Differential Disruption of Glucose and Lipid Metabolism Induced by Phthalates in Human Hepatocytes and White Adipocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture
2.3. 3D Spheroid Cell Culture
2.4. Differentiation of Human White Fat Progenitors
2.5. Cell Treatment
2.6. RNA Sequencing
2.7. Live-Cell High Content Imaging
2.8. Statistical Analysis
3. Results
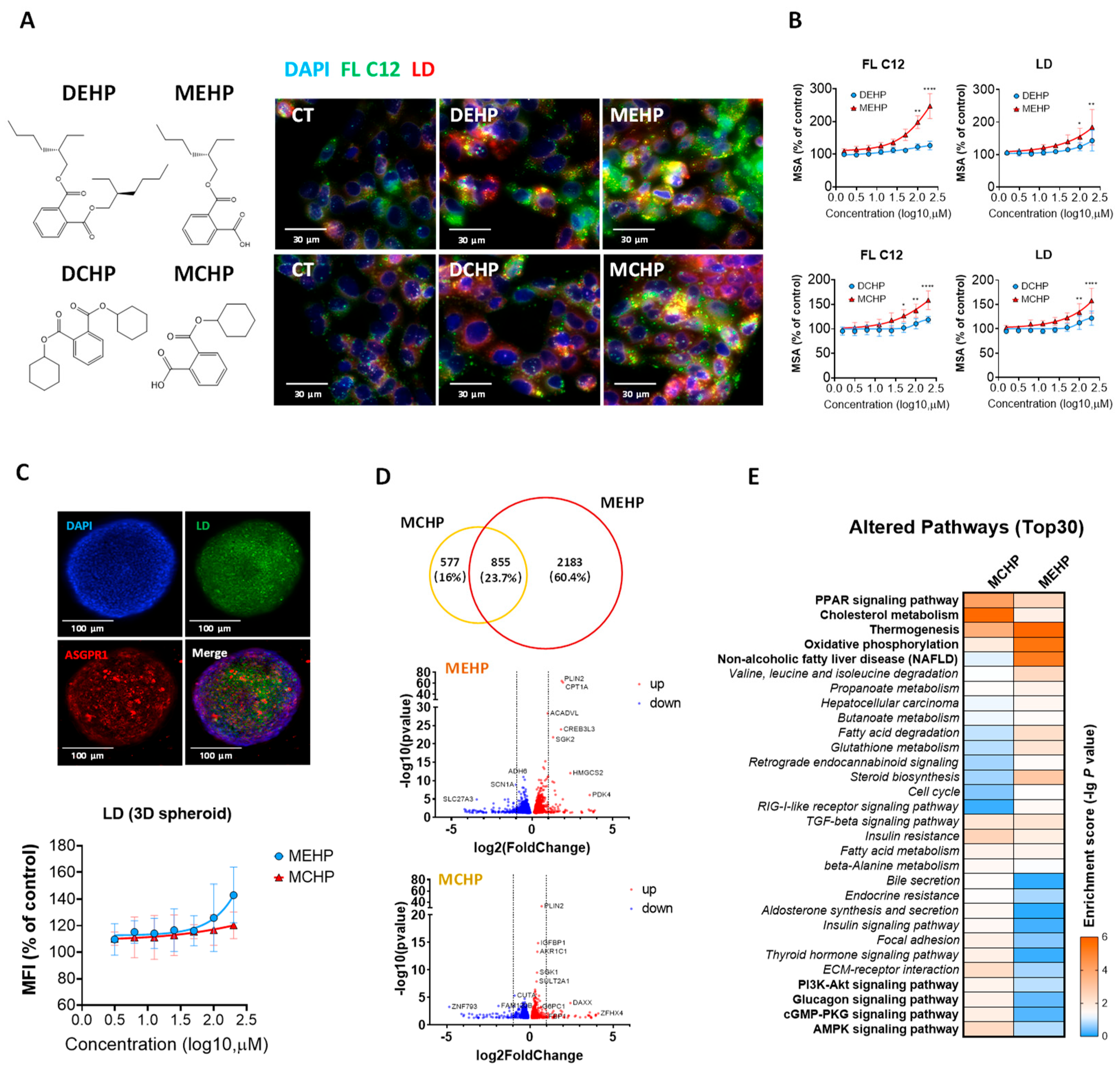
3.1. Effects of DEHP, DCHP, and Their Primary Metabolites on Hepatic Lipid Metabolism
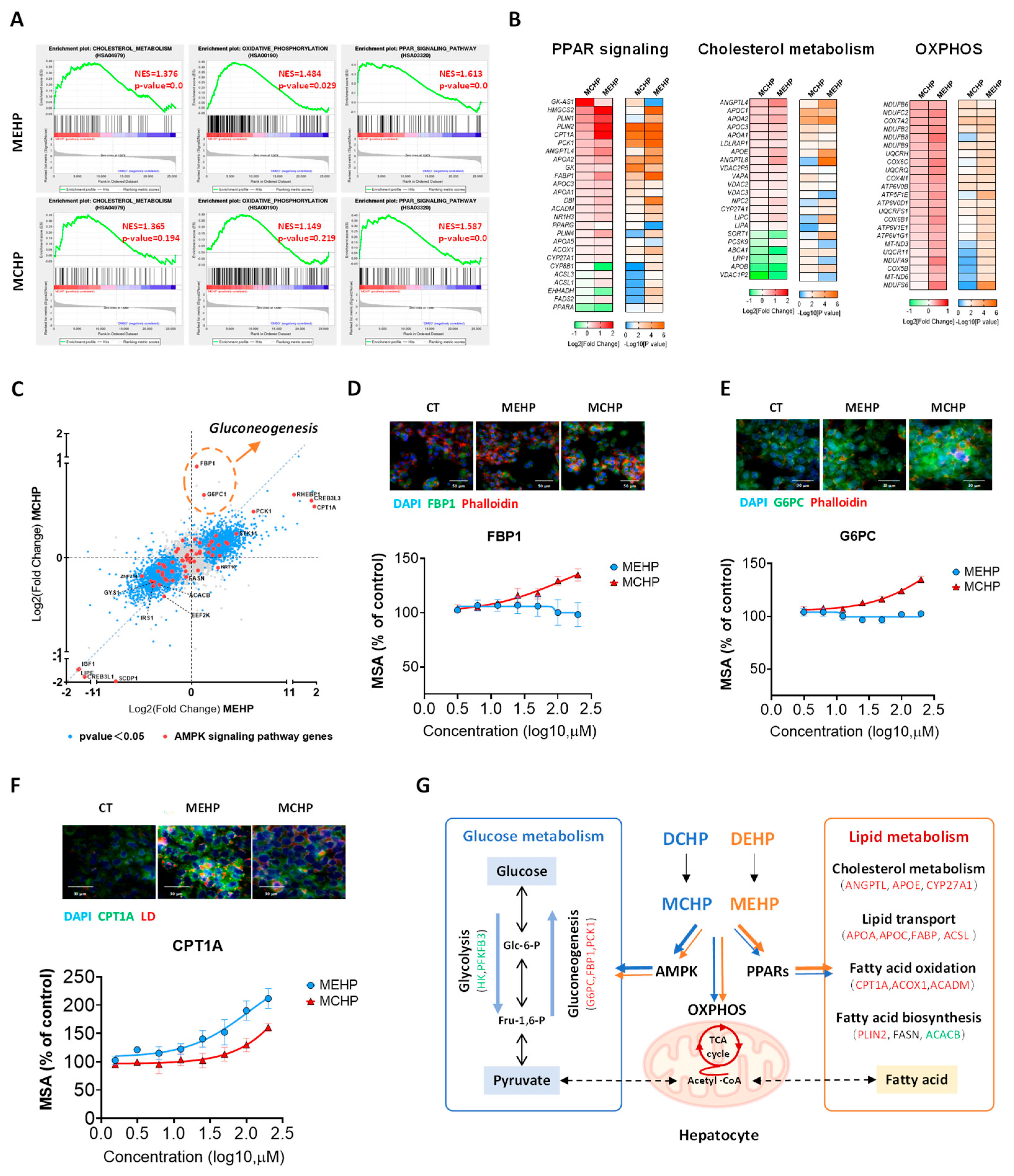
3.2. Comparative Impact of MEHP and MCHP on Lipid Metabolism in HepG2 Cells
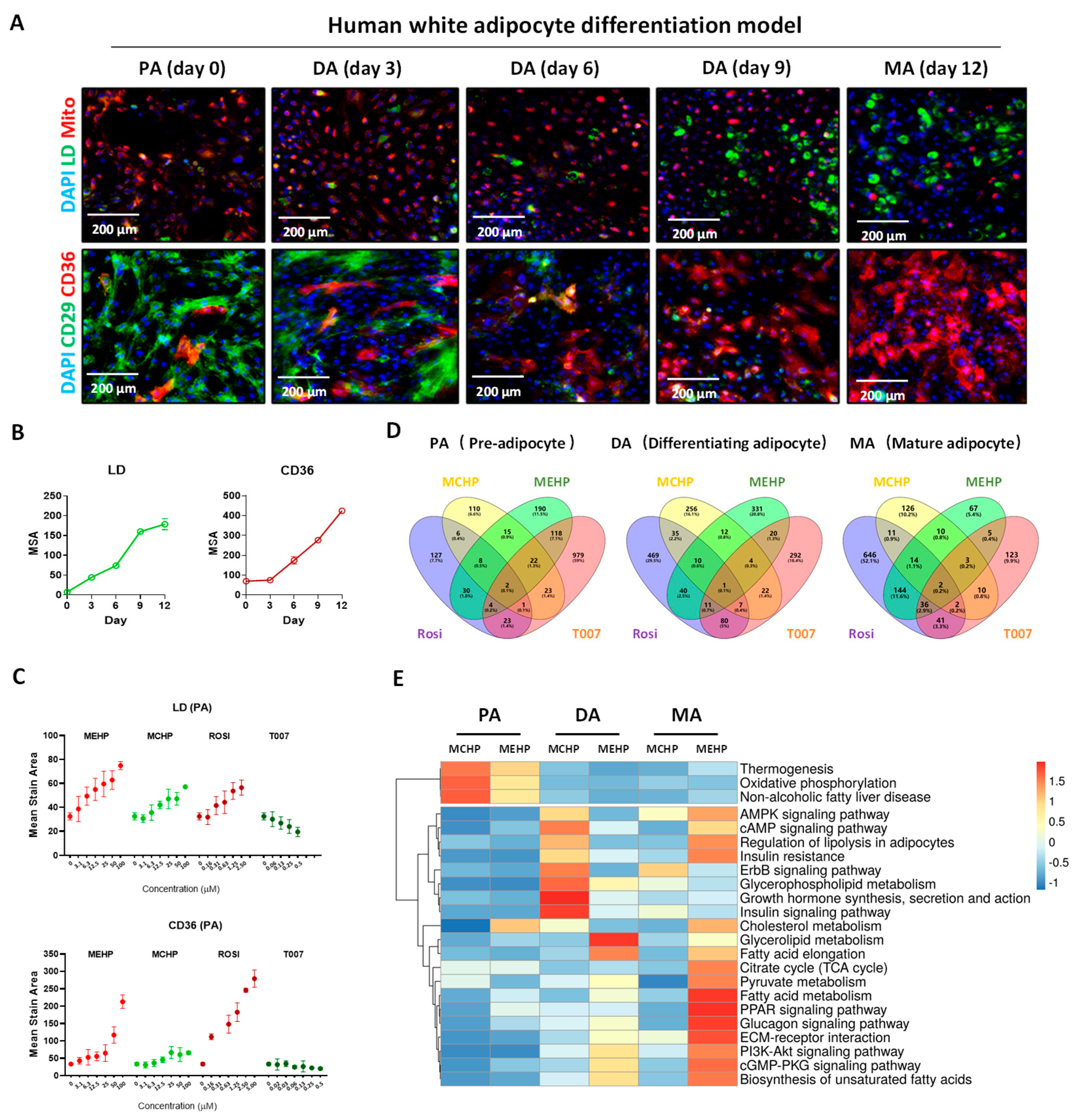
3.3. Effects of MEHP and MCHP on Lipid Metabolism in Human White Adipocytes
3.4. Distinct Impacts of MEHP and MCHP on Lipid Metabolism in Mature White Adipocytes
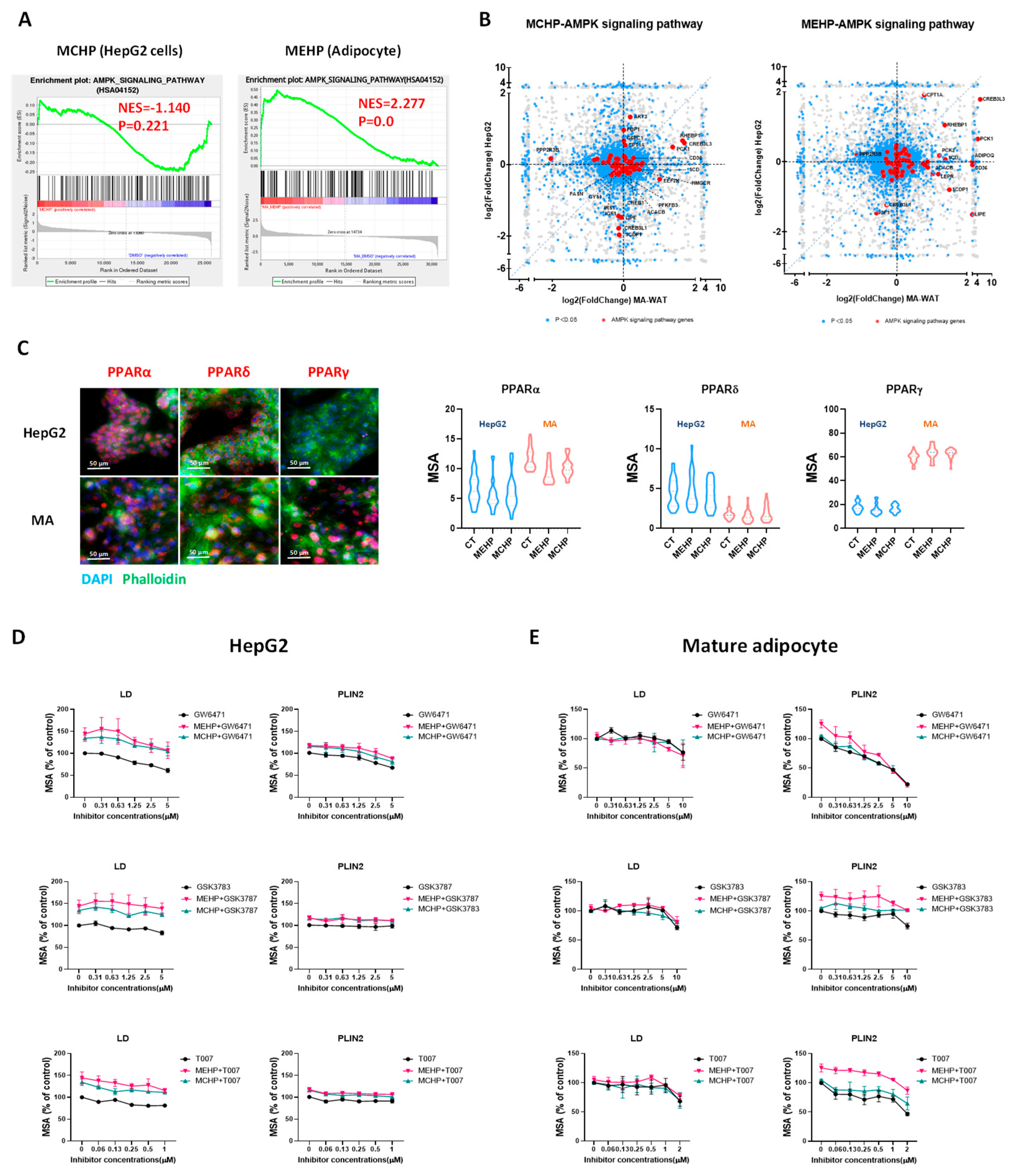
3.5. Metabolic Perturbations from MEHP and MCHP Demonstrate Cell-Type Discrepancy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schecter, A.; Lorber, M.; Guo, Y.; Wu, Q.; Yun, S.H.; Kannan, K.; Hommel, M.; Imran, N.; Hynan, L.S.; Cheng, D.; et al. Phthalate concentrations and dietary exposure from food purchased in New York State. Environ. Health Perspect. 2013, 121, 473–494. [Google Scholar] [CrossRef]
- Dodson, R.E.; Nishioka, M.; Standley, L.J.; Perovich, L.J.; Brody, J.G.; Rudel, R.A. Endocrine disruptors and asthma-associated chemicals in consumer products. Environ. Health Perspect. 2012, 120, 935–943. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; An, L.; Deng, C.; Su, H.; Wang, L.; Jiang, Z.; Zhou, J.; Wang, J.; Zhang, C.; et al. Phthalate esters in bottled drinking water and their human exposure in Beijing, China. Food Addit. Contaminants. Part B Surveill. 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Högberg, J.; Hanberg, A.; Berglund, M.; Skerfving, S.; Remberger, M.; Calafat, A.M.; Filipsson, A.F.; Jansson, B.; Johansson, N.; Appelgren, M.; et al. Phthalate Diesters and Their Metabolites in Human Breast Milk, Blood or Serum, and Urine as Biomarkers of Exposure in Vulnerable Populations. Environ. Health Perspect. 2008, 116, 334–339. [Google Scholar] [CrossRef]
- Toxicological Profile for Di(2-Ethylhexyl)Phthalate (DEHP). 2022. Available online: https://www.atsdr.cdc.gov/ToxProfiles/tp9.pdf (accessed on 20 January 2023).
- Seewoo, B.J.; Goodes, L.M.; Mofflin, L.; Mulders, Y.R.; Wong, E.V.; Toshniwal, P.; Brunner, M.; Alex, J.; Johnston, B.; Elagali, A.; et al. The plastic health map: A systematic evidence map of human health studies on plastic-associated chemicals. Environ. Int. 2023, 181, 108225. [Google Scholar] [CrossRef]
- EFSA Panel on Food Contact Materials, Enzymes and Processing Aids (CEP); Lambre, C.; Barat Baviera, J.M.; Bolognesi, C.; Chesson, A.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E.; et al. Identification and prioritisation for risk assessment of phthalates, structurally similar substances and replacement substances potentially used as plasticisers in materials and articles intended to come into contact with food. EFSA J. 2022, 20, e07231. [Google Scholar] [CrossRef]
