Single-Nucleotide Polymorphisms Associated with Mercury Levels and Neurological Symptoms: An Overview
Abstract
:1. Introduction
2. Materials and Methods
2.1. Information Source and Search Strategies
2.2. Eligibility Criteria
2.3. Data Extraction
2.4. Quality Assessment of Studies
2.5. Analysis and Illustrations
3. Results
4. Discussion
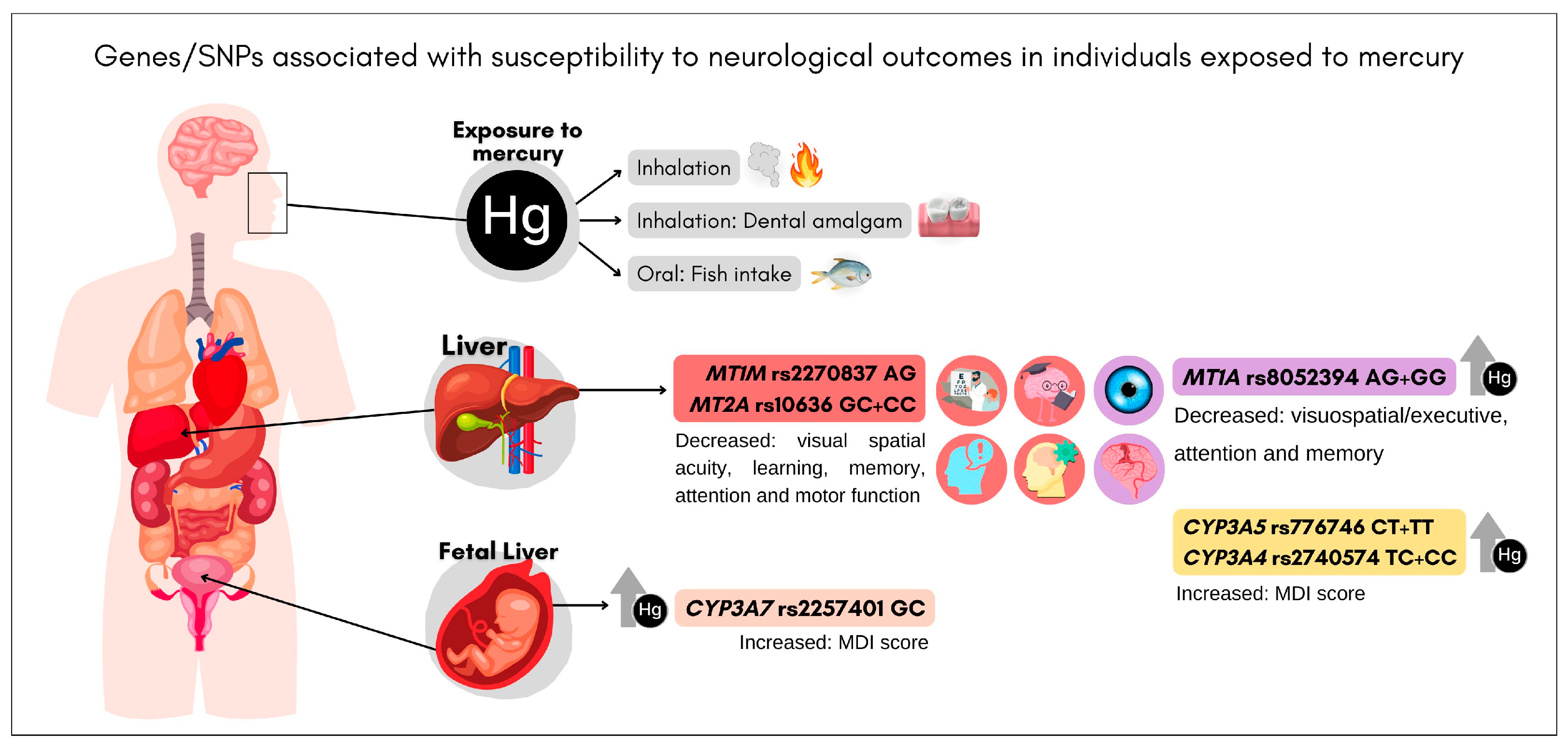
4.1. CYP3A (rs2740574, rs776746, and rs2257401)
4.2. ALAD (rs1800435)
4.3. GSTP1 (rs1695)
4.4. MTs (rs8052394, rs2270836, rs2270837, rs10636, and rs11643815)
4.5. Insights and Other Features
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Human Genome Research Institute. Available online: https://www.genome.gov/genetics-glossary (accessed on 5 January 2024).
