Detection of Salmonella in Food Matrices, from Conventional Methods to Recent Aptamer-Sensing Technologies
Abstract
1. Introduction
1.1. Salmonella and Food Contamination
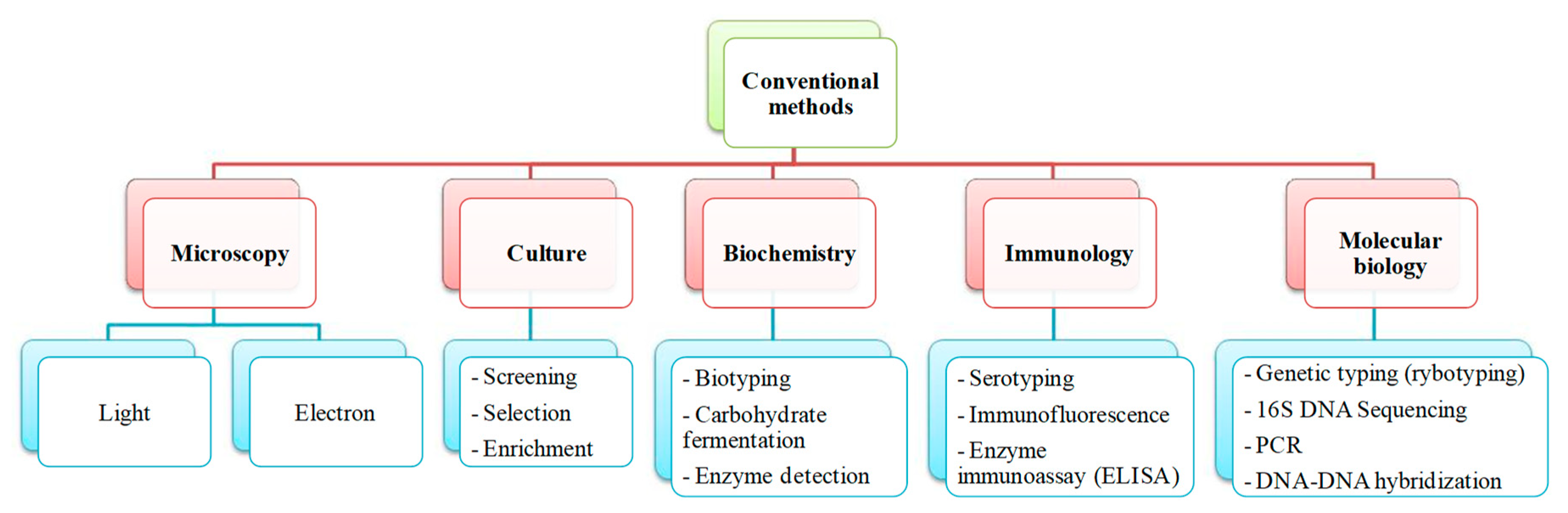
1.2. Salmonella Detection and Quantification by Conventional Methods
2. Biosensors for Salmonella Detection and Quantification
2.1. Optical Biosensors
2.1.1. Surface Plasmon Resonance Biosensors
2.1.2. Fluorescence-Based Sensors
2.1.3. Chemical Luminescence-Based Biosensors
2.2. Electrochemical Biosensors
2.2.1. Amperometry
2.2.2. Potentiometry
2.2.3. Impedimetry
2.3. Mass-Based Biosensors
3. Aptasensors for Salmonella Detection
3.1. Aptamers Selection
3.2. Aptamers as Ligands for Magnetic Separation
3.3. Optical Aptasensors
3.3.1. Surface Plasmon Resonance Aptasensors
3.3.2. Surface-Enhanced Raman Spectroscopy Aptasensors
3.3.3. Chemiluminescent Aptasensors
3.3.4. Fluorescent Aptasensors
3.3.5. Colorimetry-Based Aptasensors
3.3.6. Flat Substrate Aptasensors
3.4. Electrochemical Aptasensors
3.4.1. Potentiometry
3.4.2. Impedimetry
3.4.3. Differential Pulse Voltammetry (DPV)
3.5. Mass-Based Aptasensors
4. Conclusions
Funding
Conflicts of Interest
References
- Brandão, D.; Liébana, S.; Pividori, M.I. Multiplexed detection of foodborne pathogens based on magnetic particles. New Biotechnol. 2015, 32, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Myint, M.S.; Johnson, Y.J.; Tablante, N.L.; Heckert, R.A. The effect of pre-enrichment protocol on the sensitivity and specificity of PCR for detection of naturally contaminated Salmonella in raw poultry compared to conventional culture. Food Microbiol. 2006, 23, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.E.; Carrique-Mas, J.J.; McLaren, I.; Davies, R.H. A comparison of pooled and individual bird sampling for detection of Salmonella in commercial egg laying flocks. Prev. Vet. Med. 2011, 99, 176–184. [Google Scholar] [CrossRef] [PubMed]
- EFSA. The Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents, Antimicrobial Resistance and Foodborne Outbreaks in the European Union in 2006; EFSA: Parma, Italy, 2007. [Google Scholar]
- Zhang, H.; Ma, X.; Liu, Y.; Duan, N.; Wu, S.; Wang, Z.; Xu, B. Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella Typhimurium and Staphylococcus aureus. Biosens. Bioelectron. 2015, 74, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Pournaras, A.V.; Koraki, T.; Prodromidis, M.I. Development of an impedimetric immunosensor based on electropolymerized polytyramine films for the direct detection of Salmonella Typhimurium in pure cultures of type strains and inoculated real samples. Anal. Chim. Acta 2008, 624, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Tao, Z.; Yu, Y.; Ma, X.; Xia, Y.; Wang, L.; Wang, Z. A visual detection method for Salmonella Typhimurium based on aptamer recognition and nanogold labeling. Food Control 2014, 37, 188–192. [Google Scholar] [CrossRef]
- Sharma, H.; Mutharasan, R. Review of biosensors for foodborne pathogens and toxins. Sens. Actuators B Chem. 2013, 183, 535–549. [Google Scholar] [CrossRef]
- Huang, J.; Zong, Q.; Zhao, F.; Zhu, J.; Jiao, X.-A. Quantitative surveys of Salmonella and Campylobacter on retail raw chicken in Yangzhou, China. Food Control 2016, 59, 68–73. [Google Scholar] [CrossRef]
- Sanitaire-InVs, I.D.V. Surveillance des Toxi-Infections Alimentaires Collectives—Données TIAC 2012; Santé Publique France: Saint-Maurice, France, 2012. [Google Scholar]
- Greig, J.D.; Ravel, A. Analysis of foodborne outbreak data reported internationally for source attribution. Int. J. Food Microbiol. 2009, 130, 77–87. [Google Scholar] [CrossRef]
- D’Aoust, J.-Y. Pathogenicity of foodborne Salmonella. Int. J. Food Microbiol. 1991, 12, 17–40. [Google Scholar] [CrossRef]
- EFSA. Analysis of the Baseline Survey on the Prevalence of Campylobacter in Broiler Batches and of Campylobacter and Salmonella on Broiler Carcasses in the EU; EFSA: Parma, Italy, 2008. [Google Scholar]
- Abd-Elghany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 2015, 143, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Aydin, M.; Khatiwara, A.; Dolan, M.C.; Gilmore, D.F.; Bouldin, J.L.; Ahn, S.; Ricke, S.C. Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 2014, 38, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Santaire-InVs, I.d.V. Surveillance des Toxi-Infections Alimentaires Collectives—Données TIAC 2014; Santé Publique France: Saint-Maurice, France, 2014. [Google Scholar]
- Ohtsuka, K.; Yanagawa, K.; Takatori, K.; Hara-Kudo, Y. Detection of Salmonella enterica in Naturally Contaminated Liquid Eggs by Loop-Mediated Isothermal Amplification, and Characterization of Salmonella Isolates. Appl. Environ. Microbiol. 2005, 71, 6730–6735. [Google Scholar] [CrossRef] [PubMed]
- Tirado, C.; Schmidt, K. WHO Surveillance Programme for Control of Foodborne Infections and Intoxications: Preliminary Results and Trends Across Greater Europe. J. Infect. 2001, 43, 80–84. [Google Scholar] [CrossRef]
- Li, J.; Zhai, L.; Bie, X.; Lu, Z.; Kong, X.; Yu, Q.; Lv, F.; Zhang, C.; Zhao, H. A novel visual loop-mediated isothermal amplification assay targeting gene 62181533 for the detection of Salmonella spp. in foods. Food Control 2016, 60, 230–236. [Google Scholar] [CrossRef]
- Leon-Velarde, C.G.; Zosherafatein, L.; Odumeru, J.A. Application of an automated immunomagnetic separation–enzyme immunoassay for the detection of Salmonella enterica subspecies enterica from poultry environmental swabs. J. Microbiol. Methods 2009, 79, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Vandeplas, S.; Dubois Dauphin, R.; Beckers, Y.; Thonart, P.; Théwis, A. Salmonella in Chicken: Current and Developing Strategies to Reduce Contamination at Farm Level. J. Food Prot. 2010, 73, 774–785. [Google Scholar] [CrossRef]
- De Boer, E.; Beumer, R.R. Methodology for detection and typing of foodborne microorganisms. Int. J. Food Microbiol. 1999, 50, 119–130. [Google Scholar] [CrossRef]
- Ayçiçek, H.; Aydoğan, H.; Küçükkaraaslan, A.; Baysallar, M.; Başustaoğlu, A.C. Assessment of the bacterial contamination on hands of hospital food handlers. Food Control 2004, 15, 253–259. [Google Scholar] [CrossRef]
- Arora, P.; Sindhu, A.; Dilbaghi, N.; Chaudhury, A. Biosensors as innovative tools for the detection of food borne pathogens. Biosens. Bioelectron. 2011, 28, 1–12. [Google Scholar] [CrossRef]
- Paniel, N.; Baudart, J.; Hayat, A.; Barthelmebs, L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 2013, 64, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, R.; Mannazzu, I.; Cocolin, L.; Comi, G.; Clementi, F. Twelve-Hour PCR-Based Method for Detection of Salmonella spp. in Food. Appl. Environ. Microbiol. 2001, 67, 977–978. [Google Scholar] [CrossRef] [PubMed]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E. Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 1999, 14, 599–624. [Google Scholar] [CrossRef]