- Sakhi, A.K.; Lillegaard, I.T.; Voorspoels, S.; Carlsen, M.H.; Loken, E.B.; Brantsaeter, A.L.; Haugen, M.; Meltzer, H.M.; Thomsen, C. Concentrations of phthalates and bisphenol A in Norwegian foods and beverages and estimated dietary exposure in adults. Environ. Int. 2014, 73, 259–269. [Google Scholar] [CrossRef]
- Tang, S.; He, C.; Thai, P.; Vijayasarathy, S.; Mackie, R.; Toms, L.L.; Thompson, K.; Hobson, P.; Tscharke, B.; O’Brien, J.W.; et al. Concentrations of phthalate metabolites in Australian urine samples and their contribution to the per capita loads in wastewater. Environ. Int. 2020, 137, 105534. [Google Scholar] [CrossRef]
- Hartmann, C.; Uhl, M.; Weiss, S.; Koch, H.M.; Scharf, S.; König, J. Human biomonitoring of phthalate exposure in Austrian children and adults and cumulative risk assessment. Int. J. Hyg. Environ. Health 2015, 218, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Uhl, M.; Weiss, S.; Scharf, S.; König, J. Austrian reference values for phthalate metabolite exposure in children/adolescents and adults. Int. J. Hyg. Environ. Health 2018, 221, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, J.; Garcia, J.M.; Lin, H.; Wang, Y.; Yan, P.; Wang, L.; Tan, Y.; Luo, J.; Qiu, Z.; et al. Phthalate levels in cord blood are associated with preterm delivery and fetal growth parameters in Chinese women. PLoS ONE 2014, 9, e87430. [Google Scholar] [CrossRef]
- Proposed Designation of Dicyclohexyl Phthalate(CASRN 84-61-7) as a High-Priority Substance for Risk Evaluation. 2018. Available online: https://www.regulations.gov/document/EPA-HQ-OPPT-2018-0504-0009 (accessed on 22 August 2023).