- National Human Genome Research Institute. Polymorphism. Available online: https://www.genome.gov/genetics-glossary/Polymorphism (accessed on 5 January 2024).
- Chiarella, P.; Capone, P.; Sisto, R. Contribution of Genetic Polymorphisms in Human Health. Int. J. Environ. Res. Public Health 2023, 20, 912. [Google Scholar] [CrossRef]
- Basta, P.C.; Viana, P.V.D.S.; Vasconcellos, A.C.S.D.; Périssé, A.R.S.; Hofer, C.B.; Paiva, N.S.; Kempton, J.W.; Ciampi de Andrade, D.; Oliveira, R.A.A.D.; Achatz, R.W.; et al. Mercury Exposure in Munduruku Indigenous Communities from Brazilian Amazon: Methodological Background and an Overview of the Principal Results. Int. J. Environ. Res. Public Health 2021, 18, 9222. [Google Scholar] [CrossRef] [PubMed]
- Perini, J.A.; Silva, M.C.; Vasconcellos, A.C.S.D.; Viana, P.V.S.; Lima, M.O.; Jesus, I.M.; Kempton, J.W.; Oliveira, R.A.A.; Hacon, S.S.; Basta, P.C. Genetic Polymorphism of Delta Aminolevulinic Acid Dehydratase (ALAD) Gene and Symptoms of Chronic Mercury Exposure in Munduruku Indigenous Children within the Brazilian Amazon. Int. J. Environ. Res. Public Health 2021, 18, 8746. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; de Oliveira, R.A.A.; Vasconcellos, A.C.S.; Rebouças, B.H.; Pinto, B.D.; Lima, M.O.; de Jesus, I.M.; Machado, D.E.; Hacon, S.S.; Basta, P.C.; et al. Chronic Mercury Exposure and GSTP1 Polymorphism in Munduruku Indigenous from Brazilian Amazon. Toxics 2023, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, P.; Fu, X.; Wang, X.; Zhang, H.; Lin, C.J. Mercury pollution in China: Implications on the implementation of the Minamata Convention. Environ. Sci. Process. Impacts 2022, 24, 634–648. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.A.A.D.; Pinto, B.D.; Rebouças, B.H.; Ciampi de Andrade, D.; Vasconcellos, A.C.S.D.; Basta, P.C. Neurological Impacts of Chronic Methylmercury Exposure in Munduruku Indigenous Adults: Somatosensory, Motor, and Cognitive Abnormalities. Int. J. Environ. Res. Public Health 2021, 18, 10270. [Google Scholar] [CrossRef]
- Andreoli, V.; Sprovieri, F. Genetic aspects of susceptibility to mercury toxicity: An overview. Int. J. Environ. Res. Public Health 2017, 14, 93. [Google Scholar] [CrossRef]
- National Library of Medicine. National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/snp/ (accessed on 5 January 2024).
- SNPedia. Available online: https://www.snpedia.com/index.php (accessed on 5 January 2024).
- Von, E.E. Strengthening the reporting of observational studies in epidemiology (STROBE) statement: Guidelines for reporting observational studies. Brit. Med. J. 2014, 355, 806–808. [Google Scholar] [CrossRef]
- Cuschieri, S. The CONSORT statement. Saudi J. Anaesth. 2019, 13 (Suppl. S1), S27–S30. [Google Scholar] [CrossRef]
- Canva. Available online: https://www.canva.com/pt_br/ (accessed on 10 January 2024).
