Construction and Experimental Validation of a Quantitative Kinetic Model of Nitric Oxide Stress in Enterohemorrhagic Escherichia coli O157:H7
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Growth Media
2.2. Bacterial Strains
| Strain | Genotype | Reference |
|---|---|---|
| TUV93-0 | EHEC O157:H7 EDL933 Stx− | Leong, J.M. [44] |
| TUV hmp+/norV+ | TUV93-0 + pUA66-PnorV-norV | This work |
| TUV hmp+/norV− | TUV93-0 + pUA66 | This work |
| TUV hmp−/norV+ | TUV93-0 Δhmp + pUA66-PnorV-norV | This work |
2.3. Plasmid Construction
2.4. Glovebox Setup and Operation
2.5. Bioreactor Apparatus
2.6. NO· Treatment Assay
2.7. Respiration (O2 Consumption) Assay
2.8. Model Simulation
2.9. Parameter Optimization
2.10. BLAST Comparison of Proteins
3. Results
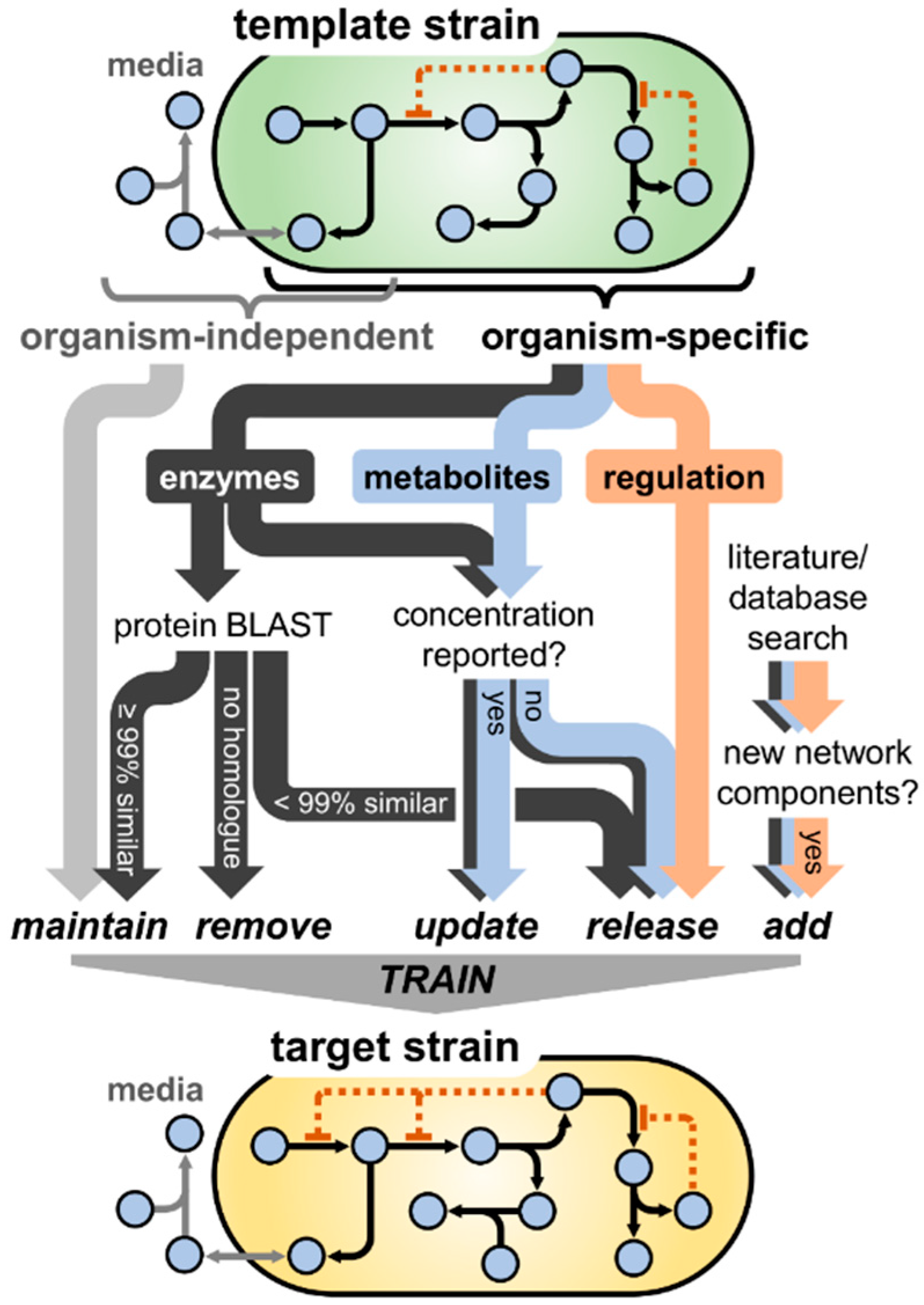
3.1. Model Translation from E. coli K-12 to Enterohemorrhagic E. coli O157:H7
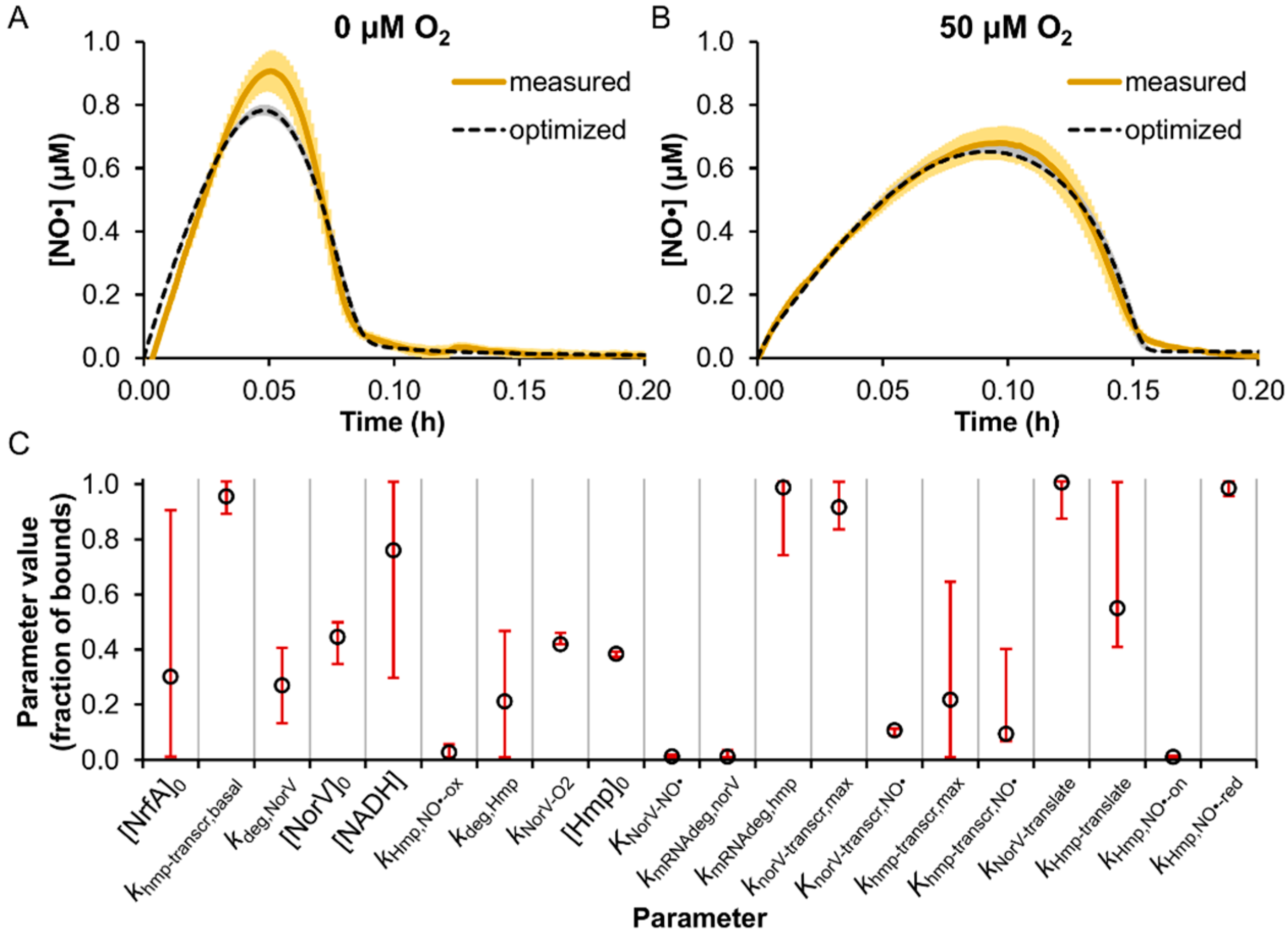
3.2. Model Parameter Training and Sensitivity Analysis
3.3. Prediction of EHEC NO· Detoxification Dynamics in the Absence of Hmp or NorV

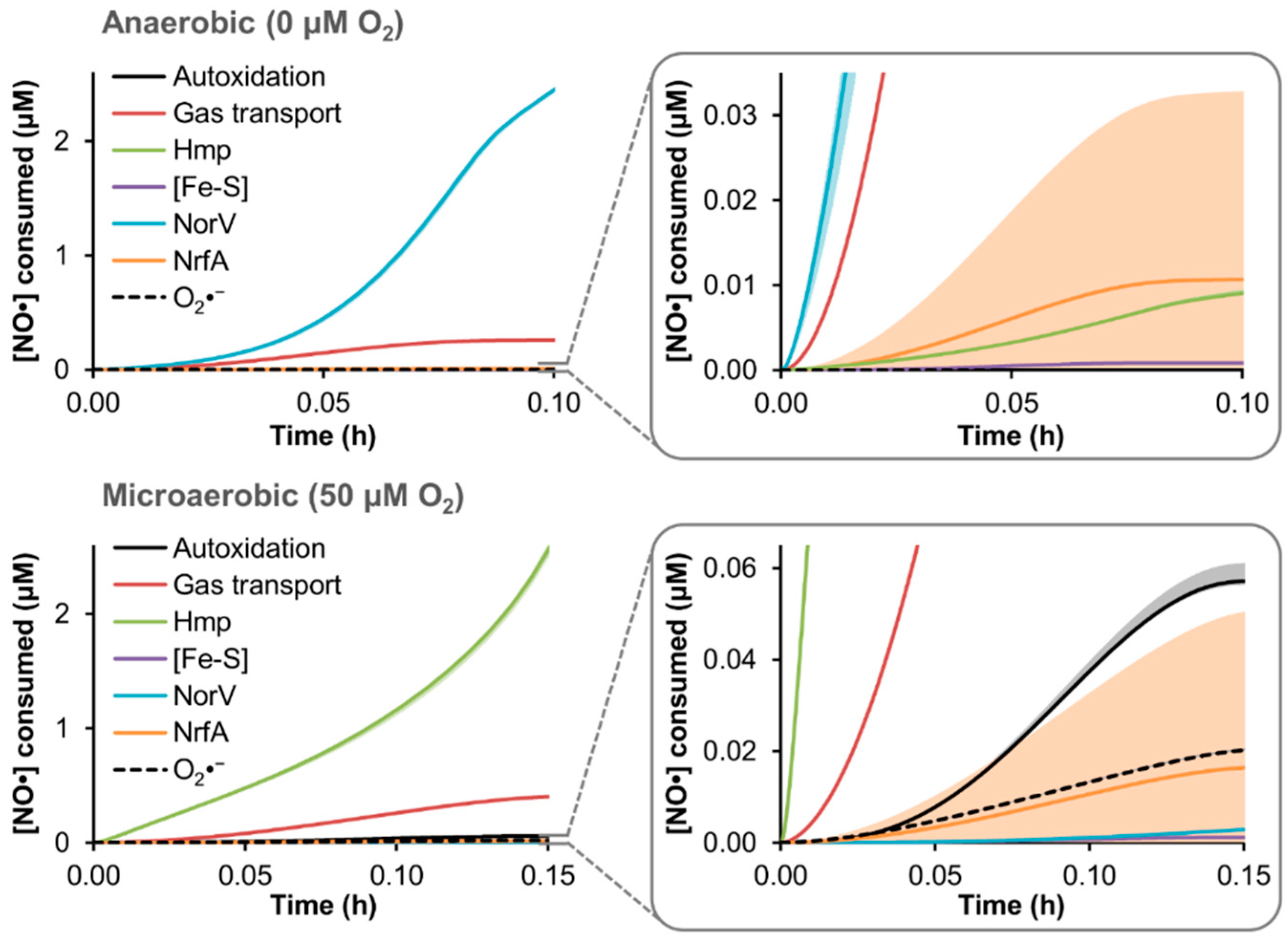
3.4. Predicted Distribution of NO· Consumption
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Russo, T.A.; Johnson, J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000, 181, 1753–1754. [Google Scholar] [CrossRef] [PubMed]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Michino, H.; Araki, K.; Minami, S.; Takaya, S.; Sakai, N.; Miyazaki, M.; Ono, A.; Yanagawa, H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 1999, 150, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Kulasekara, B.R.; Jacobs, M.; Zhou, Y.; Wu, Z.N.; Sims, E.; Saenphimmachak, C.; Rohmer, L.; Ritchie, J.M.; Radey, M.; McKevitt, M.; et al. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 2009, 77, 3713–3721. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb. Mortal. Wkly. Rep. 2006, 55, 1045–1046. [Google Scholar]
- Frank, C.; Werber, D.; Cramer, J.P.; Askar, M.; Faber, M.; an der Heiden, M.; Bernard, H.; Fruth, A.; Prager, R.; Spode, A.; et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011, 365, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- Mellmann, A.; Harmsen, D.; Cummings, C.A.; Zentz, E.B.; Leopold, S.R.; Rico, A.; Prior, K.; Szczepanowski, R.; Ji, Y.M.; Zhang, W.L.; et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 2011, 6, e22751. [Google Scholar] [CrossRef] [PubMed]
