Persistent Ventricle Partitioning in the Adult Zebrafish Heart
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry and Procedures
2.2. Lineage Labeling and Analysis of Embryonic Hearts
2.3. Imaging and Analysis of Adult Hearts and Heart Sections
3. Results
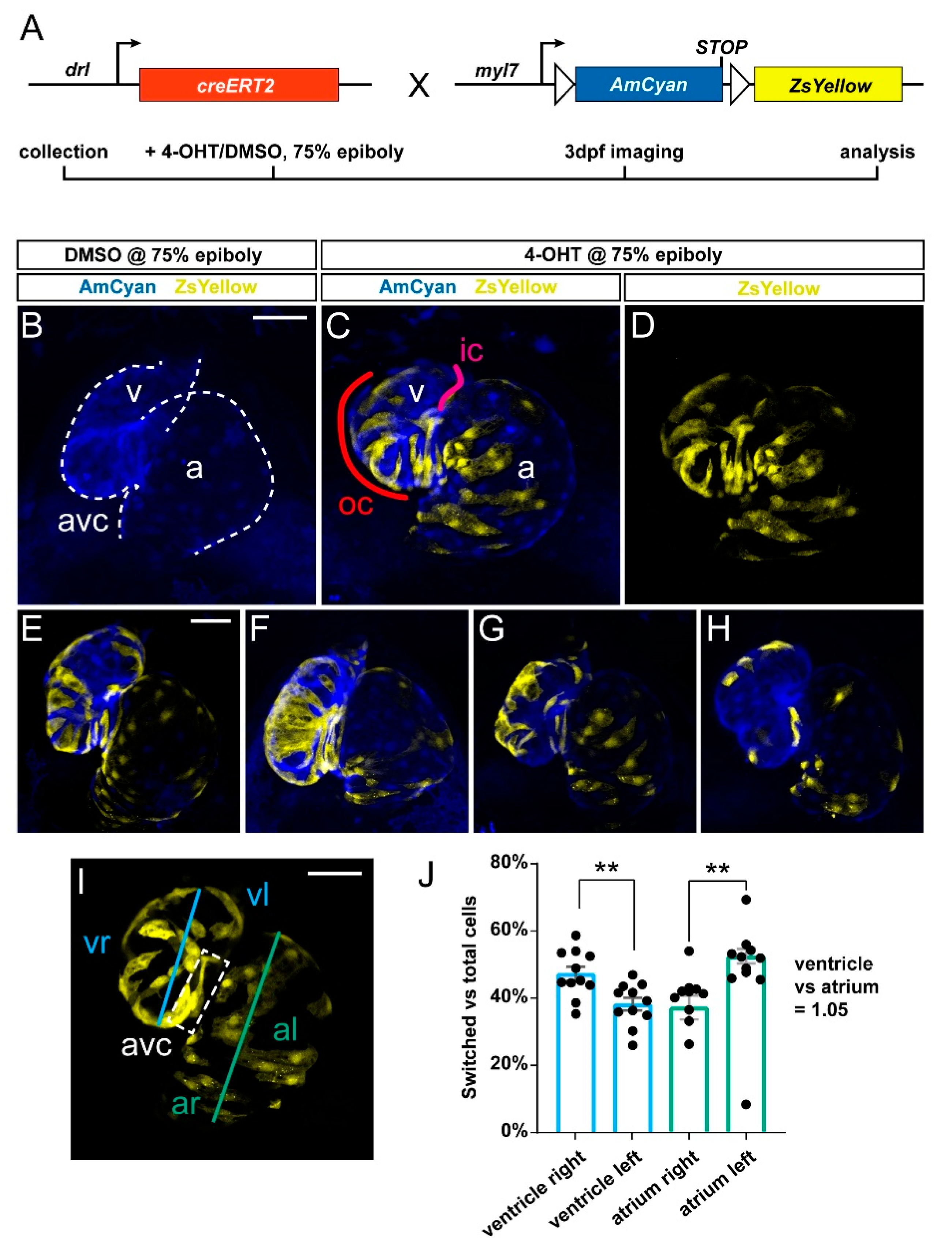
3.1. Preferentially Labeling of First Heart Field-Derived Myocardium Using drl:creERT2
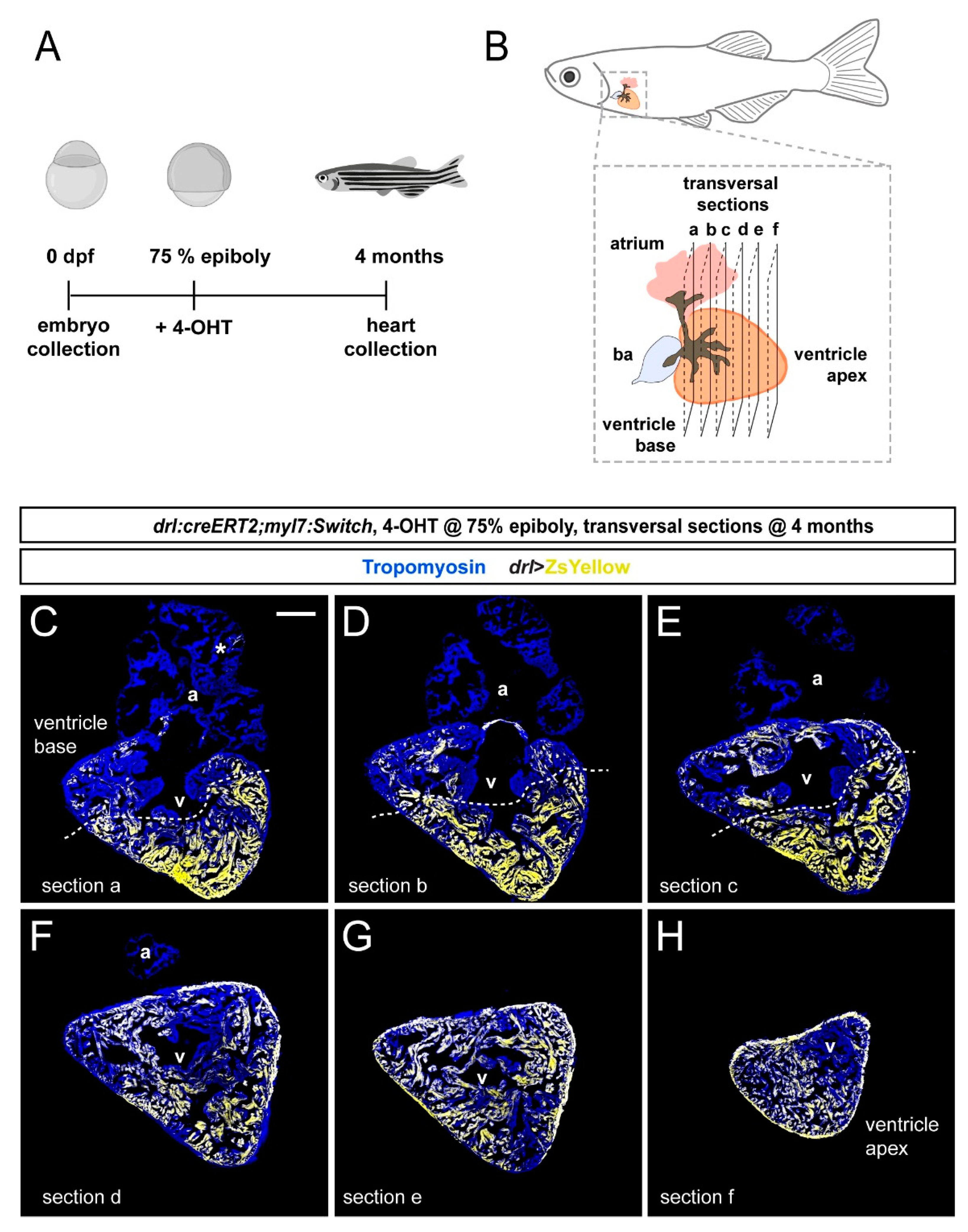
3.2. FHF Cardiomyocytes Enrich at the Atrium-Opposite Side of the Adult Ventricle
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swedlund, B.; Lescroart, F. Cardiopharyngeal Progenitor Specification: Multiple Roads to the Heart and Head Muscles. Cold Spring Harb. Perspect. Biol. 2019, 12, a036731. [Google Scholar] [CrossRef]
- Lambers, E.; Kume, T. Navigating the labyrinth of cardiac regeneration. Dev. Dyn. 2016, 245, 751–761. [Google Scholar] [CrossRef]
- Yao, Y.; Marra, A.N.; Yelon, D. Pathways Regulating Establishment and Maintenance of Cardiac Chamber Identity in Zebrafish. J. Cardiovasc. Dev. Dis. 2021, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011, 91, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felker, A.; Prummel, K.D.; Merks, A.M.; Mickoleit, M.; Brombacher, E.C.; Huisken, J.; Panáková, D.; Mosimann, C. Continuous addition of progenitors forms the cardiac ventricle in zebrafish. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Kemmler, C.L.; Riemslagh, F.W.; Moran, H.R.; Mosimann, C. From Stripes to a Beating Heart: Early Cardiac Development in Zebrafish. J. Cardiovasc. Dev. Dis. 2021, 8, 17. [Google Scholar] [CrossRef]
- De Pater, E.; Clijsters, L.; Marques, S.R.; Lin, Y.-F.F.; Garavito-Aguilar, Z.V.; Yelon, D.; Bakkers, J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 2009, 136, 1633–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hami, D.; Grimes, A.C.; Tsai, H.J.; Kirby, M.L. Zebrafish cardiac development requires a conserved secondary heart field. Development 2011, 138, 2389–2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazic, S.; Scott, I.C. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 2011, 354, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cashman, T.J.; Nevis, K.R.; Obregon, P.; Carney, S.A.; Liu, Y.; Gu, A.; Mosimann, C.; Sondalle, S.; Peterson, R.E.; et al. Latent TGF-βbinding protein 3 identifies a second heart field in zebrafish. Nature 2011, 474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosimann, C.; Panáková, D.; Werdich, A.A.; Musso, G.; Burger, A.; Lawson, K.L.; Carr, L.A.; Nevis, K.R.; Sabeh, M.K.; Zhou, Y.; et al. Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolkin, T.; Christiaen, L. Development and evolution of the ascidian cardiogenic mesoderm. Curr. Top. Dev. Biol. 2012, 100, 107–142. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.G. The Second Heart Field. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 100, pp. 33–65. [Google Scholar]
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prummel, K.D.; Nieuwenhuize, S.; Mosimann, C. The lateral plate mesoderm. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Guner-Ataman, B.; Paffett-Lugassy, N.; Adams, M.S.; Nevis, K.R.; Jahangiri, L.; Obregon, P.; Kikuchi, K.; Poss, K.D.; Burns, C.E.; Burns, C.G. Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 2013, 140, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Rydeen, A.A.B.; Waxman, J.J.S.; Bruneau, B.; Bruneau, B.; Buckingham, M.; Meilhac, S.; Zaffran, S.; Abu-Issa, R.; Kirby, M.; Lazic, S.; et al. Cyp26 Enzymes Facilitate Second Heart Field Progenitor Addition and Maintenance of Ventricular Integrity. PLoS Biol. 2016, 14, e2000504. [Google Scholar] [CrossRef]
- Zeng, X.-X.I.; Yelon, D. Cadm4 Restricts the Production of Cardiac Outflow Tract Progenitor Cells. Cell Rep. 2014, 7, 951–960. [Google Scholar] [CrossRef] [Green Version]
- Mori, A.D.; Bruneau, B.G. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr. Opin. Cardiol. 2004, 19, 211–215. [Google Scholar] [CrossRef]
- Morgenthau, A.; Frishman, W.H. Genetic Origins of Tetralogy of Fallot. Cardiol. Rev. 2018, 26, 86–92. [Google Scholar] [CrossRef]
- Vincent, S.D.; Buckingham, M.E. How to make a heart. Curr Top. Dev. Biol 2010, 90, 1–41. [Google Scholar] [CrossRef]
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835. [Google Scholar] [CrossRef]
- Koshiba-Takeuchi, K.; Mori, A.D.; Kaynak, B.L.; Cebra-Thomas, J.; Sukonnik, T.; Georges, R.O.; Latham, S.; Beck, L.; Henkelman, R.M.; Black, B.L.; et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 2009, 461, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Hanemaaijer, J.; Gregorovicova, M.; Nielsen, J.M.; Moorman, A.F.M.; Wang, T.; Planken, R.N.; Christoffels, V.M.; Sedmera, D.; Jensen, B. Identification of the building blocks of ventricular septation in monitor lizards (Varanidae). Development 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Q.; Stinnett, H.K.; Ho, R.K. Asymmetric cell convergence-driven zebrafish fin bud initiation and pre-pattern requires Tbx5a control of a mesenchymal Fgf signal. Development 2015, 142, 4329–4339. [Google Scholar] [CrossRef] [Green Version]