- Jyoti, A.; Vajpayee, P.; Singh, G.; Patel, C.B.; Gupta, K.C.; Shanker, R. Identification of Environmental Reservoirs of Nontyphoidal Salmonellosis: Aptamer-Assisted Bioconcentration and Subsequent Detection of Salmonella Typhimurium by Quantitative Polymerase Chain Reaction. Environ. Sci. Technol. 2011, 45, 8996–9002. [Google Scholar] [CrossRef] [PubMed]
- Toze, S. PCR and the detection of microbial pathogens in water and wastewater. Water Res. 1999, 33, 3545–3556. [Google Scholar] [CrossRef]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O’Kennedy, R. Advances in biosensors for detection of pathogens in food and water. Enzym. Microb. Technol. 2003, 32, 3–13. [Google Scholar] [CrossRef]
- Olivier, J.-D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Oliver, J.D. The viable but non-culturable state and its relevance in food safety. Curr. Opin. Food Sci. 2016, 8, 127–133. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Anvarian, A.H.P.; Smith, M.P.; Overton, T.W. The effects of orange juice clarification on the physiology of Escherichia coli; growth-based and flow cytometric analysis. Int. J. Food Microbiol. 2016, 219, 38–43. [Google Scholar] [CrossRef]
- Liao, H.; Zhang, L.; Hu, X.; Liao, X. Effect of high pressure CO2 and mild heat processing on natural microorganisms in apple juice. Int. J. Food Microbiol. 2010, 137, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Bi, X.; Hao, Y.; Liao, X. Induction of Viable but Nonculturable Escherichia coli O157:H7 by High Pressure CO2 and Its Characteristics. PLoS ONE 2013, 8, e62388. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mannan, K.S.B.; Andrews, S. Viable but Nonculturable Bacteria: Food Safety and Public Health Perspective. ISRN Microbiol. 2013, 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Alleron, L.; Khemiri, A.; Koubar, M.; Lacombe, C.; Coquet, L.; Cosette, P.; Jouenne, T.; Frere, J. VBNC Legionella pneumophila cells are still able to produce virulence proteins. Water Res. 2013, 47, 6606–6617. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.; Tyrrell, G.; Li, X.-F. Production of Shiga-like toxins in viable but nonculturable Escherichia coli O157:H7. Water Res. 2010, 44, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Dinu, L.-D.; Bach, S. Induction of Viable but Nonculturable Escherichia coli O157:H7 in the Phyllosphere of Lettuce: A Food Safety Risk Factor. Appl. Environ. Microbiol. 2011, 77, 8295–8302. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-S.; Durst, R.A. Simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Listeria monocytogenes with an array-based immunosorbent assay using universal protein G-liposomal nanovesicles. Talanta 2006, 69, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Magliulo, M.; Simoni, P.; Guardigli, M.; Michelini, E.; Luciani, M.; Lelli, R.; Roda, A. A Rapid Multiplexed Chemiluminescent Immunoassay for the Detection of Escherichia coli O157:H7, Yersinia enterocolitica, Salmonella Typhimurium, and Listeria monocytogenes Pathogen Bacteria. J. Agric. Food Chem. 2007, 55, 4933–4939. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Biswas, A.K.; Choi, K.; Pal, U.K. Methods for Rapid Detection of Foodborne Pathogens: An Overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef]
- Sun, Q.; Zhao, G.; Dou, W. A nonenzymatic optical immunoassay strategy for detection of Salmonella infection based on blue silica nanoparticles. Anal. Chim. Acta 2015. [Google Scholar] [CrossRef]
- Schneid, A.D.S.; Rodrigues, K.L.; Chemello, D.; Tondo, E.C.; Ayub, M.A.Z.; Aleixo, J.A.G. Evaluation of an indirect ELISA for the detection of Salmonella in chicken meat. Braz. J. Microbiol. 2006, 37, 350–355. [Google Scholar] [CrossRef]
- Abdel-Hamid, I.; Ivnitski, D.; Atanasov, P.; Wilkins, E. Highly sensitive flow-injection immunoassay system for rapid detection of bacteria. Anal. Chim. Acta 1999, 399, 99–108. [Google Scholar] [CrossRef]
- Xia, S.; Yu, Z.; Liu, D.; Xu, C.; Lai, W. Developing a novel immunochromatographic test strip with gold magnetic bifunctional nanobeads (GMBN) for efficient detection of Salmonella choleraesuis in milk. Food Control 2016, 59, 507–512. [Google Scholar] [CrossRef]
- Lynch, M.J.B.; Leon-Velarde, C.G.; McEwen, S.; Odumeru, J.A. Evaluation of an automated immunomagnetic separation method for the rapid detection of Salmonella species in poultry environmental samples. J. Microbiol. Methods 2004, 58, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139, S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Doyle, M.P. Introduction. Microbiological food safety. Microbes Infect. 2002, 4, 395–397. [Google Scholar] [CrossRef]
- Oliveira, S.D.; Rodenbusch, C.R.; Cé, M.C.; Rocha, S.L.S.; Canal, C.W. Evaluation of selective and non-selective enrichment PCR procedures for Salmonella detection. Lett. Appl. Microbiol. 2003, 36, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Uyttendaele, M.; Vanwildemeersch, K.; Debevere, J. Evaluation of real-time PCR vs automated ELISA and a conventional culture method using a semi-solid medium for detection of Salmonella. Lett. Appl. Microbiol. 2003, 37, 386–391. [Google Scholar] [CrossRef]
- Soumet, C.; Ermel, G.; Salvat, G.; Colin, P. Detection of Salmonella spp. in food products by polymerase chain reaction and hybridization assay in microplate format. Lett. Appl. Microbiol. 1997, 24, 113–116. [Google Scholar] [CrossRef]
- Wang, S.J.; Yeh, D.B. Designing of polymerase chain reaction primers for the detection of Salmonella Enteritidis in foods and faecal samples. Lett. Appl. Microbiol. 2002, 34, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mustapha, A. Evaluation of four template preparation methods for polymerase chain reaction-based detection of Salmonella in ground beef and chicken. Lett. Appl. Microbiol. 2002, 35, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Schrank, I.S.; Mores, M.A.Z.; Costa, J.L.A.; Frazzon, A.P.G.; Soncini, R.; Schrank, A.; Vainstein, M.H.; Silva, S.C. Influence of enrichment media and application of a PCR based method to detect Salmonella in poultry industry products and clinical samples. Vet. Microbiol. 2001, 82, 45–53. [Google Scholar] [CrossRef]
- Soumet, C.; Blivet, D.; Ermel, G.; Colin, P.; Salvat, G. An immunoconcentration-PCR assay to detect Salmonella in the environment of poultry houses. Int. J. Food Microbiol. 1999, 48, 221–224. [Google Scholar] [CrossRef]
- Soumet, C.; Ermel, G.; Rose, N.; Rose, V.; Drouin, P.; Salvat, G.; Colin, P. Evaluation of a Multiplex PCR assay for simultaneous identification of Salmonella sp., Salmonella Enteritidis and Salmonella Typhimurium from environmental swabs of poultry houses. Lett. Appl. Microbiol. 1999, 28, 113–117. [Google Scholar] [CrossRef]