- Goldwater, P.N.; Bettelheim, K.A. Treatment of enterohemorrhagic Escherichia coli (EHEC) infection and hemolytic uremic syndrome (HUS). BMC Med. 2012, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Zhou, X.; Kaper, J.B. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 2005, 73, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Karmali, M.A. The continuing evolution of a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2008, 105, 4535–4536. [Google Scholar] [CrossRef] [PubMed]
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086. [Google Scholar] [CrossRef]
- Obrig, T.G.; Moran, T.P.; Brown, J.E. The mode of action of Shiga toxin on peptide elongation of eukaryotic protein synthesis. Biochem. J. 1987, 244, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Schuller, S.; Heuschkel, R.; Torrente, F.; Kaper, J.B.; Phillips, A.D. Shiga toxin binding in normal and inflamed human intestinal mucosa. Microbes Infect. 2007, 9, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.P.; McDaniel, A.D.; Wolf, L.E.; Keusch, G.T.; Waldor, M.K.; Acheson, D.W.K. Quinolone antibiotics induce shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 2000, 181, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Bielaszewska, M.; Idelevich, E.A.; Zhang, W.; Bauwens, A.; Schaumburg, F.; Mellmann, A.; Peters, G.; Karch, H. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob. Agents Chemother. 2012, 56, 3277–3282. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Angulo, V.A.; Kalita, A.; Torres, A.G. Advances in the development of enterohemorrhagic Escherichia coli vaccines using murine models of infection. Vaccine 2013, 31, 3229–3235. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Tsutsuki, H.; Matsumoto, A.; Nakaya, H.; Noda, M. The nitric oxide reductase of enterohaemorrhagic Escherichia coli plays an important role for the survival within macrophages. Mol. Microbiol. 2012, 85, 492–512. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.M.; Giuffre, A.; Forte, E.; Vicente, J.B.; Saraiva, L.M.; Brunori, M.; Teixeira, M. A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J. Biol. Chem. 2002, 277, 25273–25276. [Google Scholar] [PubMed]
- Gardner, A.M.; Helmick, R.A.; Gardner, P.R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002, 277, 8172–8177. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.M.; Gardner, P.R. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli—Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J. Biol. Chem. 2002, 277, 8166–8171. [Google Scholar] [CrossRef] [PubMed]