- EFSA Panel on Food Contact Materials, Enzymes and Processing Aids (CEP); Silano, V.; Barat Baviera, J.M.; Bolognesi, C.; Chesson, A.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E.; et al. Update of the risk assessment of di-butylphthalate (DBP), butyl-benzyl-phthalate (BBP), bis(2-ethylhexyl)phthalate (DEHP), di-isononylphthalate (DINP) and di-isodecylphthalate (DIDP) for use in food contact materials. EFSA J. 2019, 17, e05838. [Google Scholar] [CrossRef]
- Sui, Y.; Meng, Z.; Chen, J.; Liu, J.; Hernandez, R.; Gonzales, M.B.; Gwag, T.; Morris, A.J.; Zhou, C. Effects of Dicyclohexyl Phthalate Exposure on PXR Activation and Lipid Homeostasis in Mice. Environ. Health Perspect. 2021, 129, 127001. [Google Scholar] [CrossRef]
- Han, H.; Lee, H.A.; Park, B.; Park, B.; Hong, Y.S.; Ha, E.H.; Park, H. Associations of phthalate exposure with lipid levels and insulin sensitivity index in children: A prospective cohort study. Sci. Total Environ. 2019, 662, 714–721. [Google Scholar] [CrossRef]
- Liu, J.; Shi, J.; Hernandez, R.; Li, X.; Konchadi, P.; Miyake, Y.; Chen, Q.; Zhou, T.; Zhou, C. Paternal phthalate exposure-elicited offspring metabolic disorders are associated with altered sperm small RNAs in mice. Environ. Int. 2023, 172, 107769. [Google Scholar] [CrossRef]
- Radke, E.G.; Galizia, A.; Thayer, K.A.; Cooper, G.S. Phthalate exposure and metabolic effects: A systematic review of the human epidemiological evidence. Environ. Int. 2019, 132, 104768. [Google Scholar] [CrossRef]