- Soumet, C.; Ermel, G.; Fach, P.; Colin, P. Evaluation of different DNA extraction procedures for the detection of Salmonella from chicken products by polymerase chain reaction. Lett. Appl. Microbiol. 1994, 19, 294–298. [Google Scholar] [CrossRef]
- Shinde, S.B.; Fernandes, C.B.; Patravale, V.B. Recent trends in in-vitro nanodiagnostics for detection of pathogens. J. Control. Release 2012, 159, 164–180. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, G.G.; Park, J.S.; Jung, Y.H.; Kwak, H.S.; Kim, S.B.; Nam, Y.S.; Kwon, S.-T. A Novel Multiplex PCR Assay for Rapid and Simultaneous Detection of Five Pathogenic Bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J. Food Prot. 2007, 70, 1656–1662. [Google Scholar] [CrossRef]
- Perry, L.; Heard, P.; Kane, M.; Kim, H.; Savikhin, S.; DomÍNguez, W.; Applegate, B. Application of multiplex polymerase chain reaction to the detection of pathogens in food. J. Rapid Methods Autom. Microbiol. 2007, 15, 176–198. [Google Scholar] [CrossRef]
- De Boer, R.F.; Ott, A.; Kesztyüs, B.; Kooistra-Smid, A.M.D. Improved Detection of Five Major Gastrointestinal Pathogens by Use of a Molecular Screening Approach. J. Clin. Microbiol. 2010, 48, 4140–4146. [Google Scholar] [CrossRef]
- Alvarez, J.; Sota, M.; Vivanco, A.B.; Perales, I.; Cisterna, R.; Rementeria, A.; Garaizar, J. Development of a Multiplex PCR Technique for Detection and Epidemiological Typing of Salmonella in Human Clinical Samples. J. Clin. Microbiol. 2004, 42, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Burtscher, C.; Fall, P.A.; Wilderer, P.A.; Wuertz, S. Detection of Salmonella spp. and Listeria monocytogenes in Suspended Organic Waste by Nucleic Acid Extraction and PCR. Appl. Environ. Microbiol. 1999, 65, 2235–2237. [Google Scholar] [PubMed]
- Mothershed, E.A.; Whitney, A.M. Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clin. Chim. Acta 2006, 363, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Kwang, J.; Littledike, E.T.; Keen, J.E. Use of the polymerase chain reaction for Salmonella detection. Lett. Appl. Microbiol. 1996, 22, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.D.; Santos, L.R.; Schuch, D.M.T.; Silva, A.B.; Salle, C.T.P.; Canal, C.W. Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol. 2002, 87, 25–35. [Google Scholar] [CrossRef]
- Pangloli, P.; Dje, Y.; Oliver, S.P.; Mathew, A.; Golden, D.A.; Taylor, W.J.; Draughon, F.A. Evaluation of Methods for Recovery of Salmonella from Dairy Cattle, Poultry, and Swine Farms. J. Food Prot. 2003, 66, 1987–1995. [Google Scholar] [CrossRef]
- Chiu, T.-H.; Chen, T.-R.; Hwang, W.-Z.; Tsen, H.-Y. Sequencing of an internal transcribed spacer region of 16S–23S rRNA gene and designing of PCR primers for the detection of Salmonella spp. in food. Int. J. Food Microbiol. 2005, 97, 259–265. [Google Scholar] [CrossRef]
- Girones, R.; Ferrús, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; de Abreu Corrêa, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef]
- Postollec, F.; Falentin, H.; Pavan, S.; Combrisson, J.; Sohier, D. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 2011, 28, 848–861. [Google Scholar] [CrossRef]
- Aabo, S.; Rasmussen, O.F.; Roseen, L.; Sørensen, P.D.; Olsen, J.E. Salmonella identification by the polymerase chain reaction. Mol. Cell. Probes 1993, 7, 171–178. [Google Scholar] [CrossRef]
- Makino, S.-I.; Kurazono, H.; Chongsanguam, M.; Hayashi, H.; Cheun, H.-I.; Suzuki, S.; Shirahata, T. Establishment of the PCR System Specific to Salmonella spp. and Its Application for the Inspection of Food and Fecal Samples. J. Vet. Med Sci. 1999, 61, 1245–1247. [Google Scholar] [CrossRef] [PubMed]
- Carlson, S.A.; Bolton, L.F.; Briggs, C.E.; Hurd, H.S.; Sharma, V.K.; Fedorka-Cray, P.J.; Jones, B.D. Detection of multiresistant Salmonella Typhimurium DT104 using multiplex and fluorogenic PCR. Mol. Cell. Probes 1999, 13, 213–222. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, C.G.; Santana, Â.P.; Da Silva, P.H.C.; Gonçalves, V.S.P.; Barros, M.d.A.F.; Torres, F.A.G.; Murata, L.S.; Perecmanis, S. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 2010, 139, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Duarte, O.G.; Bai, J.; Newell, E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2009, 63, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhai, L.; Bie, X.; Lu, Z.; Zhang, C.; Tao, T.; Li, J.; Lv, F.; Zhao, H. Survey of five food-borne pathogens in commercial cold food dishes and their detection by multiplex PCR. Food Control 2016, 59, 862–869. [Google Scholar] [CrossRef]
- Sharma, V.K.; Carlson, S.A. Simultaneous Detection of Salmonella Strains and Escherichia coli O157:H7 with Fluorogenic PCR and Single-Enrichment-Broth Culture. Appl. Environ. Microbiol. 2000, 66, 5472–5476. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.W.; Mahon, J.; Lax, A.J. Development of a probe and PCR primers specific to the virulence plasmid of Salmonella Enteritidis. Mol. Cell. Probes 1994, 8, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Song, L.; Pan, Z.; Jiao, X. Identification and Discrimination of Salmonella enterica Serovar Gallinarum Biovars Pullorum and Gallinarum Based on a One-Step Multiplex PCR Assay. Front. Microbiol. 2018, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Heymans, R.; Vila, A.; van Heerwaarden, C.A.M.; Jansen, C.C.C.; Castelijn, G.A.A.; van der Voort, M.; Biesta-Peters, E.G. Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS ONE 2018, 13, e0206316. [Google Scholar] [CrossRef]
- Chen, S.; Yee, A.; Griffiths, M.; Larkin, C.; Yamashiro, C.T.; Behari, R.; Paszko-Kolva, C.; Rahn, K.; De Grandis, S.A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int. J. Food Microbiol. 1997, 35, 239–250. [Google Scholar] [CrossRef]
- Eyigor, A.; Carli, K.T.; Unal, C.B. Implementation of real-time PCR to tetrathionate broth enrichment step of Salmonella detection in poultry. Lett. Appl. Microbiol. 2002, 34, 37–41. [Google Scholar] [CrossRef]
- De Medici, D.; Croci, L.; Delibato, E.; Di Pasquale, S.; Filetici, E.; Toti, L. Evaluation of DNA Extraction Methods for Use in Combination with SYBR Green I Real-Time PCR to Detect Salmonella enterica Serotype Enteritidis in Poultry. Appl. Environ. Microbiol. 2003, 69, 3456–3461. [Google Scholar] [CrossRef]
- Malorny, B.; Bunge, C.; Helmuth, R. A real-time PCR for the detection of Salmonella Enteritidis in poultry meat and consumption eggs. J. Microbiol. Methods 2007, 70, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Hein, I.; Flekna, G.; Krassnig, M.; Wagner, M. Real-time PCR for the detection of Salmonella spp. in food: An alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 2006, 66, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Ellingson, J.L.E.; Anderson, J.L.; Carlson, S.A.; Sharma, V.K. Twelve hour real-time PCR technique for the sensitive and specific detection of Salmonella in raw and ready-to-eat meat products. Mol. Cell. Probes 2004, 18, 51–57. [Google Scholar] [CrossRef]
