miR-27b and miR-23b Modulate Cardiomyocyte Differentiation from Mouse Embryonic Stem Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. ESC Culture and microRNA Transfection Assays
2.2. RNA Isolation, Reverse Transcriptase Reaction and Quantitative Real Time PCR
| Primers | Sequence | Annealing Temperature | Amplicon size |
|---|---|---|---|
| Wnt3a | Sense: 5'-TTCTGCAGGAACTACGTGGA-3' | 60 °C | 179 bp |
| Brachyury | Sense: 5'- CGACCACAAAGATGTAATGGAG-3' | 59.5 °C | 120 bp |
| Kcnk3 | Sense: 5'-TGTGCACCTTCACCTACCTG-3' | 59.7 °C | 203 bp |
| Kcnh1 | Sense: 5'-ACGCCCTTCAGAAAGTGCTA-3' | 62 °C | 182 bp |
| c-Troponin T | Sense: 5'-TTCGACCTGCAGGAAAAGTT-3' | 64 °C | 133bp |
| Hcn4 | Sense: 5'-CAGCGTCAGAGCGGATACTT-3' | 60 °C | 158bp |
| Cx40 | Sense: 5'-CAGAGCCTGAAGAAGCCAAC-3' | 60 °C | 178bp |
| Cx43 | Sense: 5'-GAGAGCCCGAACTCTCCTTT-3' | 60 °C | 158bp |
| miR 27b | 5'-UUCACAGUGGCUAAGUUCUGC-3' | 60 °C | 21 bp |
| miR 23b | 5'-AUCACAUUGCCAGGGAUUACC-3' | 60 °C | 21 bp |
3. Results
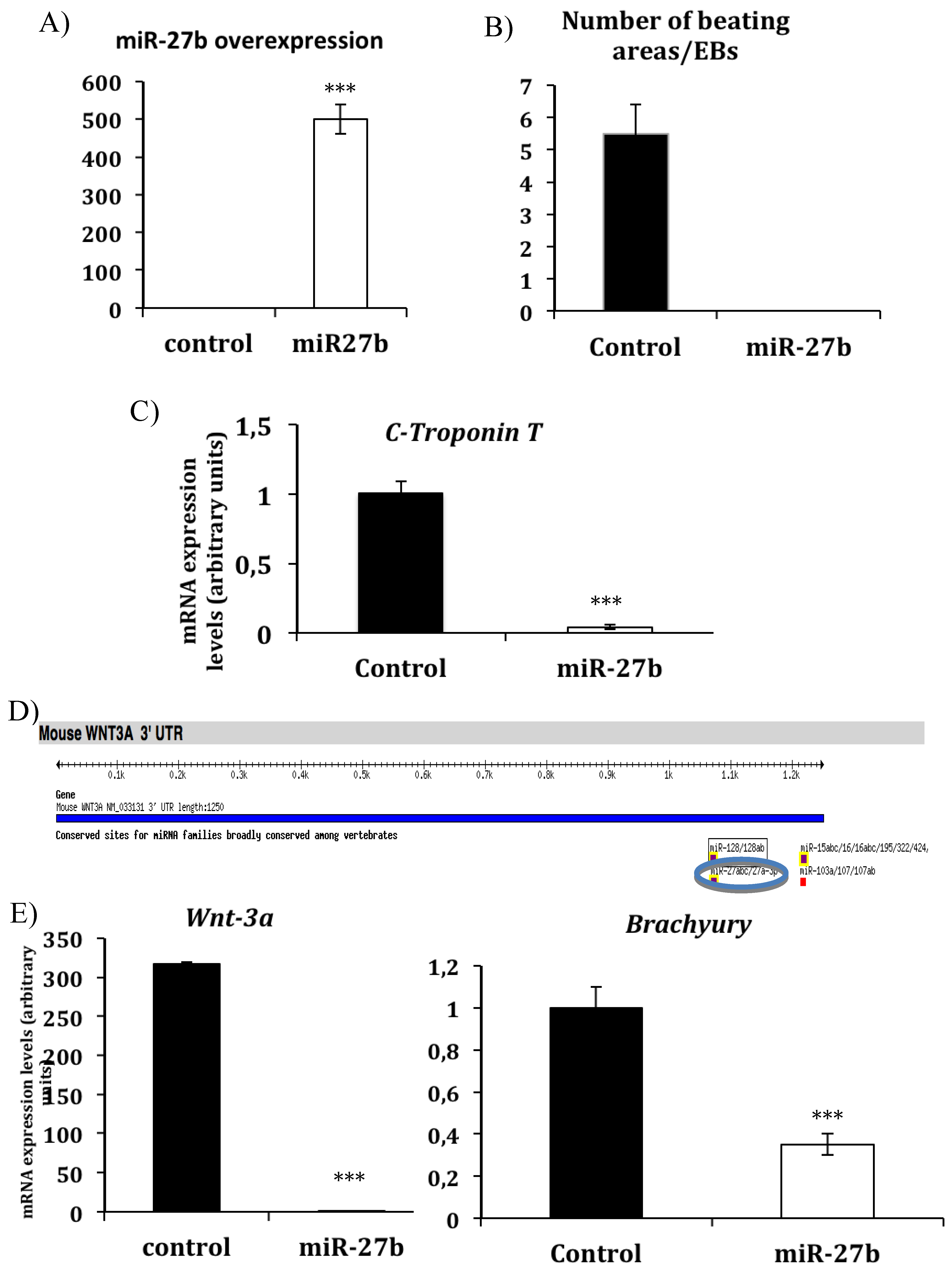
3.1. miR-27b Inhibits in Vitro Cardiac Differentiation by Modulating Wnt-Beta Catenin Canonical Pathway
3.2. miR-23b Regulates the Beating Phenotype During in Vitro Cardiac Differentiation from ESCs
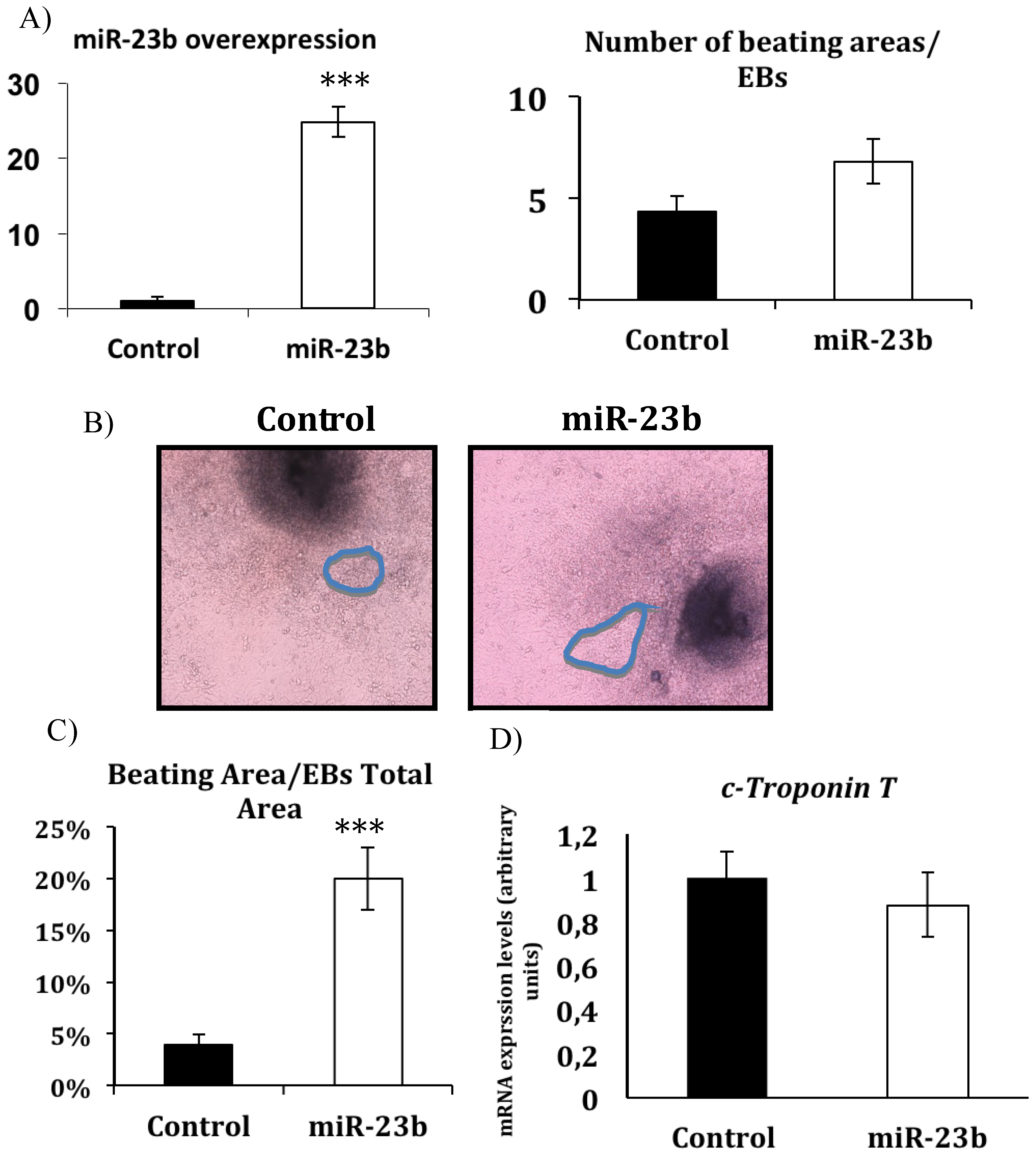

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Srivastava, D. Making or breaking the heart: From lineage determination to morphogenesis. Cell 2006, 126, 1037–1048. [Google Scholar] [CrossRef]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- da Costa Martins, P.A.; Bourajjaj, M.; Gladka, M.; Kortland, M.; van