- Barcelos, G.R.M.; Grotto, D.; de Marco, K.C.; Valentini, J.; Lengert, A.H.; de Oliveira, A.A.S.; Garcia, S.C.; Braga, G.U.L.; Engström, K.S.; Cólus, I.M.D.; et al. Polymorphisms in glutathione-related genes modify mercury concentrations and antioxidant status in subjects environmentally exposed to methylmercury. Sci. Total Environ. 2013, 463, 319–325. [Google Scholar] [CrossRef]
- Woods, J.S.; Heyer, N.J.; Ruesso, J.E.; Martin, M.D.; Pillai, P.B.; Farin, F.M. Modification of neurobehavioral effects of mercury by genetic polymorphisms of metallothionein in children. Neurotoxicol. Teratol. 2013, 39, 36–44. [Google Scholar] [CrossRef]
- Drescher, O.; Dewailly, E.; Diorio, C.; Ouellet, N.; Sidi, E.A.L.; Abdous, B.; Valera, B.; Ayotte, P. Methylmercury exposure, PON1 gene variants and serum paraoxonase activity in Eastern James Bay Cree adults. J. Expo. Sci. Environ. Epidemiol. 2014, 24, 608–614. [Google Scholar] [CrossRef]
- Barcelos, G.R.M.; de Souza, M.F.; de Oliveira, A.A.S.; Lengert, A.H.; de Oliveira, M.T.; Camargo, R.B.O.G.; Grotto, D.; Valentini, J.; Garcia, S.C.; Braga, G.U.L.; et al. Effects of genetic polymorphisms on antioxidant status and concentrations of the metals in the blood of riverside Amazonian communities co-exposed to Hg and Pb. Environ. Res. 2015, 138, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.V.; Cardoso, B.R.; Zavarize, B.; Almondes, K.; Bordon, I.; Hare, D.; Favaro, B.I.T.; Cozzolino, S.M.F. GPX1 Pro198Leu polymorphism and GSTM1 deletion do not affect selenium and mercury status in mildly exposed Amazonian women in an urban population. Sci. Total Environ. 2016, 571, 801–808. [Google Scholar] [CrossRef]
- Parajuli, R.P.; Goodrich, J.M.; Chou, H.-N.; Gruninger, S.E.; Dolinoy, D.C.; Franzblau, A.; Basu, N. Genetic Polymorphisms Are Associated with Hair, Blood, and Urine Mercury Levels in the American Dental Association (ADA) Study Participants. Environ. Res. 2016, 149, 247–258. [Google Scholar] [CrossRef]
- Lopes, A.C.B.A.; Urbano, M.R.; Souza-Nogueira, A.; Oliveira-Paula, G.H.; Michelin, A.P.; Carvalho, M.F.H.; Camargo, A.E.I.; Peixe, T.S.; Cabrera, M.A.S.; Paoliello, M.M.B. Association of lead, cadmium and mercury with paraoxonase 1 activity and malondialdehyde in a general population in Southern Brazil. Environ. Res. 2017, 156, 674–682. [Google Scholar] [CrossRef]
- Llop, S.; Tran, V.; Ballester, F.; Sofianou-Katsoulis, A.; Sunyer, J.; Engström, K.; Alhamadow, A.; Love, T.M.; Watson, G.E.; Bustamante, M.; et al. CYP3A genes and the association between prenatal methylmercury exposure and neurodevelopment. Environ. Int. 2017, 105, 34–42. [Google Scholar] [CrossRef]
- Chan, P.H.Y.; Chan, K.Y.Y.; Schooling, C.M.; Hui, L.L.; Chan, M.H.M.; Li, A.M.; Cheung, R.C.K.; Lam, H.S. Association between Genetic Variations in GSH-Related and MT Genes and Low-Dose Methylmercury Exposure in Children and Women of Childbearing Age: A Pilot Study. Environ. Res. 2020, 187, 109703. [Google Scholar] [CrossRef]
- Parajuli, R.P.; Goodrich, J.M.; Chan, H.M.; Lemire, M.; Ayotte, P.; Hegele, R.A.; Basu, N. Variation in Biomarker Levels of Metals, Persistent Organic Pollutants, and Omega-3 Fatty Acids in Association with Genetic Polymorphisms among Inuit in Nunavik, Canada. Environ. Res. 2021, 200, 111393. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, M.H.; Samms-Vaughan, M.; Saroukhani, S.; Bressler, J.; Hessabi, M.; Grove, M.L.; Shakspeare-Pellington, S.; Loveland, K.A.; Beecher, C.; Mclaughlin, W. Associations of Metabolic Genes (GSTT1, GSTP1, GSTM1) and Blood Mercury Concentrations Differ in Jamaican Children with and without Autism Spectrum Disorder. Int. J. Environ. Res. Public Health 2021, 18, 1377. [Google Scholar] [CrossRef] [PubMed]
- Sirivarasai, J.; Chaisungnern, K.; Panpunuan, P.; Chanprasertyothin, S.; Chansirikanjana, S.; Sritara, P. Role of MT1A Polymorphism and Environmental Mercury Exposure on the Montreal Cognitive Assessment (MoCA). Neuropsychiatr. Dis. Treat. 2021, 17, 2429–2439. [Google Scholar] [CrossRef]
- FAO Joint; WHO Expert Committee on Food Additives; World Health Organization. Evaluation of certain food additives and contaminants (Thirty-third report of the Joint FAO/WHO Expert Committee on Food Additives). In WHO Technical Report Series; WHO: Geneva, Switzerland, 1989; p. 776. Available online: https://iris.who.int/bitstream/handle/10665/39252/WHO_TRS_776.pdf?sequence=1 (accessed on 2 March 2024).