- Cremonesi, P.; Pisani, L.F.; Lecchi, C.; Ceciliani, F.; Martino, P.; Bonastre, A.S.; Karus, A.; Balzaretti, C.; Castiglioni, B. Development of 23 individual TaqMan® real-time PCR assays for identifying common foodborne pathogens using a single set of amplification conditions. Food Microbiol. 2014, 43, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Trinetta, V.; Shi, X.W.; Noll, L.; Magossi, G.; Zheng, W.P.; Porter, E.; Cernicchiaro, N.G.; Renter, D.; Nagaraja, T.G. A multiplex real-time PCR assay, based on inv A and pag C genes, for the detection and quantification of Salmonella enterica from cattle lymph nodes. J. Microbiol. Methods 2018, 148. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Aznar, R.; Sánchez, G. Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 2014, 116, 1–13. [Google Scholar] [CrossRef]
- Nogva, H.K.; Dromtorp, S.M.; Nissen, H.; Rudi, K. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques 2003, 34, 810, 812–813. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Sossa-Fernandez, P.; Burr, M.D.; Camper, A.K. Use of Propidium Monoazide for Live/Dead Distinction in Microbial Ecology. Appl. Environ. Microbiol. 2007, 73, 5111–5117. [Google Scholar] [CrossRef] [PubMed]
- Dinh Thanh, M.; Agustí, G.; Mader, A.; Appel, B.; Codony, F. Improved sample treatment protocol for accurate detection of live Salmonella spp. in food samples by viability PCR. PLoS ONE 2017, 12, e0189302. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [PubMed]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Raurich, S.; Garriga, M.; Aymerich, T. Effect of Amplicon Length in Propidium Monoazide Quantitative PCR for the Enumeration of Viable Cells of Salmonella in Cooked Ham. Food Anal. Methods 2013, 6. [Google Scholar] [CrossRef]
- Josefsen, M.H.; Krause, M.; Hansen, F.; Hoorfar, J. Optimization of a 12-Hour TaqMan PCR-Based Method for Detection of Salmonella Bacteria in Meat. Appl. Environ. Microbiol. 2007, 73, 3040–3048. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Taha, E.G.; Mohamed, A.; Srivastava, K.K.; Reddy, P.G. Rapid Detection of Salmonella in Chicken Meat Using Immunomagnetic Separation, CHROMagar, ELISA and Real-time Polymerase Chain Reaction (RT-PCR). Int. J. Poult. Sci. 2010, 9, 831–835. [Google Scholar] [CrossRef]
- Johnson, J.R. Development of polymerase chain reaction-based assays for bacterial gene detection. J. Microbiol. Methods 2000, 41, 201–209. [Google Scholar] [CrossRef]
- Zelada-Guillén, G.A.; Riu, J.; Düzgün, A.; Rius, F.X. Immediate Detection of Living Bacteria at Ultralow Concentrations Using a Carbon Nanotube Based Potentiometric Aptasensor. Angew. Chem. Int. Ed. 2009, 48, 7334–7337. [Google Scholar] [CrossRef]
- Taylor, A.D.; Ladd, J.; Shaoyi, J. Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems; Elwary, S., Turner, A.P.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Bell, R.L.; Jarvis, K.G.; Ottesen, A.R.; McFarland, M.A.; Brown, E.W. Recent and emerging innovations in Salmonella detection: A food and environmental perspective. Microb. Biotechnol. 2016, 9, 279–292. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Gorski, L.; Liang, A.S. Effect of enrichment medium on real-time detection of Salmonella enterica from lettuce and tomato enrichment cultures. J. Food Prot. 2010, 73, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Chua, T.; Bhagwat, A.A. A Rapid and Simple DNA Extraction Procedure to Detect Salmonella spp. and Listeria monocytogenes from Fresh Produce Using Real-time PCR. Food Anal. Methods 2009, 2, 96–101. [Google Scholar] [CrossRef]
- Brehm-Stecher, B.; Young, C.; Jaykus, L.A.; Tortorello, M.L. Sample preparation: The forgotten beginning. J. Food Prot. 2009, 72, 1774–1789. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Hoorfar, J.; Bunge, C.; Helmuth, R. Multicenter validation of the analytical accuracy of Salmonella PCR: Towards an international standard. Appl. Environ. Microbiol. 2003, 69, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Resendiz-Nava, C.; Esquivel-Hernandez, Y.; Alcaraz-Gonzalez, A.; Castaneda-Serrano, P.; Nava, G.M. PCR Assays Based on invA Gene Amplification are not Reliable for Salmonella Detection. Jundishapur J. Microbiol. 2019, 12, e68764. [Google Scholar] [CrossRef]
- Scholz, H.C.; Arnold, T.; Marg, H.; Rosler, U.; Hensel, A. Improvement of an invA-based PCR for the specific detection of Salmonella typhimurium in organs of pigs. Berl. und Munch. Tierarztl. Wochenschr. 2001, 114, 401–403. [Google Scholar]
- Kloska, F.; Casteel, M.; Kump, F.W.; Klein, G. Implementation of a Risk-Orientated Hygiene Analysis for the Control of Salmonella JAVA in the Broiler Production. Curr. Microbiol. 2017, 74, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic Real-Time PCR for Detection of Salmonella in Food. Appl. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef]
- Laube, T.; Cortés, P.; Llagostera, M.; Alegret, S.; Pividori, M.I. Phagomagnetic immunoassay for the rapid detection of Salmonella. Appl. Microbiol. Biotechnol. 2013, 98, 1795–1805. [Google Scholar] [CrossRef]
- Yang, H.; Li, H.; Jiang, X. Detection of foodborne pathogens using bioconjugated nanomaterials. Microfluid. Nanofluidics 2008, 5, 571–583. [Google Scholar] [CrossRef]
- Vohra, P.; Bugarel, M.; Turner, F.H.; Loneragan, G.; Hope, J.; Hopkins, J.P.; Stevens, M. Quantifying the survival of multiple Salmonella enterica Serovars in vivo via massively parallel whole-genome sequencing to predict zoonotic risk. Appl. Environ. Microbiol. 2018, 84, e02262-17. [Google Scholar] [CrossRef] [PubMed]
- Barthelmebs, L.; Calas-Blanchard, C.; Istamboulie, G.; Marty, J.-L.; Noguer, T. Biosensors as Analytical Tools in Food Fermentation Industry Bio-Farms for Nutraceuticals; Giardi, M.T., Rea, G., Berra, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Hayat, A.; Barthelmebs, L.; Marty, J.-L. Enzyme-linked immunosensor based on super paramagnetic nanobeads for easy and rapid detection of okadaic acid. Anal. Chim. Acta 2011, 690, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Pedrero, M.; García, J.; García, E.; García, P.; Pingarrón, J. Development of amperometric magnetogenosensors coupled to asymmetric PCR for the specific detection of Streptococcus pneumoniae. Anal. Bioanal. Chem. 2011, 399, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Lucarelli, F.; Tombelli, S.; Minunni, M.; Marrazza, G.; Mascini, M. Electrochemical and piezoelectric DNA biosensors for hybridisation detection. Anal. Chim. Acta 2008, 609, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Lopez de Alda, M.; Barceló, D. Biosensors as useful tools for environmental analysis and monitoring. Anal. Bioanal. Chem. 2006, 386, 1025–1041. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.; Fernández-Sánchez, C.; Jiménez-Jorquera, C. Ultramicroelectrode Array Based Sensors: A Promising Analytical Tool for Environmental Monitoring. Sensors 2010, 10, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.; Medlin, L.K. Electrochemical performance of a DNA-based sensor device for detecting toxic algae. Sens. Actuators B Chem. 2011, 153, 71–77. [Google Scholar] [CrossRef]