- Lee, S.; Lee, H.A.; Park, B.; Han, H.; Hong, Y.S.; Ha, E.H.; Park, H. Prospective association between phthalate exposure in childhood and liver function in adolescence: The Ewha Birth and Growth Cohort Study. Environ. Health 2023, 22, 3. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Guo, J.L.; Xue, J.C.; Bai, C.L.; Guo, Y. Phthalate metabolites: Characterization, toxicities, global distribution, and exposure assessment. Environ. Pollut. 2021, 291, 118106. [Google Scholar] [CrossRef]
- Saito, T.; Hong, P.; Tanabe, R.; Nagai, K.; Kato, K. Enzymatic hydrolysis of structurally diverse phthalic acid esters by porcine and bovine pancreatic cholesterol esterases. Chemosphere 2010, 81, 1544–1548. [Google Scholar] [CrossRef]
- Zhang, W.; Shen, X.Y.; Zhang, W.W.; Chen, H.; Xu, W.P.; Wei, W. The effects of di 2-ethyl hexyl phthalate (DEHP) on cellular lipid accumulation in HepG2 cells and its potential mechanisms in the molecular level. Toxicol. Mech. Methods 2017, 27, 245–252. [Google Scholar] [CrossRef]
- Bai, J.; He, Z.; Li, Y.; Jiang, X.; Yu, H.; Tan, Q. Mono-2-ethylhexyl phthalate induces the expression of genes involved in fatty acid synthesis in HepG2 cells. Environ. Toxicol. Pharmacol. 2019, 69, 104–111. [Google Scholar] [CrossRef]
- Xue, R.; Lynes, M.D.; Dreyfuss, J.M.; Shamsi, F.; Schulz, T.J.; Zhang, H.; Huang, T.L.; Townsend, K.L.; Li, Y.; Takahashi, H.; et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat. Med. 2015, 21, 760–768. [Google Scholar] [CrossRef]
