Mis-Expression of a Cranial Neural Crest Cell-Specific Gene Program in Cardiac Neural Crest Cells Modulates HAND Factor Expression, Causing Cardiac Outflow Tract Phenotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transgenic Mice
2.2. Cloning
2.3. Bone and Cartilage Staining, X-Gal Staining and Histology
2.4. Lysotracker and TUNEL
2.5. Immunohistochemistry
2.6. In Situ Hybridization
2.7. Quantitative RT-PCR
3. Results
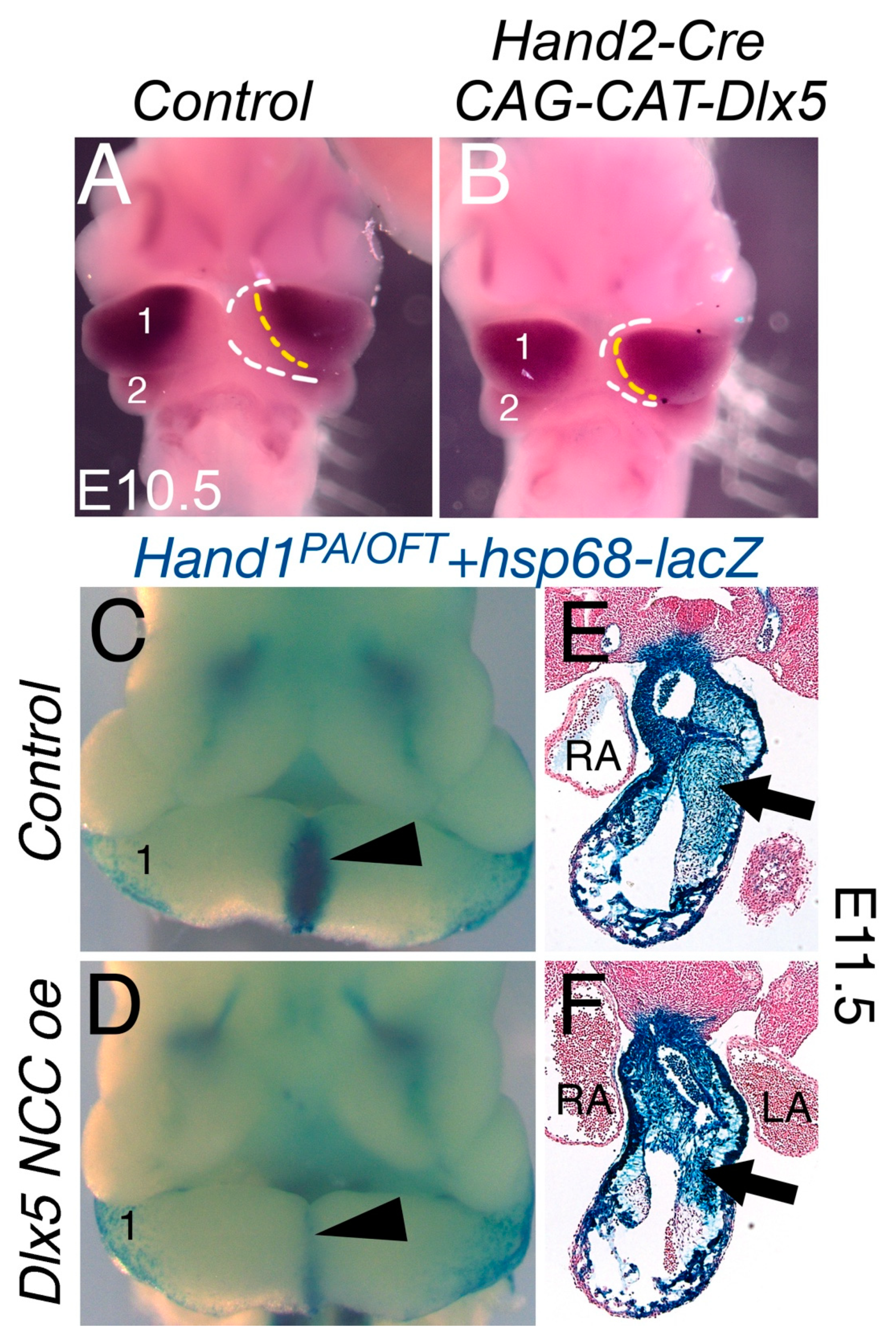
3.1. NCC Expression of CAG-CAT-Dlx5 Results in Midface Clefting
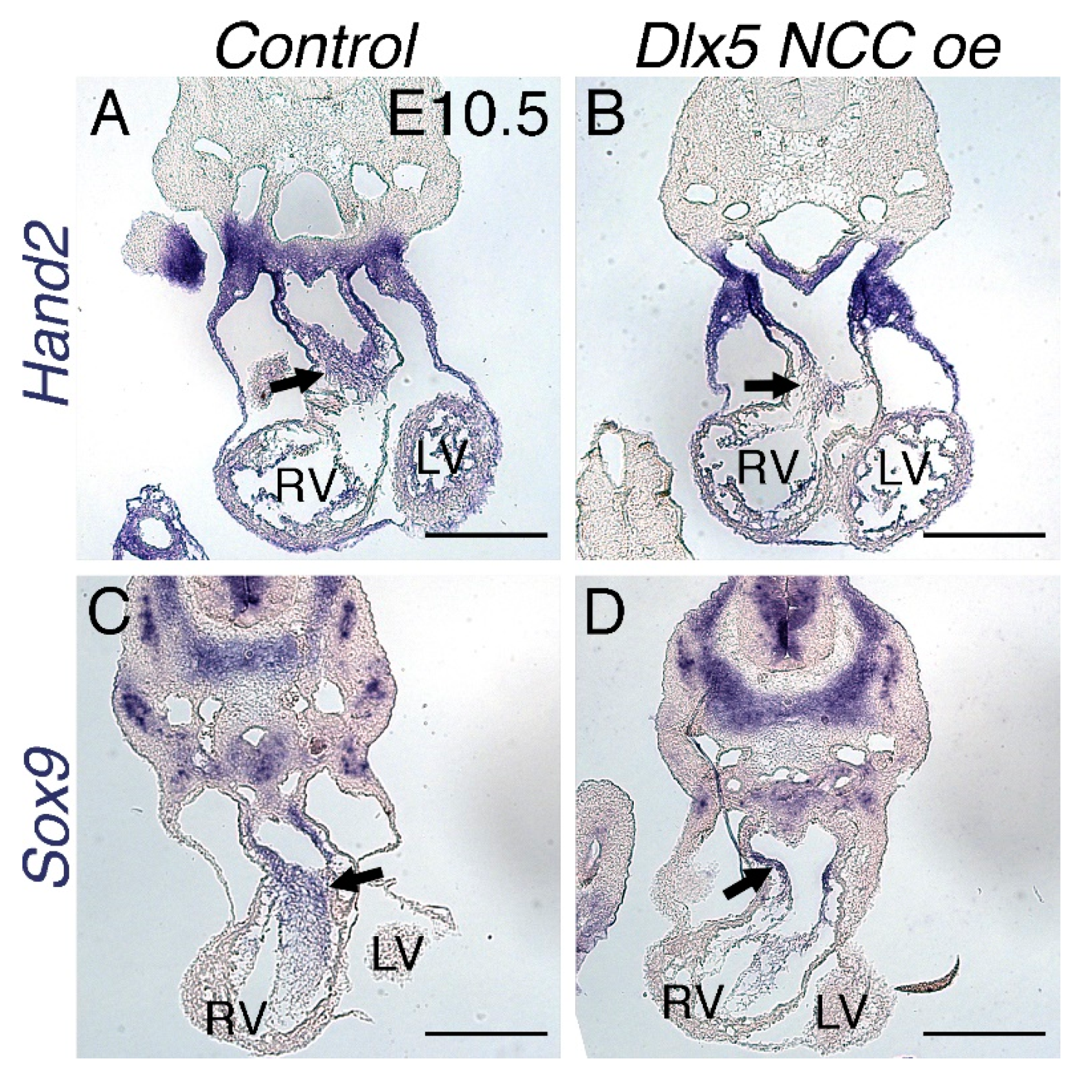
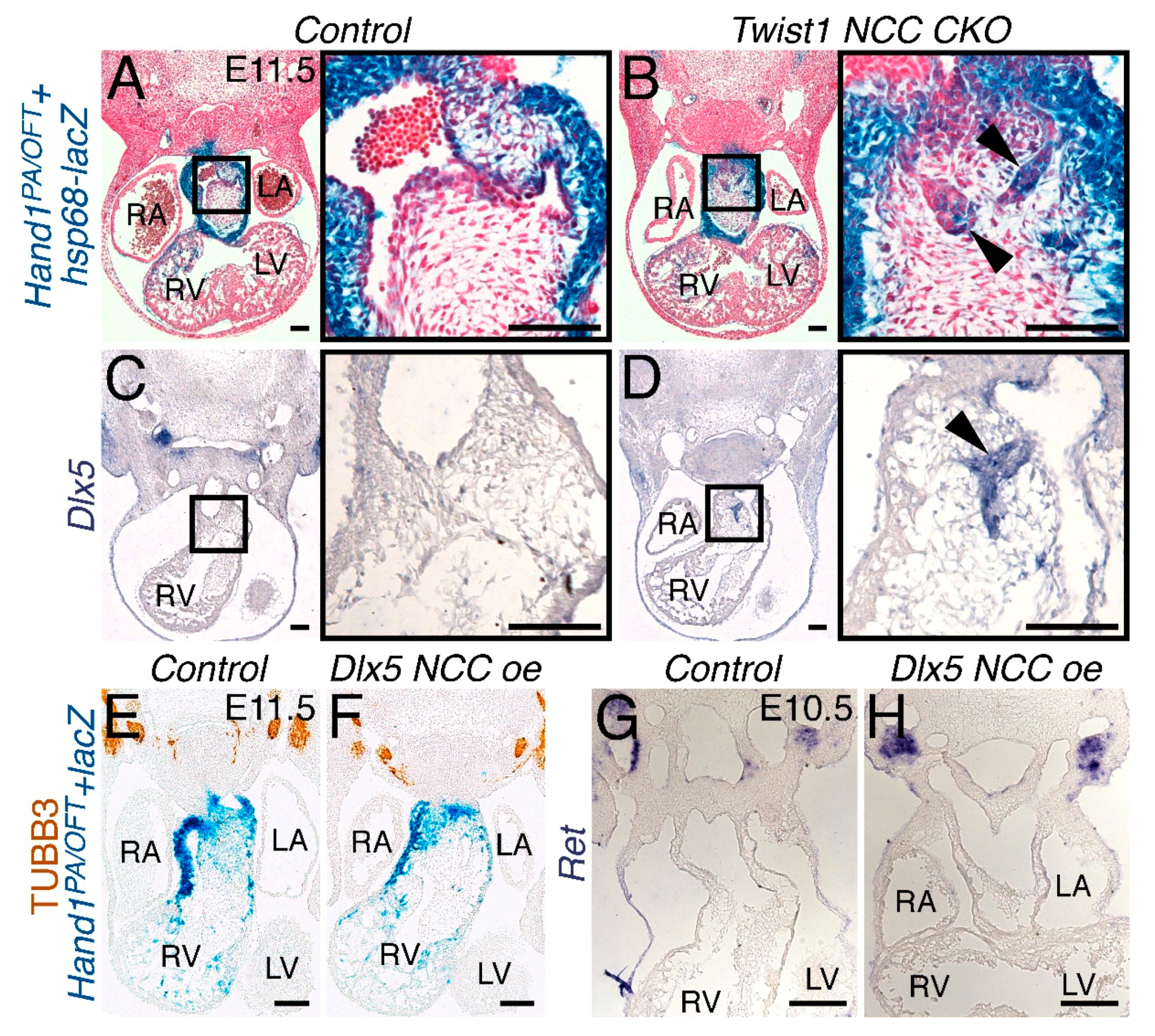
3.2. NCC Expression of CAG-CAT-Dlx5 Down Regulates Hand1 Ventral Cap Expression
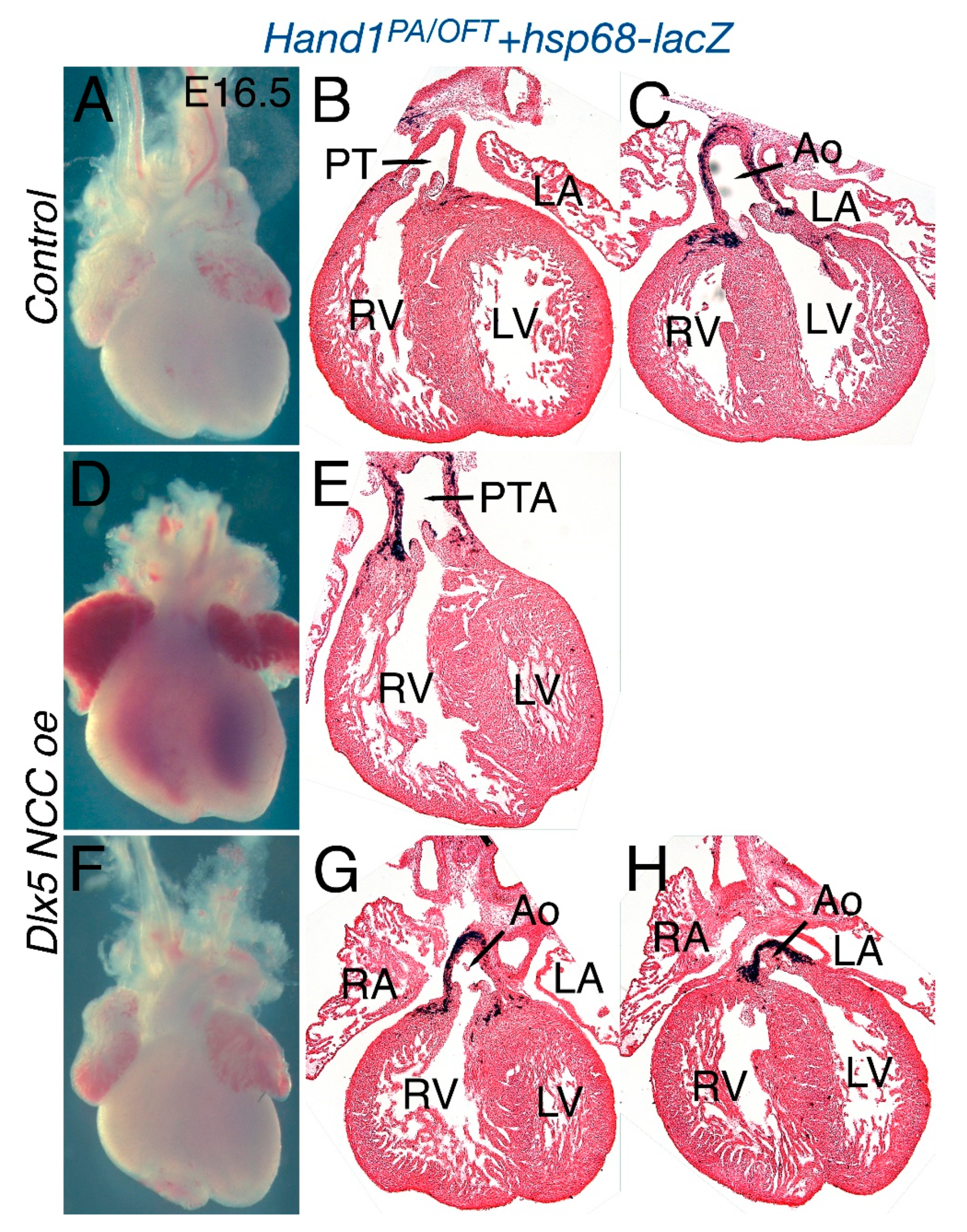
3.3. Dlx5 NCC oe OFTs Present Persistent Truncus Arteriosus (PTA)
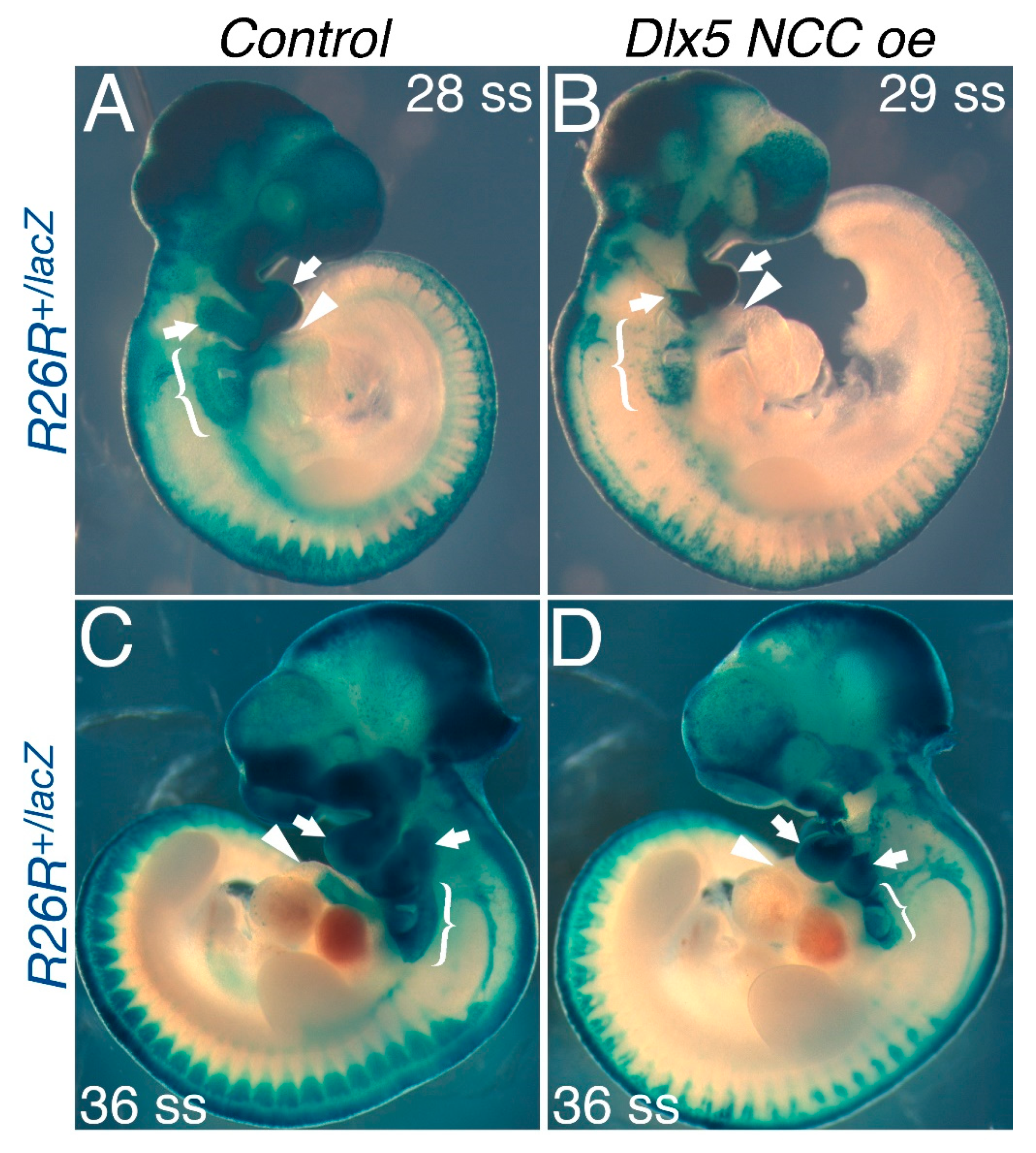
3.4. NCC Migration into the OFT Is Limited in Dlx5 NCC oe Mice
3.5. Dlx5 NCC oe Embryos Exhibit NCC Cell Death Within the Neural Tube, Pharyngeal Arches but Minimally Within the OFT
3.6. Persistent Dlx5 Expression Does Not Induce NCCs to Adopt a Neuronal Cell Fate
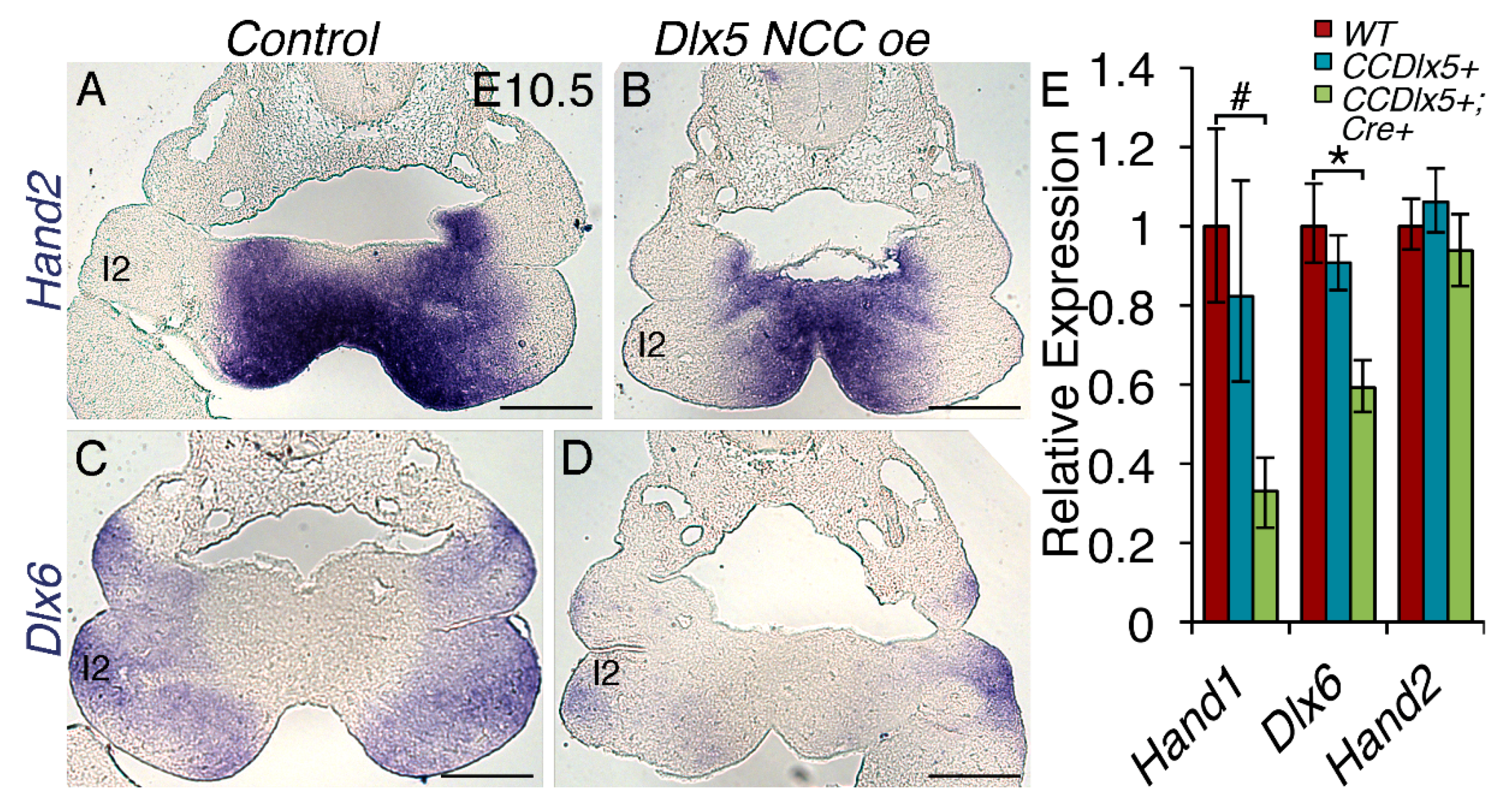
3.7. Persistent Dlx5 Expression within NCCs Downregulates Dlx6 Expression
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoffman, J.I.E. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr. Cardiol. 1995, 16, 155–165. [Google Scholar] [CrossRef]