- Legrand, M.; Feeley, M.; Tikhonov, C.; Schoen, D.; Li-Muller, A. Methylmercury blood guidance values for Canada. Can. J. Public Health 2010, 101, 28–31. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Guidance for Identifying Populations at Risk from Mercury Exposure; UNEP DTIE Chemicals Branch and WHO Department of Food Safety, Zoonoses and Foodborne Diseases: Geneva, Switzerland, 2008; p. 176. Available online: http://www.who.int/foodsafety/publications/chem/mercuryexposure.pdf (accessed on 2 March 2024).
- Llop, S.; Balallester, F.; Broberg, K. Effect of gene-mercury interactions on mercury toxicokinetics and neurotoxicity. Curr. Environ. Health Rep. 2015, 2, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, P.; Nagini, S. Cytochrome P450 structure, function and clinical significance: A review. Curr. Drug Targets 2017, 19, 38–54. [Google Scholar] [CrossRef]
- Klyushova, L.S.; Perepechaeva, M.L.; Grishanova, A.Y. The role of CYP3A in health and disease. Biomedicines 2022, 10, 2686. [Google Scholar] [CrossRef]
- Lamba, J.K.; Lin, Y.S.; Schuetz, E.G. Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv. Drug Deliv. Rev. 2002, 54, 1271–1294. [Google Scholar] [CrossRef]
- Rodríguez-Antona, C.; Jande, M.; Rane, A.; Ingelman-Sundberg, M. Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers. Clin. Pharmacol. Ther. 2005, 77, 259–270. [Google Scholar] [CrossRef]
- Suarez-Kurtz, G.; Perini, J.A.; Bastos-Rodrigues, L.; Pena, S.D.; Struchiner, C. Impact of population admixture on the distribution of the CYP3A5*3 polymorphism. Pharmacogenomics 2007, 8, 1299–1306. [Google Scholar] [CrossRef]
- Sim, S.C.; Ingelman-Sundberg, M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: A peer-reviewed database of CYP variants and their associated effects. Hum. Genom. 2010, 4, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Chernyak, Y.I.; Merinova, A.P. CYP3A Polymorphism and Chronic Mercury Intoxication. Bull. Exp. Biol. Med. 2020, 168, 492–495. [Google Scholar] [CrossRef]
- Smith, C.M.; Wang, X.; Hu, H.; Kelsey, K.T. A polymorphism in the delta-aminolevulinic aciddehydratase gene may modify thepharmacokinetics and toxicity of lead. Environ. Health Perspect. 1995, 103, 248–253. [Google Scholar] [CrossRef]
- Ballatori, N.; Clarkson, W. Biliary Secretion of Glutathione and of Glutathione-Metal Complexes. Fundam. Appl. Toxicol. 1985, 5, 816–831. [Google Scholar] [CrossRef] [PubMed]
- Gundacker, C.; Wittmann, K.J.; Kukuckova, M.; Komarnicki, G.; Hikkel, I.; Gencik, M. Genetic Background of Lead and Mercury Metabolism in a Group of Medical Students in Austria. Environ. Res. 2009, 109, 786–796. [Google Scholar] [CrossRef]