- Scholtes, C.; Giguere, V. Transcriptional control of energy metabolism by nuclear receptors. Nat. Rev. Mol. Cell Biol. 2022, 23, 750–770. [Google Scholar] [CrossRef]
- Maradonna, F.; Carnevali, O. Lipid Metabolism Alteration by Endocrine Disruptors in Animal Models: An Overview. Front. Endocrinol. 2018, 9, 654. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Zhao, T.; Yang, L.; Guo, S.; Shi, Y.; Zhang, X.; Zhou, L.; Ye, L. Mono-2-ethylhexyl phthalate (MEHP) promoted lipid accumulation via JAK2/STAT5 and aggravated oxidative stress in BRL-3A cells. Ecotoxicol. Environ. Saf. 2019, 184, 109611. [Google Scholar] [CrossRef]
- Xu, M.; Li, Y.; Wang, X.; Zhang, Q.; Wang, L.; Zhang, X.; Cui, W.; Han, X.; Ma, N.; Li, H.; et al. Role of Hepatocyte- and Macrophage-Specific PPARgamma in Hepatotoxicity Induced by Diethylhexyl Phthalate in Mice. Environ. Health Perspect. 2022, 130, 17005. [Google Scholar] [CrossRef]
- Chiang, H.C.; Kuo, Y.T.; Shen, C.C.; Lin, Y.H.; Wang, S.L.; Tsou, T.C. Mono(2-ethylhexyl)phthalate accumulation disturbs energy metabolism of fat cells. Arch. Toxicol. 2016, 90, 589–601. [Google Scholar] [CrossRef]
- Ribeiro, C.; Mendes, V.; Peleteiro, B.; Delgado, I.; Araújo, J.; Aggerbeck, M.; Annesi-Maesano, I.; Sarigiannis, D.; Ramos, E. Association between the exposure to phthalates and adiposity: A meta-analysis in children and adults. Environ. Res. 2019, 179, 108780. [Google Scholar] [CrossRef]
- Ribeiro, C.M.; Beserra, B.T.S.; Silva, N.G.; Lima, C.L.; Rocha, P.R.S.; Coelho, M.S.; Neves, F.A.R.; Amato, A.A. Exposure to endocrine-disrupting chemicals and anthropometric measures of obesity: A systematic review and meta-analysis. BMJ Open 2020, 10, e033509. [Google Scholar] [CrossRef]