- Vine, C.E.; Cole, J.A. Nitrosative stress in Escherichia coli: Reduction of nitric oxide. Biochem. Soc. Trans. 2011, 39, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.R.; Gardner, A.M.; Martin, L.A.; Salzman, A.L. Nitric oxide dioxygenase: An enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 1998, 95, 10378–10383. [Google Scholar] [CrossRef] [PubMed]
- Hausladen, A.; Gow, A.J.; Stamler, J.S. Nitrosative stress: Metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 1998, 95, 14100–14105. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Brynildsen, M.P. A kinetic platform to determine the fate of nitric oxide in Escherichia coli. PLoS Comput. Biol. 2013, 9, e1003049. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Adolfsen, K.J.; Brynildsen, M.P. Deciphering nitric oxide stress in bacteria with quantitative modeling. Curr. Opin. Microbiol. 2014, 19, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Svensson, L.; Poljakovic, M.; Save, S.; Gilberthorpe, N.; Schon, T.; Strid, S.; Corker, H.; Poole, R.K.; Persson, K. Role of flavohemoglobin in combating nitrosative stress in uropathogenic Escherichia coli—Implications for urinary tract infection. Microb. Pathog. 2010, 49, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Bang, I.S.; Liu, L.; Vazquez-Torres, A.; Crouch, M.L.; Stamler, J.S.; Fang, F.C. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 2006, 281, 28039–28047. [Google Scholar] [CrossRef] [PubMed]
- Stevanin, T.M.; Poole, R.K.; Demoncheaux, E.A.G.; Read, R.C. Flavohemoglobin Hmp protects Salmonella enterica serovar typhimurium from nitric oxide-related killing by human macrophages. Infect. Immun. 2002, 70, 4399–4405. [Google Scholar] [CrossRef] [PubMed]
- Sebbane, F.; Lemaitre, N.; Sturdevant, D.E.; Rebeil, R.; Virtaneva, K.; Porcella, S.F.; Hinnebusch, B.J. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. USA 2006, 103, 11766–11771. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.R.; Dunman, P.M.; Fang, F.C. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 2006, 61, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.M.; Hay, A.J.; Liu, Z.; Desland, F.A.; Zhang, J.; Zhong, Z.T.; Zhu, J. The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. mBio 2012, 3, e00013-12. [Google Scholar] [CrossRef] [PubMed]