- Schilling, T.F.; Le pabic, P. Neural Crest Cells; Academic Press: London, UK, 2014. [Google Scholar] [CrossRef]
- Ruest, L.-B.; Dager, M.; Yanagisawa, H.; Charité, J.; Hammer, R.E.; Olson, E.N.; Yanagisawa, M.; Clouthier, D.E. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Boil. 2003, 257, 263–277. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, A.C.; Funato, N.; Chapman, S.; McKee, M.D.; Richardson, J.A.; Olson, E.N.; Yanagisawa, H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev. Boil. 2007, 310, 154–168. [Google Scholar] [CrossRef] [Green Version]
- Hendershot, T.J.; Liu, H.; Clouthier, D.E.; Shepherd, I.T.; Coppola, E.; Studer, M.; Firulli, A.B.; Pittman, U.L.; Howard, M.J. Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev. Boil. 2008, 319, 179–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barron, F.; Woods, C.; Kuhn, K.; Bishop, J.; Howard, M.J.; Clouthier, D.E. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development 2011, 138, 2249–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, R.M.; Firulli, A.B. Hand Factors in Cardiac Development. Anat. Rec. Adv. Integr. Anat. Evol. Boil. 2018, 302, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincentz, J.W.; Casasnovas, J.J.; Barnes, R.M.; Que, J.; Clouthier, D.E.; Wang, J.; Firulli, A.B. Exclusion of Dlx5/6 expression from the distal-most mandibular arches enables BMP-mediated specification of the distal cap. Proc. Natl. Acad. Sci. USA 