- Schaffert, A.; Karkossa, I.; Ueberham, E.; Schlichting, R.; Walter, K.; Arnold, J.; Bluher, M.; Heiker, J.T.; Lehmann, J.; Wabitsch, M.; et al. Di-(2-ethylhexyl) phthalate substitutes accelerate human adipogenesis through PPARgamma activation and cause oxidative stress and impaired metabolic homeostasis in mature adipocytes. Environ. Int. 2022, 164, 107279. [Google Scholar] [CrossRef]
- Hsu, J.W.; Yeh, S.C.; Tsai, F.Y.; Chen, H.W.; Tsou, T.C. Fibroblast growth factor 21 secretion enhances glucose uptake in mono(2-ethylhexyl)phthalate-treated adipocytes. Toxicol. Vitr. 2019, 59, 246–254. [Google Scholar] [CrossRef]
- Casals-Casas, C.; Feige, J.N.; Desvergne, B. Interference of pollutants with PPARs: Endocrine disruption meets metabolism. Int. J. Obes. 2008, 32 (Suppl. S6), S53–S61. [Google Scholar] [CrossRef]
- Useini, A.; Engelberger, F.; Künze, G.; Sträter, N. Structural basis of the activation of PPARγ by the plasticizer metabolites MEHP and MINCH. Environ. Int. 2023, 173, 107822. [Google Scholar] [CrossRef]
- Sargis, R.M.; Johnson, D.N.; Choudhury, R.A.; Brady, M.J. Environmental endocrine disruptors promote adipogenesis in the 3T3-L1 cell line through glucocorticoid receptor activation. Obesity 2010, 18, 1283–1288. [Google Scholar] [CrossRef]
- Leng, Y.; Sun, Y.; Huang, W.; Lv, C.; Cui, J.; Li, T.; Wang, Y. Identification of dicyclohexyl phthalate as a glucocorticoid receptor antagonist by molecular docking and multiple in vitro methods. Mol. Biol. Rep. 2021, 48, 3145–3154. [Google Scholar] [CrossRef]