Oort, R.J.; Pinto, Y.M.; Molkentin, J.D.; De Windt, L.J. Conditional dicer gene deletion in the postnatalmyocardium provokes spontaneous cardiac remodeling. Circulation 2008, 118, 1567–1576. [Google Scholar] [CrossRef]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef]
- Liu, N.; Bezprozvannaya, S.; Williams, A.H.; Qi, X.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008, 22, 3242–3254. [Google Scholar] [CrossRef]
- Wilson, K.D.; Hu, S.; Venkatasubrahmanyam, S.; Fu, J.D.; Sun, N.; Abilez, O.J.; Baugh, J.J.; Jia, F.; Ghosh, Z.; Li, R.A.; Butte, A.J.; Wu, J.C. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: Role for miR-499. Circ. Cardiovasc. Genet. 2010, 3, 426–435. [Google Scholar] [CrossRef]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar]
- Ivey, K.N.; Muth, A.; Arnold, J.; King, F.W.; Yeh, R.F.; Fish, J.E.; Hsiao, E.C.; Schwartz, R.J.; Conklin, B.R.; Bernstein, H.S.; Srivastava, D. MicroRNA regulation of cell lineages in mouse and humanem bryonic stem cells. Cell. Stem Cell. 2008, 6, 219–229. [Google Scholar]
- Chinchilla, A.; Lozano, E.; Daimi, H.; Esteban, F.J.; Crist, C.; Aranega, A.E.; Franco, D. MicroRNA profiling during mouse ventricular maturation: A role for miR-27 modulating Mef2c expression. Cardiovasc. Res. 2011, 89, 98–108. [Google Scholar] [CrossRef]
- Lozano, E.; Chinchilla, A.; Martínez, S.; Hernández, F.; Navarro, F.; Lyons, G.E.; Franco, D.; Aránega, A.E. Pitx2c modulates cardiac-specific transcription factors networks in differentiating cardiomyocytes from murine embryonic stem cells. Cells Tissues Organs 2011, 194, 349–362. [Google Scholar] [CrossRef]