2016, 113, 7563–7568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firulli, B.A.; Fuchs, R.K.; Vincentz, J.W.; Clouthier, D.E.; Firulli, A.B. Hand1 phosphoregulation within the distal arch neural crest is essential for craniofacial morphogenesis. Development 2014, 141, 3050–3061. [Google Scholar] [CrossRef] [Green Version]
- Bonilla-Claudio, M.; Wang, J.; Bai, Y.; Klysik, E.; Selever, J.; Martin, J.F. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development 2012, 139, 709–719. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Selever, J.; Murali, D.; Sun, X.; Brugger, S.M.; Ma, L.; Schwartz, R.J.; Maxson, R.; Furuta, Y.; Martin, J.F. Threshold-specific requirements for Bmp4 in mandibular development. Dev. Boil. 2005, 283, 282–293. [Google Scholar] [CrossRef] [Green Version]
- Soldatov, R.; Kaucká, M.; Kastriti, M.E.; Petersen, J.; Chontorotzea, T.; Englmaier, L.; Akkuratova, N.; Yang, Y.; Häring, M.; Dyachuk, V.; et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 2019, 364, eaas9536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincentz, J.W.; Firulli, B.A.; Lin, A.; Spicer, U.B.; Howard, M.J.; Firulli, A.B. Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest. PLoS Genet. 2013, 9, e1003405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincentz, J.W.; VanDusen, N.J.; Fleming, A.B.; Rubart, M.; Firulli, B.A.; Howard, M.J.; Firulli, A.B. A Phox2- and Hand2-dependent Hand1 cis-regulatory element reveals a unique gene dosage requirement for Hand2 during sympathetic neurogenesis. J. Neurosci. 2012, 32, 2110–2120. [Google Scholar] [CrossRef] [PubMed]