- Vareille, M.; de Sablet, T.; Hindre, T.; Martin, C.; Gobert, A.P. Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 10199–10204. [Google Scholar] [CrossRef] [PubMed]
- Branchu, P.; Matrat, S.; Vareille, M.; Garrivier, A.; Durand, A.; Crepin, S.; Harel, J.; Jubelin, G.; Gobert, A.P. NsrR, GadE, and GadX interplay in repressing expression of the Escherichia coli O157: H7 LEE pathogenicity island in response to nitric oxide. PLoS Pathog. 2014, 10, e1003874. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, I.; Chang, A.; Placzek, S.; Sohngen, C.; Rother, M.; Lang, M.; Munaretto, C.; Ulas, S.; Stelzer, M.; Grote, A.; et al. BRENDA in 2013: Integrated reactions, kinetic data, enzyme function data, improved disease classification: New options and contents in BRENDA. Nucleic Acids Res. 2013, 41, D764–D772. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.R.; Gardner, A.M.; Martin, L.A.; Dou, Y.; Li, T.; Olson, J.S.; Zhu, H.; Riggs, A.F. Nitric-oxide dioxygenase activity and function of flavohemoglobins. sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 2000, 275, 31581–31587. [Google Scholar] [CrossRef] [PubMed]
- Bowman, L.A.; McLean, S.; Poole, R.K.; Fukuto, J.M. The diversity of microbial responses to nitric oxide and agents of nitrosative stress close cousins but not identical twins. Adv. Microb. Physiol. 2011, 59, 135–219. [Google Scholar] [PubMed]
- Lancaster, J.R., Jr. Nitroxidative, nitrosative, and nitrative stress: Kinetic predictions of reactive nitrogen species chemistry under biological conditions. Chem. Res. Toxicol. 2006, 19, 1160–1174. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.H.; Dedon, P.C.; Deen, W.M. Kinetic analysis of intracellular concentrations of reactive nitrogen species. Chem. Res. Toxicol. 2008, 21, 2134–2147. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Brynildsen, M.P. Model-driven identification of dosing regimens that maximize the antimicrobial activity of nitric oxide. Metab. Eng. Commun. 2014, 1, 12–18. [Google Scholar] [CrossRef]
- Robinson, J.L.; Brynildsen, M.P. An ensemble-guided approach Identifies ClpP as a major regulator of transcript levels in nitric oxide-stressed Escherichia coli. Metab. Eng. 2015, 31, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Campellone, K.G.; Giese, A.; Tipper, D.J.; Leong, J.M. A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol. Microbiol. 