- Palchetti, I.; Mascini, M. Electroanalytical biosensors and their potential for food pathogen and toxin detection. Anal. Bioanal. Chem. 2008, 391, 455–471. [Google Scholar] [CrossRef]
- Setterington, E.B.; Alocilja, E.C. Electrochemical Biosensor for Rapid and Sensitive Detection of Magnetically Extracted Bacterial Pathogens. Biosensors 2012, 2, 15–31. [Google Scholar] [CrossRef]
- Uliana, C.V.; Tognolli, J.O.; Yamanaka, H. Application of Factorial Design Experiments to the Development of a Disposable Amperometric DNA Biosensor. Electroanalysis 2011, 23, 2607–2615. [Google Scholar] [CrossRef]
- Muhammad-Tahir, Z.; Alocilja, E.C. Fabrication of a disposable biosensor for Escherichia coli O157:H7 detection. Sens. J. IEEE 2003, 3, 345–351. [Google Scholar] [CrossRef]
- Luo, C.; Lei, Y.; Yan, L.; Yu, T.; Li, Q.; Zhang, D.; Ding, S.; Ju, H. A Rapid and Sensitive Aptamer-Based Electrochemical Biosensor for Direct Detection of Escherichia coli O111. Electroanalysis 2012, 24, 1186–1191. [Google Scholar] [CrossRef]
- Cho, I.-H.; Irudayaraj, J. In-situ immuno-gold nanoparticle network ELISA biosensors for pathogen detection. Int. J. Food Microbiol. 2013, 164, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Wu, W.; Lu, X.; Zeng, L. Lateral flow biosensor for DNA extraction-free detection of Salmonella based on aptamer mediated strand displacement amplification. Biosens. Bioelectron. 2014, 56, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-H.; Park, M.-K. Immunosensors combined with a light microscopic imaging system for rapid detection of Salmonella. Food Control 2016, 59, 780–786. [Google Scholar] [CrossRef]
- Lafleur, J.P.; Jönsson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E.; Stricker, S. Application of Electrochemical Biosensors for Detection of Food Pathogenic Bacteria. Electroanalysis 2000, 12, 317–325. [Google Scholar] [CrossRef]
- Lan, Y.-B.; Wang, S.-Z.; Yin, Y.-G.; Hoffmann, W.C.; Zheng, X.-Z. Using a Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Chicken Carcass. J. Bionic Eng. 2008, 5, 239–246. [Google Scholar] [CrossRef]
- Seo, K.H.; Brackett, R.E.; Hartman, N.F.; Campbell, D.P. Development of a Rapid Response Biosensor for Detection of Salmonella Typhimurium. J. Food Prot. 1999, 62, 431–437. [Google Scholar] [CrossRef]
- Meeusen, C.; Alocilja, E.C.; Osburn, W. Evaluation of a surface plasmon resonance biosensor in the detection of Salmonella spp. ASAE 2001. [Google Scholar] [CrossRef]
- Ko, S.; Grant, S.A. A novel FRET-based optical fiber biosensor for rapid detection of Salmonella Typhimurium. Biosens. Bioelectron. 2006, 21, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Abdelhaseib, M.U.; Singh, A.K.; Bailey, M.; Singh, M.; El-Khateib, T.; Bhunia, A.K. Fiber optic and light scattering sensors: Complimentary approaches to rapid detection of Salmonella enterica in food samples. Food Control 2016, 61, 135–145. [Google Scholar] [CrossRef]
- Kim, G.; Moon, J.-H.; Moh, C.-Y.; Lim, J.-G. A microfluidic nano-biosensor for the detection of pathogenic Salmonella. Biosens. Bioelectron. 2015, 67, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Viter, R.; Tereshchenko, A.; Smyntyna, V.; Ogorodniichuk, J.; Starodub, N.; Yakimova, R.; Khranovskyy, V.; Ramanavicius, A. Toward development of optical biosensors based on photoluminescence of TiO2 nanoparticles for the detection of Salmonella. Sens. Actuators B Chem. 2017, 252, 95–102. [Google Scholar] [CrossRef]
- Song, Y.; Li, W.; Duan, Y.; Li, Z.; Deng, L. Nicking enzyme-assisted biosensor for Salmonella Enteritidis detection based on fluorescence resonance energy transfer. Biosens. Bioelectron. 2014, 55, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.-H.; Bhunia, A.K. Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol. 2013, 33, 166–171. [Google Scholar] [CrossRef]
- Guler Gokce, Z.; Akalın, P.; Kok, F.N.; Sarac, A.S. Impedimetric DNA biosensor based on polyurethane/poly(m-anthranilic acid) nanofibers. Sens. Actuators B Chem. 2018, 254, 719–726. [Google Scholar] [CrossRef]
- Moongkarndi, P.; Rodpai, E.; Kanarat, S. Evaluation of an immunochromatographic assay for rapid detection of Salmonella enterica serovars Typhimurium and Enteritidis. J. Vet. Diagn. Investig. 2011, 23, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.; Stanker, L.H.; Young, C.R. Detection of Salmonella in poultry using a silicon chip-based biosensor. J. Biochem. Biophys. Methods 1999, 41, 61–67. [Google Scholar] [CrossRef]
- Mutreja, R.; Jariyal, M.; Pathania, P.; Sharma, A.; Sahoo, D.K.; Suri, C.R. Novel surface antigen based impedimetric immunosensor for detection of Salmonella Typhimurium in water and juice samples. Biosens. Bioelectron. 2016, 85, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Om, A.S.; Mun, J.H. Nano-particle enhanced impedimetric biosensor for detedtion of foodborne pathogens. J. Phys. Conf. Ser. 2007, 61, 555–559. [Google Scholar] [CrossRef]
- Pal, N.; Sharma, S.; Gupta, S. Sensitive and rapid detection of pathogenic bacteria in small volumes using impedance spectroscopy technique. Biosens. Bioelectron. 2016, 77, 270–276. [Google Scholar] [CrossRef]
- Prusak-Sochaczewski, E.; Luong, J.H.T.; Guilbault, G.G. Development of a piezoelectric immunosensor for the detection of Salmonella Typhimurium. Enzym. Microb. Technol. 1990, 12, 173–177. [Google Scholar] [CrossRef]
- König, B.; Grätzel, M. Detection of Viruses and Bacteria with Piezoelectric Immunosensors. Anal. Lett. 1993, 26, 1567–1585. [Google Scholar] [CrossRef]
- Ye, J.; Letcher, S.V.; Rand, A.G. Piezoelectric Biosensor for Detection of Salmonella Typhimurium. J. Food Sci. 1997, 62, 1067–1086. [Google Scholar] [CrossRef]
- Pathirana, S.T.; Barbaree, J.; Chin, B.A.; Hartell, M.G.; Neely, W.C.; Vodyanoy, V. Rapid and sensitive biosensor for Salmonella. Biosens. Bioelectron. 2000, 15, 135–141. [Google Scholar] [CrossRef]
- Su, X.-L.; Li, Y. A QCM immunosensor for Salmonella detection with simultaneous measurements of resonant frequency and motional resistance. Biosens. Bioelectron. 2005, 21, 840–848. [Google Scholar] [CrossRef]
- Salam, F.; Uludag, Y.; Tothill, I.E. Real-time and sensitive detection of Salmonella Typhimurium using an automated quartz crystal microbalance (QCM) instrument with nanoparticles amplification. Talanta 2013, 115, 761–767. [Google Scholar] [CrossRef]