- Liu, J.; Hernandez, R.; Li, X.; Meng, Z.; Chen, H.; Zhou, C. Pregnane X Receptor Mediates Atherosclerosis Induced by Dicyclohexyl Phthalate in LDL Receptor-Deficient Mice. Cells 2022, 11, 1125. [Google Scholar] [CrossRef]
- Wyde, M.E.; Kirwan, S.E.; Zhang, F.; Laughter, A.; Hoffman, H.B.; Bartolucci-Page, E.; Gaido, K.W.; Yan, B.; You, L. Di-n-butyl phthalate activates constitutive androstane receptor and pregnane X receptor and enhances the expression of steroid-metabolizing enzymes in the liver of rat fetuses. Toxicol. Sci. 2005, 86, 281–290. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Xu, M.; Shang, H.; You, L.; Yang, J.; Jia, X.; Yang, H.; Wu, Y.; Yang, X.; Wan, Y. Differential Disruption of Glucose and Lipid Metabolism Induced by Phthalates in Human Hepatocytes and White Adipocytes. Toxics 2024, 12, 214. https://doi.org/10.3390/toxics12030214
Tian Y, Xu M, Shang H, You L, Yang J, Jia X, Yang H, Wu Y, Yang X, Wan Y. Differential Disruption of Glucose and Lipid Metabolism Induced by Phthalates in Human Hepatocytes and White Adipocytes. Toxics. 2024; 12(3):214. https://doi.org/10.3390/toxics12030214
Chicago/Turabian StyleTian, Yaru, Miao Xu, Hailin Shang, Lijuan You, Jing Yang, Xudong Jia, Hui Yang, Yongning Wu, Xingfen Yang, and Yi Wan. 2024. "Differential Disruption of Glucose and Lipid Metabolism Induced by Phthalates in Human Hepatocytes and White Adipocytes" Toxics 12, no. 3: 214. https://doi.org/10.3390/toxics12030214