2002, 43, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Zaslaver, A.; Bren, A.; Ronen, M.; Itzkovitz, S.; Kikoin, I.; Shavit, S.; Liebermeister, W.; Surette, M.G.; Alon, U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 2006, 3, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Adolfsen, K.J.; Brynildsen, M.P. A kinetic platform to determine the fate of hydrogen peroxide in Escherichia coli. PLoS Comput. Biol. 2015, 11, e1004562. [Google Scholar] [CrossRef] [PubMed]
- Denicola, A.; Souza, J.M.; Radi, R.; Lissi, E. Nitric oxide diffusion in membranes determined by fluorescence quenching. Arch. Biochem. Biophys. 1996, 328, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Kelm, M. Nitric oxide metabolism and breakdown. Biochim. Biophys. Acta 1999, 1411, 273–289. [Google Scholar] [CrossRef]
- Zamora-Sillero, E.; Hafner, M.; Ibig, A.; Stelling, J.; Wagner, A. Efficient characterization of high-dimensional parameter spaces for systems biology. BMC Syst. Biol. 2011, 5, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akaike, H. Information theory and extension of the maximum likelihood principle. In Proceedings of the 2nd International Symposium on Information Theory, Tsahkadsor, Armenia, 2–8 September 1971; Petrov, B.N., Csaki, F., Eds.; Akadémiai Kiadó: Budapest, Hungary, 1973; pp. 267–281. [Google Scholar]
- Hurvich, C.M.; Tsai, C.L. Regression and time-series model selection in small samples. Biometrika 1989, 76, 297–307. [Google Scholar] [CrossRef]
- Turkheimer, F.E.; Hinz, R.; Cunningham, V.J. On the undecidability among kinetic models: From model selection to model averaging. J. Cereb. Blood Flow Metab. 2003, 23, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2015, 36, D623–D631. [Google Scholar] [CrossRef] [PubMed]
- Hausladen, A.; Gow, A.; Stamler, J.S. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. USA 2001, 98, 10108–10112. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.E.; Sedelnikova, S.; Soballe, B.; Hughes, M.N.; Poole, R.K. Escherichia coli flavohaemoglobin (Hmp) with equistoichiometric FAD and haem contents has a low affinity for dioxygen in the absence or presence of nitric oxide. Biochem. J. 2001, 353, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Stevanin, T.M.; Ioannidis, N.; Mills, C.E.; Kim, S.O.; Hughes, M.N.; Poole, R.K. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo’ or bd, from nitric oxide. J. Biol. Chem. 2000, 275, 35868–35875. [Google Scholar] [CrossRef] [PubMed]