- Wahlberg, K.; Love, T.M.; Pineda, D.; Engström, K.; Watson, G.E.; Thurston, S.W.; Yeates, A.J.; Mulhern, M.S.; McSorley, E.M.; Strain, J.J.; et al. Maternal Polymorphisms in Glutathione-Related Genes Are Associated with Maternal Mercury Concentrations and Early Child Neurodevelopment in a Population with a Fish-Rich Diet. Environ. Int. 2018, 115, 142–149. [Google Scholar] [CrossRef]
- Medina Pérez, O.M.; Flórez-Vargas, O.; Rincón Cruz, G.; Rondón González, F.; Rocha Muñoz, L.; Sánchez Rodríguez, L.H. Glutathione-Related Genetic Polymorphisms Are Associated with Mercury Retention and Nephrotoxicity in Gold-Mining Settings of a Colombian Population. Sci. Rep. 2021, 11, 8716. [Google Scholar] [CrossRef] [PubMed]
- Lozano, M.; Murcia, M.; Soler-Blasco, R.; González, L.; Iriarte, G.; Rebagliato, M.; Lopez-Espinosa, M.J.; Esplugues, A.; Ballester, F.; Llop, S. Exposure to mercury among 9-year-old children and neurobehavioural function. Environ. Int. 2021, 146, 106173. [Google Scholar] [CrossRef]
- Irvine, G.W.; Pinter, T.B.; Stillman, M.J. Defining the metal binding pathways of human metallothionein 1A: Balancing zinc availability and cadmium seclusion. Metallomics 2016, 8, 71–81. [Google Scholar] [CrossRef]
- Babula, P.; Masarik, M.; Adam, V.; Eckschlager, T.; Stiborova, M.; Trnkova, L.; Skutkova, H.; Provaznik, I.; Hubalek, J.; Kizek, R. Mammalian metallothioneins: Properties and functions. Metallomics 2012, 4, 739–750. [Google Scholar] [CrossRef]
- Wang, Y.; Goodrich, J.M.; Gillespie, B.; Werner, R.; Basu, N.; Franzblau, A. An investigation of modifying effects of metallothionein single-nucleotide polymorphisms on the association between mercury exposure and biomarker levels. Environ. Health Perspect. 2012, 120, 530–534. [Google Scholar] [CrossRef]
- Engstrom, K.S.; Stromberg, U.; Lundh, T.; Johannsson, I.; Vessby, B.; Hallmans, G.; Skerfving, S.; Broberg, K. Genetic variation in glutathione-related genes and body burden of methylmercury. Environ. Health Perspect. 2008, 116, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Gundacker, C.; Komarnicki, G.; Jagiello, P.; Gencikova, A.; Dahmen, N.; Wittmann, K.J.; Gencik, M. Glutathione-S-transferase polymorphism, metallothionein expression, and mercury levels among students in Austria. Sci. Total Environ. 2007, 385, 37–47. [Google Scholar] [CrossRef]
- Goodrich, J.M.; Chou, H.N.; Gruninger, S.E.; Franzblau, A.; Basu, N. Exposures of dental professionals to elemental mercury and methylmercury. J. Expo. Sci. Environ. Epidemiol. 2016, 26, 78–85. [Google Scholar] [CrossRef]
- Romo, L.; Findlay, S.D.; Burge, C.B. Regulatory features aid interpretation of 3′UTR variants. Am. J. Hum. Genet. 2024, 10, 350–363. [Google Scholar] [CrossRef]
- Moleirinho, A.; Carneiro, J.; Matthiesen, R.; Silva, R.M.; Amorim, A.; Azevedo, L. Gains, losses and changes of function after gene duplication: Study of the metallothionein family. PLoS ONE 2011, 6, e18487. [Google Scholar] [CrossRef]