- Roda, A.; Mirasoli, M.; Michelini, E.; Di Fusco, M.; Zangheri, M.; Cevenini, L.; Roda, B.; Simoni, P. Progress in chemical luminescence-based biosensors: A critical review. Biosens. Bioelectron. 2016, 76, 164–179. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. TrAC Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar] [CrossRef]
- Sajid, M.; Kawde, A.-N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2014. [Google Scholar] [CrossRef]
- Singh, J.; Sharma, S.; Nara, S. Evaluation of gold nanoparticle based lateral flow assays for diagnosis of Enterobacteriaceae members in food and water. Food Chem. 2015, 170, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.M.; Lee, S.Y. Optical Biosensors for the Detection of Pathogenic Microorganisms. Trends Biotechnol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Mak, W.C.; Beni, V.; Turner, A.P.F. Lateral-flow technology: From visual to instrumental. TrAC Trends Anal. Chem. 2016, 79, 297–305. [Google Scholar] [CrossRef]
- Kang, Y.; Feng, K.-J.; Chen, J.-W.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q.; Key, S. Electrochemical detection of thrombin by sandwich approach using antibody and aptamer. Bioelectrochemistry 2008, 73, 76–81. [Google Scholar] [CrossRef]
- Liao, J.C.; Mastali, M.; Li, Y.; Gau, V.; Suchard, M.A.; Babbitt, J.; Gornbein, J.; Landaw, E.M.; McCabe, E.R.B.; Churchill, B.M.; et al. Development of an Advanced Electrochemical DNA Biosensor for Bacterial Pathogen Detection. J. Mol. Diagn. 2007, 9, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.N.; Gal, D.; Cristler, N.; Banditrat, C.; Heller, A. Enzyme-Amplified Amperometric Sandwich Test for RNA and DNA. Anal. Chem. 2001, 74, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Paniel, N.; Baudart, J. Colorimetric and electrochemical genosensors for the detection of Escherichia coli DNA without amplification in seawater. Talanta 2013, 115, 133–142. [Google Scholar] [CrossRef]
- Tombelli, S.; Minunni, M.; Mascini, M. Aptamers-based assays for diagnostics, environmental and food analysis. Mol. Eng. 2007, 24, 191–200. [Google Scholar] [CrossRef]
- Barthelmebs, L.; Hayat, A.; Limiadi, A.W.; Marty, J.-L.; Noguer, T. Electrochemical DNA aptamer-based biosensor for OTA detection, using superparamagnetic nanoparticles. Sens. Actuators B Chem. 2011, 156, 932–937. [Google Scholar] [CrossRef]
- Hamula, C.L.A.; Zhang, H.; Li, F.; Wang, Z.; Chris Le, X.; Li, X.-F. Selection and analytical applications of aptamers binding microbial pathogens. TrAC Trends Anal. Chem. 2011, 30, 1587–1597. [Google Scholar] [CrossRef]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Seok Kim, Y.; Ahmad Raston, N.H.; Bock Gu, M. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Janagama, H.; Dwivedi, H.P.; Senthil Kumar, T.M.A.; Jaykus, L.-A.; Schefers, J.; Sreevatsan, S. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes 2009, 23, 20–28. [Google Scholar] [CrossRef]
- Pan, Q.; Zhang, X.; Wu, H.; He, P.; Wang, F.; Zhang, M.; Hu, J.; Xia, B.; Wu, J. Aptamers that preferentially bind type IVB pili and inhibit human monocytic-cell invasion by Salmonella enterica serovar Typhi. Antimicrob. Agents Chemother. 2005, 49, 4052–4060. [Google Scholar] [CrossRef]
- Bruno, J.G.; Kiel, J.L. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens. Bioelectron. 1999, 14, 457–464. [Google Scholar] [CrossRef]
- Labib, M.; Zamay, A.S.; Kolovskaya, O.S.; Reshetneva, I.T.; Zamay, G.S.; Kibbee, R.J.; Sattar, S.A.; Zamay, T.N.; Berezovski, M.V. Aptamer-Based Viability Impedimetric Sensor for Bacteria. Anal. Chem. 2012, 84, 8966–8969. [Google Scholar] [CrossRef]
- Labib, M.; Zamay, A.S.; Kolovskaya, O.S.; Reshetneva, I.T.; Zamay, G.S.; Kibbee, R.J.; Sattar, S.A.; Zamay, T.N.; Berezovski, M.V. Aptamer-Based Impedimetric Sensor for Bacterial Typing. Anal. Chem. 2012, 84, 8114–8117. [Google Scholar] [CrossRef]
- Dwivedi, H.; Smiley, R.D.; Jaykus, L.-A. Selection of DNA aptamers for capture and detection of Salmonella Typhimurium using a whole-cell SELEX approach in conjunction with cell sorting. Appl. Microbiol. Biotechnol. 2013, 97, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Chen, X.; Huang, Y.; Xia, Y.; Ma, X.; Wang, Z. Selection and Characterization of Aptamers against Salmonella Typhimurium Using Whole-Bacterium Systemic Evolution of Ligands by Exponential Enrichment (SELEX). J. Agric. Food Chem. 2013, 61, 3229–3234. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Kim, G.; Lee, S.; Park, S. Identification of Salmonella Typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J. Microbiol. Methods 2013, 95, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-C.; Baig, I.; Lee, S.-C.; Moon, J.-Y.; Yoon, M.-Y. Development of ssDNA Aptamers for the Sensitive Detection of Salmonella Typhimurium and Salmonella Enteritidis. Appl. Biochem. Biotechnol. 2014, 174, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.-Y.; Chon, J.-W.; Choi, I.-S.; Park, C.; Kim, D.-E.; Seo, K.-H. Development of RNA aptamers for detection of Salmonella Enteritidis. J. Microbiol. Methods 2012, 89, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, S.; Shahrokhian, S.; Nurmohammadi, F. Nanoporous gold as a suitable substrate for preparation of a new sensitive electrochemical aptasensor for detection of Salmonella Typhimurium. Sens. Actuators B Chem. 2018, 255, 1536–1544. [Google Scholar] [CrossRef]
- Dinshaw, I.J.; Muniandy, S.; Teh, S.J.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J. Electroanal. Chem. 2017, 806, 88–96. [Google Scholar] [CrossRef]
- Yang, M.; Peng, Z.; Ning, Y.; Chen, Y.; Zhou, Q.; Deng, L. Highly Specific and Cost-Efficient Detection of Salmonella Paratyphi a Combining Aptamers with Single-Walled Carbon Nanotubes. Sensors 2013, 13, 6865–6881. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-H.; Li, M.; Wang, Y.; Ouyang, H.-X.; Wang, L.; Li, C.-X.; Cao, Y.-C.; Meng, Q.-H.; Lu, J.-X. Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella Typhimurium. Nanoscale Res. Lett. 2012, 7, 1–7. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; Chamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens. Bioelectron. 2016. [Google Scholar] [CrossRef]
- Bagheryan, Z.; Raoof, J.-B.; Golabi, M.; Turner, A.P.F.; Beni, V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella Typhimurium in food sample. Biosens. Bioelectron. 2016, 80, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-X.; Ye, Z.-Z.; Si, C.-Y.; Ying, Y.-B. Application of Aptamer Based Biosensors for Detection of Pathogenic Microorganisms. Chin. J. Anal. Chem. 2012, 40, 634–642. [Google Scholar] [CrossRef]
- Yoo, S.M.; Kim, D.-K.; Lee, S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 2015, 132, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, S.P.; Wang, Y.; Irudayaraj, J. SERS driven cross-platform based multiplex pathogen detection. Sens. Actuators B Chem. 2011, 152, 183–190. [Google Scholar] [CrossRef]