- Guerra, A.; Germano, R.F.; Stone, O.; Arnaout, R.; Guenther, S.; Ahuja, S.; Uribe, V.; Vanhollebeke, B.; Stainier, D.Y.; Reischauer, S. Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish. Elife 2018, 7, e32833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prummel, K.D.; Hess, C.; Nieuwenhuize, S.; Parker, H.J.; Rogers, K.W.; Kozmikova, I.; Racioppi, C.; Brombacher, E.C.; Czarkwiani, A.; Knapp, D.; et al. A conserved regulatory program initiates lateral plate mesoderm emergence across chordates. Nat. Commun. 2019, 10, 3857. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Iranzo, H.; Galardi-Castilla, M.; Minguillón, C.; Sanz-Morejón, A.; González-Rosa, J.M.J.M.; Felker, A.; Ernst, A.; Guzmán-Martínez, G.; Mosimann, C.; Mercader, N. Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat. Commun. 2018, 9, 428. [Google Scholar] [CrossRef] [Green Version]
- Carney, T.J.; Mosimann, C. Switch and Trace: Recombinase Genetics in Zebrafish. Trends Genet. 2018, 34, 362–378. [Google Scholar] [CrossRef]
- Sauer, B.; Henderson, N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 1988, 85, 5166–5170. [Google Scholar] [CrossRef] [Green Version]
- Branda, C.S.; Dymecki, S.M. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 2004, 6, 7–28. [Google Scholar] [CrossRef] [Green Version]
- Metzger, D.; Clifford, J.; Chiba, H.; Chambon, P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA 1995, 92, 6991–6995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feil, R.; Brocard, J.; Mascrez, B.; LeMeur, M.; Metzger, D.; Chambon, P. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. USA 1996, 93, 10887–10890. [Google Scholar] [CrossRef] [Green Version]
- Hans, S.; Kaslin, J.; Freudenreich, D.; Brand, M. Temporally-controlled site-specific recombination in zebrafish. PLoS ONE 2009, 4, e4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hans, S.; Freudenreich, D.; Geffarth, M.; Kaslin, J.; Machate, A.; Brand, M. Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev. Dyn. 2011, 240, 108–115. [Google Scholar] [CrossRef]