- Antunes Dos Santos, A.; Appel Hort, M.; Culbreth, M.; López-Granero, C.; Farina, M.; Rocha, J.B.; Aschner, M. Methylmercury and brain development: A review of recent literature. J. Trace Elem. Med. Biol. 2016, 38, 99–107. [Google Scholar] [CrossRef]
- Grandjean, P.; Weihe, P.; White, R.F.; Debes, F.; Araki, S.; Yokoyama, K.; Murata, K.; Sørensen, N.; Dahl, R.; Jørgensen, P.J. Cognitive deficit in 7-year-old children with prenatal exposure to methylmercury. Neurotoxicol. Teratol. 1997, 19, 417–428. [Google Scholar] [CrossRef]
- Debes, F.; Budtz-Jørgensen, E.; Weihe, P.; White, R.F.; Grandjean, P. Impact of prenatal methylmercury exposure on neurobehavioral function at age 14 years. Neurotoxicol. Teratol. 2006, 28, 536–547. [Google Scholar] [CrossRef]
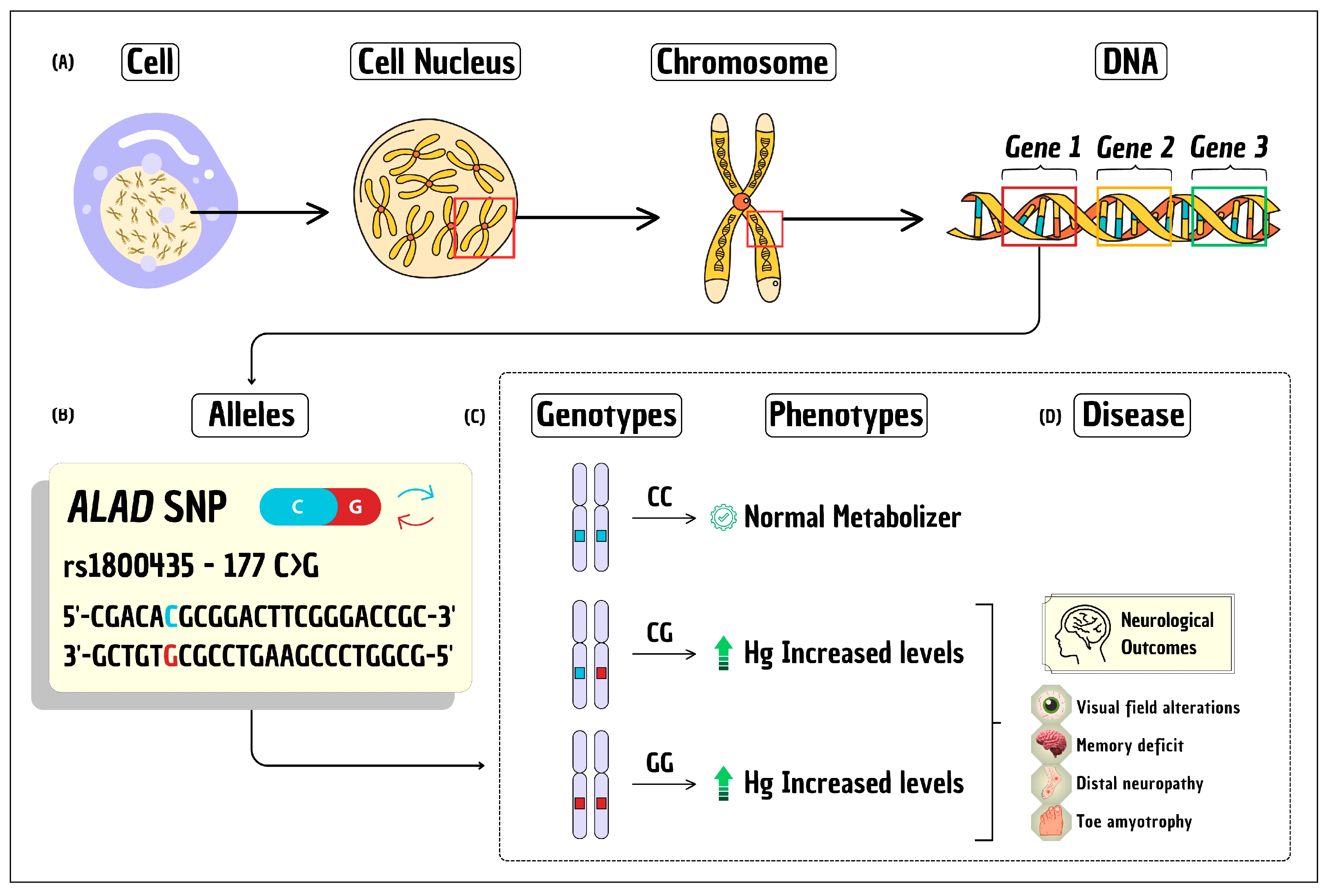

| Reference | Population | Number | Sex (Male) | Age (Years) | BMI (kg/m2) | Quality Assessment (%) a |
|---|---|---|---|---|---|---|
| [15] | South America (Brazil) | 400 | 51.7 | 41.8 ± 16 | 25.7 ± 4.2 | 91 |
| [18] | South America (Brazil) | 395 | 52.4 | 40.5 (18–87) | 24.0 | 91 |
| [19] | South America (Brazil) | 149 | 0 | 26.6 ± 7.8 (18–48) | 22.2 ± 2.6 (17–30) | 91 |
| [5] | South America (Brazil) | 103 | 39.8 | 6.6 ± 4.5 (0–14) | 16.0± 1.3 (14–22) b 18.8 ± 1.9 (15–22) c 0.2 ± 0.8 (−1.7–1.8) d | 95 |
| [21] | South America (Brazil) | 959 | 43.9 | 54.7 ± 10.4 | 28.0 ± 5.2 | 82 |
| [17] | North America (Canada) | 881 | 41.9 | 40.1 | 33.8 | 100 |
| [24] | North America (Canada) | 665 | 100 | 37.2 ± 14.1 (18–76) | 27.4 ± 5.7 (17–47) | 95 |
| [20] | North America (USA) | 380 | 62.6 | 54.8 ± 11.4 (25–82) | - | 95 |
| [22] | Europe (Spain) | 625 | 52.6 | 1.1 ± 1.3 | - | 95 |
| [22] | Europe (Italy) | 573 | 51.6 | 1.6 ± 0.9 | - | 95 |
| [22] | Europe (Greece) | 281 | 44.7 | 1.5 ± 1.1 | - | 95 |
| [16] | Europe (Portugal) | 507 | 49.7 | 10.2 ± 0.8 (8–12) | - | 100 e |
| [22] | Africa (Seychelles) | 211 | 48.8 | 2.3 ± 1.2 | - | 95 |
| [22] | Africa (Seychelles) | 949 | 50.8 | 1.7 ± 1.3 | 95 | |
| [25] | Africa (Jamaica) | 266 | 81.6 | (2–8) f | - | 95 |
| [23] | Asia (China—Children) | 179 | 54.3 | 4.6 (4–5) | 14.9 (14–16) | 95 |
| Asia (China—Mothers) | 165 | 0 | 36.0 (33–40) | 22.0 (20–25) | 95 | |
| [26] | Asia (Thailand) | 436 | 65.1 | 58.8 ± 3.0 | 24.8 ± 3.7 | 86 |
| Reference | Hg-Exposure Route | Population Group | Cord Blood (µg/L) | Blood (µg/L) | Hair (μg/g) | Urinary (μg/L) | ≥Hg-Reference Limit (Yes or No) f |
|---|---|---|---|---|---|---|---|
| [15] | Fish consumption | Riverine—Adults | - | 48.5 ± 36.5 | 13.8 ± 10.2 | - | Yes |
| [17] | Fish consumption | Indigenous—Adults | - | 3.3 (3.0–3.7) | - | - | No |