- Connerney, J.; Andreeva, V.; Leshem, Y.; Muentener, C.; Mercado, M.A.; Spicer, D.B. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev. Dyn. 2006, 235, 1334–1346. [Google Scholar] [CrossRef]
- Barnes, R.M.; Firulli, B.A.; Conway, S.J.; Vincentz, J.W.; Firulli, A.B. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev. Dyn. 2010, 239, 3086–3097. [Google Scholar] [CrossRef] [Green Version]
- VanDusen, N.J.; Casanovas, J.; Vincentz, J.W.; Firulli, B.A.; Osterwalder, M.; Lopez-Rios, J.; Zeller, R.; Zhou, B.; Grego-Bessa, J.; De La Pompa, J.L.; et al. Hand2 is an essential regulator for two Notch-dependent functions within the embryonic endocardium. Cell Rep. 2014, 9, 2071–2083. [Google Scholar] [CrossRef] [Green Version]
- Firulli, B.A.; Milliar, H.; Toolan, K.P.; Harkin, J.; Fuchs, R.K.; Robling, A.G.; Firulli, A.B. Defective Hand1 phosphoregulation uncovers essential roles for Hand1 in limb morphogenesis. Development 2017, 144, 2480–2489. [Google Scholar] [CrossRef] [Green Version]
- Vincentz, J.; Toolan, K.P.; Zhang, W.; Firulli, A.B. Hand factor ablation causes defective left ventricular chamber development and compromised adult cardiac function. PLoS Genet. 2017, 13, e1006922. [Google Scholar] [CrossRef]