- Duan, N.; Wu, S.; Zhu, C.; Ma, X.; Wang, Z.; Yu, Y.; Jiang, Y. Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella Typhimurium and Staphylococcus aureus. Anal. Chim. Acta 2012, 723, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Yu, Y.; Ma, X.; Xia, Y.; Chen, X.; Huang, Y.; Wang, Z. A dual-color flow cytometry protocol for the simultaneous detection of Vibrio parahaemolyticus and Salmonella Typhimurium using aptamer conjugated quantum dots as labels. Anal. Chim. Acta 2013, 804, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, Y.; Wu, S.; Duan, N.; Xu, B.; Wang, Z. Simultaneous detection of Staphylococcus aureus and Salmonella Typhimurium using multicolor time-resolved fluorescence nanoparticles as labels. Int. J. Food Microbiol. 2016, 237, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Jiang, Y.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Methods 2014, 98, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Ozalp, V.C.; Bayramoglu, G.; Erdem, Z.; Arica, M.Y. Pathogen detection in complex samples by quartz crystal microbalance sensor coupled to aptamer functionalized core–shell type magnetic separation. Anal. Chim. Acta 2014. [Google Scholar] [CrossRef] [PubMed]
- Bayraç, C.; Eyidoğan, F.; Avni Öktem, H. DNA aptamer-based colorimetric detection platform for Salmonella Enteritidis. Biosens. Bioelectron. 2017, 98, 22–28. [Google Scholar] [CrossRef]
- Zheng, M.; Jagota, A.; Strano, M.S.; Santos, A.P.; Barone, P.; Chou, S.G.; Diner, B.A.; Dresselhaus, M.S.; Mclean, R.S.; Onoa, G.B.; et al. Structure-Based Carbon Nanotube Sorting by Sequence-Dependent DNA Assembly. Science 2003, 302, 1545–1548. [Google Scholar] [CrossRef] [PubMed]





| Culture and Colony-Based Methods | Immunology Based Methods | Polymerase Chain Reaction | DNA Based Methods | |
|---|---|---|---|---|
| Advantages | Low coast Sensitivity Selectivity with chromogenic media | Fast Robust Specificity “Real time” analyses | Specific Sensitive Rapid Accuracy Detection of small amounts of target nucleic acid | Specific Sensitive Rapid Reusability Stability Detection of small amounts of target |
| Drawbacks | Labor intensiveness Time-consuming Low sensitivity Microbial contamination VBNC | Low sensitivity Low affinity of the antibody to the pathogen or other analyte Interference from contaminants | False negative PCR results No distinction between dead or alive cells | No distinction between dead or alive cells |
| Progress | Association with DNA, antibody, or biochemical-based methods | Association with other methods: Immunomagnetic separation on magnetic beads coupled with matrix-assisted laser desorption ionization time-of-flight mass spectrometry, combination of immunomagnetic separation with flow cytometry | Reverse Transcriptase PCR (RT-PCR) to distinguish live and dead cells Association with another method, the biosensors | Design of aptamers |
| Microorganism | Sample Matrix | Bioreceptor | Immobilization Method | Transducer | Limit of Detection | Analyze Time | Working Range | References |
|---|---|---|---|---|---|---|---|---|
| S. Typhimurium | Chicken carcass | Antibody to Common Structural Antigens (CSA-1) | Succinimidyl-6-(biotinamido) hexanoate (HS-LC-Bioin) | SPR | 106 CFU/mL | - | - | [135] |
| - | - | 107 CFU/mL | - | - | [137] | |||
| Chicken carcass wash fluid | Direct reductive amination | Integrated optic interferometer | Direct assay: 107 CFU/mL Sandwich assay: 105 CFU/mL | 10 min | - | [136] | ||
| S. Typhimurium | Phosphate buffered saline (PBS) Pork | Antibody to CSA-1 | Protein G | FRET | 103 cells/mL 105 cells/mL | 5 min | - | [138] |
| S. Typhimurium S. Enteritidis | Poultry | - Capture: rabbit polyclonal pAb-anti-Salmonella antibody - Reporter: rabbit pAb-3238 and mouse anti-S. Enteritidis mAb-2F11 | Sulfo- N-hydroxysuccinimide (NHS)-LC-Biotin | BARDOT (bacterial rapid detection using optical scattering technology) | 103 CFU/mL | 12 h | - | [139] |
| S. Typhimurium | Borate buffer & chicken extract | anti-Salmonella polyclonal antibodies | Covalent | Quantum dot nanoparticles | 103 CFU/mL | 30 min | 0 to 106 CFU/mL, | [140] |
| S. Typhimurium | phosphate buffer saline | Antibodies against Salmonella antigens | Glass/TiO2/anti-S-Ab | Titanium dioxide (TiO2) nanoparticles Photoluminescence | - | - | 103 to 105 cell/mL | [141] |
| S. Enteritidis | Water Milk | DNA | NHS | FRET | 102 CFU/mL 1.5 × 102 CFU/mL | 2 h | 102 to 3 × 103 CFU/mL 1.5 × 102 to 3 × 103 CFU/mL | [142] |
| PBS Shredded beef Chicken Turkey breast | - Capture: rabbit polyclonal pAb-anti-Salmonella antibody - Reporter: mouse monoclonal antibodies | Sulfo-NHS-LC-Biotin | Evanescent-based fiber optic sensor | 103 CFU/mL 107 to 108 CFU/mL after 18 h of enrichment | 2 h | - | [143] | |
| Salmonella spp. | Buffer | DNA | Covalent | EIS | - | - | 0.1 µM–10 µM | [144] |
| Salmonella | Chicken | Anti-Salmonella rabbit pAbs | Dithio-bis-succinimidyl propionate (DSP) | Immunosensors combined with light microscopic imaging system (LMIS) | 103 CFU/chicken | - | - | [132] |
| S. Choleraesuis | PBS Whole milk (Test yes/no) | - Capture: 5F11-B11 monoclonal antibody - Detection: 11D8-D4 monoclonal antibody | Capture antibody: deposition onto the LFA Colloidal gold particles: sodium citrate chemical reduction | LFA | 5 × 105 CFU/mL - | 20 h | - | [47] |
| S. Typhimurium S. Enteritidis | PBS Chicken (Test of specificity) | - Anti-Salmonella rabbit pAbs - Mouse anti–S. Typhimurium - Mouse anti–S. Enteritidis | Colloidal gold particles Mousse antibodies were applied onto the nitrocellulose membrane | LFA | 104 CFU/mL 106 CFU/mL 100% 100% | 5–15 min | - | [145] |
| S. Typhimurium | Buffer | Antibody to CSA-1 | Carbodiimide | Flow injection amperometry immunofiltration assay | 50 cells/mL | 35 min | 50–200 cells/mL | [46] |
| S. Typhimurium | Chicken carcass washing samples | - Monoclonal fluorescein isothiocyanate labeled anti-Salmonella antibody - Polyclonal rabbit anti-Salmonella antibody | Biotin | Potentiommetry | 119 CFU/mL | 15 min | - | [146] |
| S. Typhimurium | Water | Outer membrane porin protein (OmpD) | Carboxilated graphen-graphen oxide | Impedimetry | 10 CFU/mL | - | - | [147] |
| S. Enteritidis | Buffer Milk | Biotinylated rabbit anti-Salmonella polyclonal antibody | Neutravidin | 106 CFU/mL 104 CFU/mL (with nanoparticles) 105 CFU/mL (with nanoparticles) | 3 min | - | [148] | |
| S. Typhi | Buffer | Rabbit anti-Salmonella spp. polyclonal antibody | Covalent | 100 CFU/mL | 5 min | - | [149] | |
| S. Typhimurium | Buffer | Anti-Salmonella antibody | Polyethyleneimine | QCM | 105 CFU/mL | 5 h | 105 to 109 CFU/mL | [150] |
| Protein A | 106 CFU/mL | - | 106 to 108 CFU/mL | [151] | ||||