| [18] | Fish consumption | Riverine—Adults | - | 39.8 | - | - | Yes |
| [20] | Occupational and fish consumption | Urban—Adults | - | 3.7 ± 3.9 | 0.6 ± 1.0 | 1.3 ± 1.8 | No |
| [19] | Fish consumption | Urban—Adults | - | - | 0.6 ± 0.7 (0.0–4.4) | - | No |
| [21] | Fish consumption | Urban—Adults | - | 1.4 (1.3–1.5) | - | - | No |
| [23] | Fish consumption | Urban—Adults | - | - | 0.9 (0.7–1.4) b | - | No |
| [24] | Fish consumption | Indigenous—Adults | - | - | 1.0 (0.7–1.4) | - | No |
| [26] | Dental amalgam and fish consumption | Urban—Adults | - | 6.3 (0.8–27.6) | - | - | No |
| [16] | Dental amalgam | Urban—Children | - | - | - | 1.3 ± 3.0 c 1.8 ± 2.3 d (0.0–31.7) e | No |
| [22] | Fish consumption | Urban—Children | 39.3 ± 25 (Seychelles) | - | 5.8 (Seychelles) | - | Yes |
| 11.3 ± 9 (Spain) | - | - | - | Yes | |||
| 7.5 ± 5 (Greece) | - | - | - | No | |||
| 5.6 ± 4 (Italy) | - | - | - | No | |||
| [23] | Fish consumption | Urban—Children | - | - | 1.0 (0.7–1.5) a | - | No |
| [25] | Fish consumption | Urban—Children | - | 1.0 ± 1.3 | - | - | No |
| [5] | Fish consumption | Indigenous—Children | - | - | 7.0 ± 4.5 (1.4–23.9) | - | Yes |
| Gene | Chr | dnSNP ID | Gene Locus | SNP Details | Minor Allele | Frequency | References |
|---|---|---|---|---|---|---|---|
| ALAD | 9q32 | rs1800435 | Exon 4 | 177 C>G (Lys59Asn) | G | 1 and 3 | [5,18] |
| ATP7B | 13q14.3 | rs1061472 | Exon 10 | 2495 C>T (Lys832Arg) | T | 27 and 48 | [20,24] |
| ATP7B | 13q14.3 | rs732774 | Exon 12 | 2855 T>C (Arg952Lys) | C | 47 | [20] |
| CYP3A4 | 7q22.1 | rs2740574 | Promoter | −392T>C | C | 1–54 (range) a | [22] |
| CYP3A5 | 7q22.1 | rs776746 | Exon 14 | 6986 C>T (Splice Defect) | T | 6–55 (range) a | [22] |
| CYP3A7 | 7q22.1 | rs2257401 | Exon 11 | 26041 G>C (Thr409Arg) | C | 46–92 (range) a | [22] |
| FADS2 | 11q12.2 | rs174602 | Intron | T>C (Asp6Asp) | C | 18 | [24] |
| FADS2 | 11q12.2 | rs74771917 | Intron | C>T | T | 27 | [24] |
| FADS3 | 11q12.2 | rs7115739 | Intron | 1287-380 G>T | T | 27 | [24] |
| GCLC | 6p12.1 | rs17883901 | Promoter | −129G>A | A | 8 f | [18,20,23] |
| GCLM | 6p12.1 | rs41303970 | Promoter | −588 G>A | A | 12 and 30 | [15,23] |
| GPX1 | 3p21.3 | rs1050450 | Exon 2 | 559G>A (Pro198Leu) | A | 3 and 25 | [19,23] |
| GPX4 | 19p13.3 | rs713041 | 3′UTR | 718 C>T | T | 11 | [24] |
| GSTP1 | 11p13.2 | rs1695 | Exon 5 | 313 A>G (Ile105Val) | G | 18–47 (range) b | [15,18,20,23,25] |
| MT1A | 16q13 | rs8052394 | Exon | 152 A>G (Lys51Arg) | G | 22 | [26] |
| MT1M | 16q13 | rs2270836 | Intron 2 | 95-49 C>T | T | 22 and 35 | [20,23] |
| MT1M | 16q13 | rs2270837 | 3′UTR | A>G | G | 17–18 c (range)/24 d | [16] |
| MT1M | 16q13 | rs9936741 | Intron 2 | 31 T>C | C | 13 | [23] |
| MT2A | 16q13 | rs10636 | 3′UTR | +838 G>C | C | 23–25 c/23–25 d (range) e | [16,20,23] |