- Black, B.L.; Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998, 14, 167–196. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Gill, J.G.; Kyba, M.; Murphy, T.L.; Murphy, K.M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 2006, 133, 3787–3796. [Google Scholar] [CrossRef]
- Stengel, R.; Rivera-Milla, E.; Sahoo, N.; Ebert, C.; Bollig, F.; Heinemann, S.H.; Schönherr, R.; Englert, C. Kcnh1 voltage-gated potassium channels are essential for early zebrafish development. J. Biol. Chem. 2012, 287, 35565–35575. [Google Scholar] [CrossRef]
- Putzke, C.; Wemhöner, K.; Sachse, F.B.; Rinné, S.; Schlichthörl, G.; Li, X.T.; Jaé, L.; Eckhardt, I.; Wischmeyer, E.; Wulf, H.; Preisig-Müller, R.; Daut, J.; Decher, N. The acid-sensitive potassium channel TASK-1 in rat cardiac muscle. Cardiovasc. Res. 2007, 75, 59–68. [Google Scholar] [CrossRef]
- Boyett, M.R.; Honjo, H.; Kodama, I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 2000, 47, 658–687. [Google Scholar] [CrossRef]
- Porrello, E.R. microRNAs in cardiac development and regeneration. Clin. Sci. 2013, 125, 151–166. [Google Scholar] [CrossRef]
- Huelsken, J.; Vogel, R.; Brinkmann, V.; Erdmann, B.; Birchmeier, C.; Birchmeier, W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000, 148, 567–578. [Google Scholar] [CrossRef]
- van den Heuvel, N.H.; van Veen, T.A.; Lim, B.; Jonsson, M.K. Lessons from the heart: Mirroring electrophysiological characteristics during cardiac development to in vitro differentiation of stem cell derived cardiomyocytes. J. Mol. Cell. Cardiol. 2013, 67, 12–25. [Google Scholar] [CrossRef]