- Vincentz, J.W.; Barnes, R.M.; Rodgers, R.; Firulli, B.A.; Conway, S.J.; Firulli, A.B. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev. Boil. 2008, 320, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Barnes, R.M.; Firulli, B.A.; VanDusen, N.J.; Morikawa, Y.; Conway, S.J.; Cserjesi, P.; Vincentz, J.W.; Firulli, A.B. Hand2 loss-of-function in Hand1-expressing cells reveals distinct roles in epicardial and coronary vessel development. Circ. Res. 2011, 108, 940–949. [Google Scholar] [CrossRef] [Green Version]
- Danielian, P.; Muccino, D.; Rowitch, D.H.; Michael, S.K.; McMahon, A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Boil. 1998, 8, 1323-S2. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, M.; Narboux-Nême, N.; Gitton, Y.; De Lombares, C.; Fontaine, A.; Alfama, G.; Kitazawa, T.; Kawamura, Y.; Heude, E.; Marshall, L.; et al. Probing the origin of matching functional jaws: Roles of Dlx5/6 in cranial neural crest cells. Sci. Rep. 2018, 8, 14975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robledo, R.F.; Rajan, L.; Li, X.; Lufkin, T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genome Res. 2002, 16, 1089–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavares, A.; Clouthier, D.E. Cre recombinase-regulated Endothelin1 transgenic mouse lines: Novel tools for analysis of embryonic and adult disorders. Dev. Boil. 2015, 400, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Tani-Matsuhana, S.; Vieceli, F.; Gandhi, S.; Inoue, K.; E Bronner, M. Transcriptome profiling of the cardiac neural crest reveals a critical role for MafB. Dev. Boil. 2018, 444, S209–S218. [Google Scholar] [CrossRef]