| MT4 | 16q13 | rs11643815 | 3′UTR | G>A Gly48Asp | A | 0.6 and 9 | [20,23] |
| MTHFR | 1p36.22 | rs2274976 | Exon 11 | 1793 C>T (Arg594Gln) | T | 9 and 49 | [20,24] |
| PON1 | 7q21.3 | rs705379 | Exon 6 | −108 G>A | A | 37 | [17] |
| Gene | Biological Pathways | SNP | Allele or Genotypes a | Hg Levels | Neurological Outcomes | Reference |
|---|---|---|---|---|---|---|
| ALAD | Heme Biosynthesis | rs1800435 | CG | High | - | [18] |
| ATP7B | Copper Transport | rs1061472 | CT and TT | Low | - | [20] |
| ATP7B | rs732774 | TC and CC | Low | - | [20] | |
| CYP3A4 | Xenobiotics Metabolism | rs2740574 | TC + CC | High b | Yes e | [22] |
| CYP3A5 | rs776746 | CT + TT | High c | Yes e | [22] | |
| CYP3A7 | rs2257401 | GC | High d | Yes e | [22] | |
| FADS2 | Fatty Acids Synthesis | rs174602 | CC | Low | - | [24] |
| FADS2 | rs74771917 | TT | Low | - | [24] | |
| FADS3 | rs7115739 | TT | Low | - | [24] | |
| GCLC | Glutathione Synthesis | rs17883901 | A | Low | - | [23] |
| GCLM | rs41303970 | AA | Low | - | [15] | |
| GPX1 | Detoxification of hydrogen peroxide by glutathione peroxidase | rs1050450 | A | Low | - | [23] |
| GPX4 | rs713041 | T and TT | Low | - | [24] | |
| GSTP1 | Glutathione | rs1695 | G | High | - | [23] |
| GSTP1 | rs1695 | AG | High f | - | [25] | |
| MT1A | Metallothionein | rs8052394 | AG + GG | High g | Yes h | [26] |
| MT1M | rs9936741 | C | Low | - | [23] | |
| MT1M | rs2270836 | CT and TT | High | - | [20] | |
| MT1M | rs2270837 | AG | No | Yes i | [16] | |
| MT2A | rs10636 | GC + CC | No | Yes i | [16] | |
| MT4 | rs11643815 | GA and AA | High | - | [20] | |
| MTHFR | Metionina Biosynthesis | rs2274976 | T and TT | Low | - | [24] |
| PON1 | Lactonase and ester hydrolase activity | rs705379 | GA and AA | Low | - | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perini, J.A.; Cardoso, J.V.; Knesse, A.d.O.; Pessoa-Silva, F.O.; Vasconcellos, A.C.S.d.; Machado, D.E.; Basta, P.C. Single-Nucleotide Polymorphisms Associated with Mercury Levels and Neurological Symptoms: An Overview. Toxics 2024, 12, 226. https://doi.org/10.3390/toxics12030226
Perini JA, Cardoso JV, Knesse AdO, Pessoa-Silva FO, Vasconcellos ACSd, Machado DE, Basta PC. Single-Nucleotide Polymorphisms Associated with Mercury Levels and Neurological Symptoms: An Overview. Toxics. 2024; 12(3):226. https://doi.org/10.3390/toxics12030226
Chicago/Turabian StylePerini, Jamila Alessandra, Jessica Vilarinho Cardoso, Alana de Oliveira Knesse, Felipe Oliveira Pessoa-Silva, Ana Claudia Santiago de Vasconcellos, Daniel Escorsim Machado, and Paulo Cesar Basta. 2024. "Single-Nucleotide Polymorphisms Associated with Mercury Levels and Neurological Symptoms: An Overview" Toxics 12, no. 3: 226. https://doi.org/10.3390/toxics12030226