- Mosimann, C.; Kaufman, C.K.; Li, P.; Pugach, E.K.; Tamplin, O.J.; Zon, L.I. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 2011, 138, 169–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felker, A.; Nieuwenhuize, S.; Dolbois, A.; Blazkova, K.; Hess, C.; Low, L.W.L.; Burger, S.; Samson, N.; Carney, T.J.; Bartunek, P.; et al. In Vivo Performance and Properties of Tamoxifen Metabolites for CreERT2 Control. PLoS ONE 2016, 11, e0152989. [Google Scholar] [CrossRef] [Green Version]
- Felker, A.; Mosimann, C. Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 2016, 135, 219–244. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Bise, T.; Sallin, P.; Pfefferli, C.; Jaźwińska, A. Multiple cryoinjuries modulate the efficiency of zebrafish heart regeneration. Sci. Rep. 2020, 10, 11551. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Pfefferli, C.; Jaźwińska, A. The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Verzi, M.P.; McCulley, D.J.; De Val, S.; Dodou, E.; Black, B.L. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 2005, 287, 134–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.-M.; Black, B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 2004, 131, 3931–3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolfi, A.; Gainous, T.B.; Young, J.J.; Mori, A.; Levine, M.; Christiaen, L. Early chordate origins of the vertebrate second heart field. Science 2010, 329, 565–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diogo, R.; Kelly, R.G.; Christiaen, L.; Levine, M.; Ziermann, J.M.; Molnar, J.L.; Noden, D.M.; Tzahor, E. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature 2015, 520, 466–473. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfefferli, C.; Moran, H.R.; Felker, A.; Mosimann, C.; Jaźwińska, A. Persistent Ventricle Partitioning in the Adult Zebrafish Heart. J. Cardiovasc. Dev. Dis. 2021, 8, 41. https://doi.org/10.3390/jcdd8040041
Pfefferli C, Moran HR, Felker A, Mosimann C, Jaźwińska A. Persistent Ventricle Partitioning in the Adult Zebrafish Heart. Journal of Cardiovascular Development and Disease. 2021; 8(4):41. https://doi.org/10.3390/jcdd8040041
Chicago/Turabian StylePfefferli, Catherine, Hannah R. Moran, Anastasia Felker, Christian Mosimann, and Anna Jaźwińska. 2021. "Persistent Ventricle Partitioning in the Adult Zebrafish Heart" Journal of Cardiovascular Development and Disease 8, no. 4: 41. https://doi.org/10.3390/jcdd8040041