| Polyethylenimine-glutaraldehyde and dithiobissuccinimidylpropionate coupling | - | 25 min | 5.3 × 105 to 1.2 × 109 CFU/mL | [152] | ||||
| Polyvalent somatic O antibody of Salmonella spp. | Langmuir-Blodgett | AWD | 350+/−150 cells/mL | 100 s | 102 to 107 CFU/mL | [153] | ||
| Chicken breast | Antibody to CSA-1 | Protein A | QCM | 102 cells/mL (with anti-Salmonella-magnetic beads) | ΔF 105–108 cells/mL ΔR 106–108 cells/mL | [154] | ||
| PBS Chicken meat | Mouse monoclonal antibody against S. Typhimurium | EDC-NHS | 10–20 CFU/mL Validation: good sensitivity | 12 min | [155] |
| Optical | Lateral Flow Assays | Electrochemical | Mass Based | |
|---|---|---|---|---|
| Advantages | - Easy to use - High sensitivity | - Good reproducibility - Very low shelf life - Rapid - Portable - User-friendly - Less interferences - Adequate specificity | - User-friendly - Miniaturization | - High sensitivity - Portable - Rapid - Simple - Stable output |
| Drawbacks | - Pretreatment of sample may be required | - Poor quantitative discrimination - Reproducibility may vary from lot to lot - Low signal intensity - Pretreatment of sample may be required - Mostly qualitative or semi-quantitative | - Low selectivity | - Low sensitivity with liquid samples - Interference induces by nonspecific binding |
| Microorganism | Aptamers Name | Target for the SELEX | Aptamer Sequences (5′-3′) | Size (Base) | Kd | References |
|---|---|---|---|---|---|---|
| DNA Aptamers | ||||||
| S. Typhimurium | 33 | OMPs | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG | 40 | - | [172] |
| 45 | GAGGAAAGTCTATAGCAGAGGAGATGTGTGAACCGAGTAA | |||||
| 33 | OMPs | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG (from Joshi et al. [172]) | 40 | - | [182] | |
| - | [183] | |||||
| S8-7 | Whole cell | CTGATGTGTGGGTAGGTGTCGTTGATTTCTTCTGGTGGGG | 40 | 1.73 ± 0.54 μM | [177] | |
| ST2P | Whole cell | CAAAGATGAGTAGGAAAAGATATGTGCGTCTACCTCTTGACTAAT | 87 | 6.33 × 10−3 ± 0.58 × 10−3 µM | [178] | |
| C4 | Whole cell | ACGGGCGTGGGGGCAATGCCTGCTTGTAGGCTTCCCCTGTGCGCG | 45 | - | [179] | |
| S. Typhimurium | St1 | Whole cell | CCGATGTCCGTTAGGGCTCCTCCATAGAT | 29 | 0.530 ± 0.01 μM | [180] |
| S. Enteritidis | Se-1 | CACACCGGAAGGGATGCCACCTAAACCCC | 30 | 4.66 ± 0.35 μM | ||
| Se-2 | CACAGATGACGTCTGGCACATAATTAACAC | 30 | 3.83 ± 0.10 μM | |||
| S. Paratyphi A | Apt 22 | Whole cell | ATGGACGAATATCGTCTCCCAGTGAATTCAGTCGGACAGCG | 41 | 47 × 10−3 ± 3 × 10−3 µM | [184] |
| S. Typhimurium | A2 | - | CCAAAGGCTACGCGTTAACGTGGTGTTGG | 29 | - | [185] |
| S. Enteritidis | - | OMPs | TCGGCAACAAGGTCACCCGGAGAAGATCGGTGGTCAAACTGCATAGGTAGTCCAGAAGCCGAACAAGCTGAGGATGAAGAACAACGGCT | 89 | - | [131] |
| S. Typhi | - | IVB Pili | GGGAACAGUCCGAGCCUCACUGUUAUCCGAUAGCAGCGCGGGAUGAGGGUCAAUGCGUCAUAGGAUCCCGC | 71 | - | [102] |
| S. Enteritidis | SENT-9 | Whole cell | CTCCTCTGACTGTAACCACGCACAAAGGCTCGCGCATGGTGTGTACGTTCTTACAGAGGT | 60 | 7 × 10−3 µM | [176] |
| S. Typhimurium | STYP-3 | Whole cell | GAGTTAATCAATACAAGGCGGGAACATCCTTGGCGGTGC | 39 | 25 × 10−3 µM | [175] |
| - | OMPs | TTTGGTCCTTGTCTTATGTCCAGAATGCGAGGAAAGTCTATAGCAGAGGAGATGTGTGAACCGAGTAAATTTCTCCTACTGGGATAGGTGGATTAT (modified from Aptamer 45 of Joshi et al. [172]) | 96 | - | [186,187] | |
| RNA Aptamers | ||||||
| S. Typhi | S-PS8.4 | IVB pili | UCACUGUUAUCCGAUAGCAGCGCGGGAUGA | 30 | 8.56 × 10−3 µM | [173] |
| S. Enteritidis | S 25 | Whole cell | GGGUUCACUGCAGACUUGACGAAGCUUGAGAGAUGCCCCCUGAUGTGCAUUCUUGUUGUGUUGCGGCAAUGGAUCCACAUCTACGAAUUC | 90 | - | [181] |
| Microorganism | Sample Matrix | Aptamer Reference | Immobilization Method | Transducer | Limit of Detection | Analyze Time | Working Range | References |
|---|---|---|---|---|---|---|---|---|
| S. Typhimurium | Buffer | 33 from Joshi et al. [172] | Gold surface Thiolated aptamers | SPR | 30 CFU/mL | - | 104–109 CFU/mL | [189] |
| Unknown: obtained from Dr. Srinand Sreevatsan’s group | Gold nanoparticles thiolated aptamers | SERS | 102 CFU/mL | 45 min | 102–103 CFU/mL | [190] | ||
| S. Paratyphi A | City water | Apt22 | Free: DNAzyme | Chemiluminescence | 104 CFU/mL | - | 104–108 CFU/mL | [184] |
| S. Typhimurium | Buffer | 33 from Joshi et al. [172] | Avidin-biotin | Fluorescent | 5 CFU/mL | - | 101–105 CFU/mL | [191] |
| ST2P | 25 CFU/mL | - | 50–106 CFU/mL | [178] | ||||
| Buffer Shrimp samples (Validation) | Free: Flow cytometry | 5 × 103 CFU/mL | - | 3.8 × 104–3.8 × 107 CFU/mL | [192] | |||
| Buffer Water from Tai Lake (Validation) | 33 from Joshi et al. [172] | Streptavidin-biotin | Optical-UV | 7 CFU/mL | - | 50–106 CFU/mL | [7] | |
| Buffer | A2 | Adsorption | 105 CFU/mL | 20 min | - | [185] | ||
| Buffer Milk (Validation) | 33 from Joshi et al. [172] | Avidin-biotin | Fluorescent | 15 CFU/mL | - | 102–105 CFU/mL | [193] | |
| S. Enteritidis | Milk | - | Streptavidin-biotin | LFA | 101 CFU/mL | - | - | [131] |
| S. Typhi | Phosphate buffer | - | EDC-NHS-amine | Potentiometry | - | 60 s | 0.2–106 CFU/mL | [102] |
| S. Enteritidis | Buffer | SENT-9 | Self-assembled monolayer (SAM) | Impedimetry | 600 cells/mL | 10 min | 103–105 CFU/mL | [176] |
| S. Typhimurium | STYP-3 | - | [175] | |||||
| S. Typhimurium | Buffer Pork (Validation) | 33 from Joshi et al., [172] | Gold nanoparticles thiolated aptamers | 3 CFU/mL | - | 2.4–2.4 × 103 CFU/mL | [194] | |
| Buffer | 33 from Joshi et al., [172] | Self-assembled monolayer (SAM) | 1 CFU/mL | 40 min | 6.5 × 102 to 6.5 × 108 CFU/mL | [182] | ||
| Eggs | 6.5 × 103 to 6.5 × 107 CFU/mL | |||||||
| Buffer Apple Juice (Validation) | Aptamer 45 from Joshi et al., [172] with length modification | Covalent | 3 CFU/mL | - | 102–108 CFU/mL | [186] | ||
| EDC-NHS-amine | 6 CFU/mL | - | 101–108 CFU/mL | [187] | ||||
| S. Typhimurium | Milk | S8-7 from Dwivedi et al. [177] | Amine | QCM | 100 CFU/mL | 10 min | 100–4 × 104 CFU/mL | [195] |
| Buffer Chicken meat | 33 from Joshi et al., [172] | Thiolated aptamers – glutaraldehyde - rGO-CHI | DPV | 101 CFU/mL | - | 101 to 106 CFU/mL | [183] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paniel, N.; Noguer, T. Detection of Salmonella in Food Matrices, from Conventional Methods to Recent Aptamer-Sensing Technologies. Foods 2019, 8, 371. https://doi.org/10.3390/foods8090371
Paniel N, Noguer T. Detection of Salmonella in Food Matrices, from Conventional Methods to Recent Aptamer-Sensing Technologies. Foods. 2019; 8(9):371. https://doi.org/10.3390/foods8090371
Chicago/Turabian StylePaniel, Nathalie, and Thierry Noguer. 2019. "Detection of Salmonella in Food Matrices, from Conventional Methods to Recent Aptamer-Sensing Technologies" Foods 8, no. 9: 371. https://doi.org/10.3390/foods8090371
APA StylePaniel, N., & Noguer, T. (2019). Detection of Salmonella in Food Matrices, from Conventional Methods to Recent Aptamer-Sensing Technologies. Foods, 8(9), 371. https://doi.org/10.3390/foods8090371