- Minoux, M.; Rijli, F.M. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 2010, 137, 2605–2621. [Google Scholar] [CrossRef] [Green Version]
- Holler, K.L.; Hendershot, T.J.; Troy, S.E.; Vincentz, J.W.; Firulli, A.B.; Howard, M.J. Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev. Boil. 2010, 341, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Tsuchihashi, T.; Maeda, J.; Shin, C.H.; Ivey, K.N.; Black, B.L.; Olson, E.N.; Yamagishi, H.; Srivastava, D. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev. Boil. 2010, 351, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Cheung, M.; Briscoe, J. Neural crest development is regulated by the transcription factor Sox9. Development 2003, 130, 5681–5693. [Google Scholar] [CrossRef] [Green Version]
- Sahar, D.E.; Longaker, M.T.; Quarto, N. Sox9 neural crest determinant gene controls patterning and closure of the posterior frontal cranial suture. Dev. Boil. 2005, 280, 344–361. [Google Scholar] [CrossRef]
- Sullivan, K.; Cleveland, D.W. Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc. Natl. Acad. Sci. USA 1986, 83, 4327–4331. [Google Scholar] [CrossRef] [Green Version]
- Beck, K.D.; Valverde, J.; Alexi, T.; Poulsen, K.; Moffat, B.; Vandlen, R.A.; Rosenthal, A.; Hefti, F. Mesencephalic dopaminergic neurons protected by GDNF from axotomy-induced degeneration in the adult brain. Nature 1995, 373, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Ruest, L.-B.; Xiang, X.; Lim, K.-C.; Levi, G.; Clouthier, D.E. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development 2004, 131, 4413–4423. [Google Scholar] [CrossRef] [Green Version]
- Vincentz, J.W.; Firulli, A.B. The Cardiac Neural Crest and Their Role in Development and Disease; Elsevier BV: London, UK, 2014; Volume 1, pp. 205–229. [Google Scholar]
- Firulli, A.B.; McFadden, D.G.; Lin, Q.; Srivastava, D.; Olson, E.N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 1998, 18, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Thomas, T.; Lin, Q.; Kirby, M.L.; Brown, R.; Olson, E.N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997, 16, 154–160. [Google Scholar] [CrossRef] [PubMed]
- McFadden, D.G.; Barbosa, A.C.; Richardson, J.A.; Schneider, M.D.; Srivastava, D.; Olson, E.N. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 2004, 132, 189–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charité, J.; McFadden, D.G.; Merlo, G.; Levi, G.; Clouthier, D.E.; Yanagisawa, M.; Richardson, J.A.; Olson, E.N. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genome Res. 2001, 15, 3039–3049. [Google Scholar] [CrossRef] [Green Version]
