhorst, D.; Männer, J.; Abdelilah-Seyfried, S. Morphogenetic control of zebrafish cardiac looping by Bmp signaling. Development 2019, 146, dev.180091. [Google Scholar] [CrossRef] [PubMed]
- Grimes, D.T.; Patterson, V.L.; Luna-Arvizu, G.; Schottenfeld-Roames, J.; Irons, Z.H.; Burdine, R.D. Left-right asymmetric heart jogging increases the robustness of dextral heart looping in zebrafish. Dev. Biol. 2020, 459, 79–86. [Google Scholar] [CrossRef] [PubMed]
- de Pater, E.; Clijsters, L.; Marques, S.R.; Lin, Y.; Garavito-Aguilar, Z.V.; Yelon, D.; Bakkers, J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 2009, 136, 1633–1641. [Google Scholar] [CrossRef]
- Zhou, Y.; Cashman, T.; Nevis, K.R.; Obregon, P.; Carney, S.A.; Liu, Y.; Gu, A.; Mosimann, C.; Sondalle, S.; Peterson, R.E.; et al. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 2011, 474, 645–648. [Google Scholar] [CrossRef]
- Bayraktar, M.; Männer, J. Cardiac looping may be driven by compressive loads resulting from unequal growth of the heart and pericardial cavity. Observations on a physical simulation model. Front. Physiol. 2014, 5, 112. [Google Scholar] [CrossRef]
- Guner-Ataman, B.; Paffett-Lugassy, N.; Adams, M.S.; Nevis, K.R.; Jahangiri, L.; Obregon, P.; Kikuchi, K.; Poss, K.D.; Burns, C.E. Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 2013, 140, 1353–1363. [Google Scholar] [CrossRef]
- Jahangiri, L.; Sharpe, M.; Novikov, N.; González-Rosa, J.M.; Borikova, A.; Nevis, K.; Paffett-Lugassy, N.; Zhao, L.; Adams, M.; Guner-Ataman, B.; et al. The AP-1 transcription factor component Fosl2 potentiates the rate of myocardial differentiation from the zebrafish second heart field. Development 2016, 143, 113–122. [Google Scholar] [CrossRef]
- Zeng, X.-X.I.; Yelon, D. Cadm4 Restricts the Production of Cardiac Outflow Tract Progenitor Cells. Cell Rep. 2014, 7, 951–960. [Google Scholar] [CrossRef]
- Duong, T.B.; Holowiecki, A.; Waxman, J.S. Retinoic acid signaling restricts the size of the first heart field within the anterior lateral plate mesoderm. Dev. Biol. 2021, 473, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, A.J.; Huq, A.; Weinstein, B.M.; Walker, C.; Fishman, M.; Stainier, D.Y.R. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002, 31, 106–110. [Google Scholar] [CrossRef]
- Steed, E.; Faggianelli, N.; Roth, S.; Ramspacher, C.; Concordet, J.-P.; Vermot, J. klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nat. Commun. 2016, 7, 11646. [Google Scholar] [CrossRef] [PubMed]
- Govindan, J.; Iovine, M.K. Hapln1a Is Required for Connexin43-Dependent Growth and Patterning in the Regenerating Fin Skeleton. PLoS ONE 2014, 9, e88574. [Google Scholar] [CrossRef]
- Pfefferli, C.; Moran, H.; Felker, A.; Mosimann, C.; Jaźwińska, A. Persistent Ventricle Partitioning in the Adult Zebrafish Heart. J. Cardiovasc. Dev. Dis. 2021, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Kalogirou, S.; Malissovas, N.; Moro, E.; Argenton, F.; Stainier, D.; Beis, D. Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc. Res. 2014, 104, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Maerker, M.; Getwan, M.; Dowdle, M.E.; Pelliccia, J.L.; McSheene, J.C.; Yartseva, V.; Minegishi, K.; Vick, P.; Giraldez, A.J.; Hamada, H.; et al. Bicc1 and dicer regulate left-right patterning through post-transcriptional control of the Nodal-inhibitor dand5. BioRxiv 2020. [Google Scholar] [CrossRef]
- Chrystal, P.W.; French, C.R.; Jean, F.; Havrylov, S.; van Baarle, S.; Peturson, A.-M.; Xu, P.; Crump, J.G.; Pilgrim, D.B.; Lehmann, O.J.; et al. The Axenfeld-Rieger syndrome gene FOXC1 contributes to left-right patterning. Genes 2021, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Tessadori, F.; Kruse, F.; van den Brink, S.C.; van den Boogaard, M.; Christoffels, V.M.; Bakkers, J. Twisting of the heart tube during cardiac looping is a tbx5-dependent and tissue-intrinsic process. BioRxiv 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, K.A.; Uribe, V. Getting to the Heart of Left–Right Asymmetry: Contributions from the Zebrafish Model. J. Cardiovasc. Dev. Dis. 2021, 8, 64. https://doi.org/10.3390/jcdd8060064
Smith KA, Uribe V. Getting to the Heart of Left–Right Asymmetry: Contributions from the Zebrafish Model. Journal of Cardiovascular Development and Disease. 2021; 8(6):64. https://doi.org/10.3390/jcdd8060064
Chicago/Turabian StyleSmith, Kelly A., and Veronica Uribe. 2021. "Getting to the Heart of Left–Right Asymmetry: Contributions from the Zebrafish Model" Journal of Cardiovascular Development and Disease 8, no. 6: 64. https://doi.org/10.3390/jcdd8060064
APA StyleSmith, K. A., & Uribe, V. (2021). Getting to the Heart of Left–Right Asymmetry: Contributions from the Zebrafish Model. Journal of Cardiovascular Development and Disease, 8(6), 64. https://doi.org/10.3390/jcdd8060064