- Tilden, J.; Young, W.; McNamara, A.M.; Custer, C.; Boesel, B.; LambertFair, M.; Majkowski, J.; Vugia, D.; Werner, S.B.; Hollingsworth, J.; et al. A new route of transmission for Escherichia coli: Infection from dry fermented salami. Am. J. Public Health 1996, 86, 1142–1145. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, J.; Gomez, T.; Doyle, M.P.; Wells, J.G.; Zhao, T.; Tauxe, R.V.; Griffin, P.M. Lessons from a large outbreak of Escherichia coli O157:H7 infections: Insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 1999, 122, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.S.; Jelacic, S.; Habeeb, R.L.; Watkins, S.L.; Tarr, P.I. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 2000, 342, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Orth, D.; Grif, K.; Zimmerhackl, L.B.; Wurzner, R. Prevention and treatment of enterohemorrhagic Escherichia coli infections in humans. Expert Rev. Anti Infect Ther. 2008, 6, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.M.; Martin, L.A.; Gardner, P.R.; Dou, Y.; Olson, J.S. Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J. Biol. Chem. 2000, 275, 12581–12589. [Google Scholar] [CrossRef] [PubMed]
- Hyduke, D.R.; Jarboe, L.R.; Tran, L.M.; Chou, K.J.; Liao, J.C. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 8484–8489. [Google Scholar] [CrossRef] [PubMed]
- Pullan, S.T.; Gidley, M.D.; Jones, R.A.; Barrett, J.; Stevanin, T.A.; Read, R.C.; Green, J.; Poole, R.K. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: Unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 2007, 189, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, J.L.; Brynildsen, M.P. Construction and Experimental Validation of a Quantitative Kinetic Model of Nitric Oxide Stress in Enterohemorrhagic Escherichia coli O157:H7. Bioengineering 2016, 3, 9. https://doi.org/10.3390/bioengineering3010009
Robinson JL, Brynildsen MP. Construction and Experimental Validation of a Quantitative Kinetic Model of Nitric Oxide Stress in Enterohemorrhagic Escherichia coli O157:H7. Bioengineering. 2016; 3(1):9. https://doi.org/10.3390/bioengineering3010009
Chicago/Turabian StyleRobinson, Jonathan L., and Mark P. Brynildsen. 2016. "Construction and Experimental Validation of a Quantitative Kinetic Model of Nitric Oxide Stress in Enterohemorrhagic Escherichia coli O157:H7" Bioengineering 3, no. 1: 9. https://doi.org/10.3390/bioengineering3010009