- Belevych, A.E.; Sansom, S.E.; Terentyeva, R.; Ho, H.T.; Nishijima, Y.; Martin, M.M.; Jindal, H.K.; Rochira, J.A.; Kunitomo, Y.; Abdellatif, M.; Carnes, C.A.; Elton, T.S.; Györke, S.; Terentyev, D. MicroRNA-1 and -133 increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Spitzner, M.; Ousingsawat, J.; Scheidt, K.; Kunzelmann, K.; Schreiber, R. Voltage-gated K+ channels support proliferation of colonic carcinoma cells. FASEB J. 2007, 21, 35–44. [Google Scholar]
- Hashem, S.I.; Lam, M.L.; Mihardja, S.S.; White, S.M.; Lee, R.J.; Claycomb, W.C. Shox2 regulates the pacemaker gene program in embryoid bodies. Stem Cells Dev. 2013, 22, 2915–2926. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vilches, J.M.; Pulido, A.; Hernández-Torres, F.; Franco, D.; Aránega, A. miR-27b and miR-23b Modulate Cardiomyocyte Differentiation from Mouse Embryonic Stem Cells. J. Cardiovasc. Dev. Dis. 2014, 1, 41-51. https://doi.org/10.3390/jcdd1010041
Vilches JM, Pulido A, Hernández-Torres F, Franco D, Aránega A. miR-27b and miR-23b Modulate Cardiomyocyte Differentiation from Mouse Embryonic Stem Cells. Journal of Cardiovascular Development and Disease. 2014; 1(1):41-51. https://doi.org/10.3390/jcdd1010041
Chicago/Turabian StyleVilches, José Manuel, Antonio Pulido, Francisco Hernández-Torres, Diego Franco, and Amelia Aránega. 2014. "miR-27b and miR-23b Modulate Cardiomyocyte Differentiation from Mouse Embryonic Stem Cells" Journal of Cardiovascular Development and Disease 1, no. 1: 41-51. https://doi.org/10.3390/jcdd1010041