- Minoux, M.; Holwerda, S.; Vitobello, A.; Kitazawa, T.; Kohler, H.; Stadler, M.B.; Rijli, F.M. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science 2017. [Google Scholar] [CrossRef]


| Genotype | n | PTA + VSD | DORV + VSD | Phenotypically Normal |
|---|---|---|---|---|
| Wnt1-Cre | 1 | 0 (0%) | 0 (0%) | 1 (100%) |
| CAG-CAT-Dlx5 | 3 | 0 (0%) | 0 (0%) | 3 (100%) |
| CAG-CAT-Dlx5; Wnt1-Cre | 10 | 6 (60%) | 3 (30%) | 1 (10%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincentz, J.W.; Clouthier, D.E.; Firulli, A.B. Mis-Expression of a Cranial Neural Crest Cell-Specific Gene Program in Cardiac Neural Crest Cells Modulates HAND Factor Expression, Causing Cardiac Outflow Tract Phenotypes. J. Cardiovasc. Dev. Dis. 2020, 7, 13. https://doi.org/10.3390/jcdd7020013
Vincentz JW, Clouthier DE, Firulli AB. Mis-Expression of a Cranial Neural Crest Cell-Specific Gene Program in Cardiac Neural Crest Cells Modulates HAND Factor Expression, Causing Cardiac Outflow Tract Phenotypes. Journal of Cardiovascular Development and Disease. 2020; 7(2):13. https://doi.org/10.3390/jcdd7020013
Chicago/Turabian StyleVincentz, Joshua W., David E. Clouthier, and Anthony B. Firulli. 2020. "Mis-Expression of a Cranial Neural Crest Cell-Specific Gene Program in Cardiac Neural Crest Cells Modulates HAND Factor Expression, Causing Cardiac Outflow Tract Phenotypes" Journal of Cardiovascular Development and Disease 7, no. 2: 13. https://doi.org/10.3390/jcdd7020013





