A Robust DNA Isolation Protocol from Filtered Commercial Olive Oil for PCR-Based Fingerprinting
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oil Samples and DNA Extraction
- One mL per sample was vigorously mixed for 5 min and transferred into a 2 mL microcentrifuge tube, then it was shaken with 500 µL of hexane for 5 min and centrifuged (15 min, 4 °C, 12 krpm).
- The oily supernatant was transferred to a fresh 2 mL tube and the pellet was mixed with 400 µL of lysis buffer (50 mM Tris pH 8.5, 1 mM EDTA, 0.5% v/v Tween 20).
- Samples were incubated at 48 °C for 1 h and then centrifuged (15 min, 4 °C, 12 krpm).
- The two aqueous phases from the oil/buffer mix and the pellet were transferred into separate 1.5 mL microcentrifuge tubes, being careful to avoid contact with the interface between oil and buffer.
- The pellet derived from treatment of the aqueous phase at point 4 was discarded.
- The three phases were transferred into separate 1.5 mL microcentrifuge tubes and the samples were gently mixed with 500 µL of cold isopropanol, held 30 min at −80 °C, and centrifuged again (30 min, 4 °C, 12 krpm).
- The pellet was washed by adding 500 µL of absolute ethanol, held 30 min at −80 °C and centrifuged (30 min, 4 °C, 14 krpm).
- The pellet was washed once more by adding 500 µL of 70% ethanol and centrifuged (30 min, 4 °C, 14 krpm).
- After removing the supernatant, the pellet was dried in vacuum pump for 30 min and finally resuspended in 50 µL of 1× TE buffer pH 8 (10 mM Tris, 1 mM EDTA).
2.2. SSRs and SNPs Assay
3. Results
3.1. Set Up of An Effective Protocol for DNA Extraction from Filtered Oils and EVOO Traceability
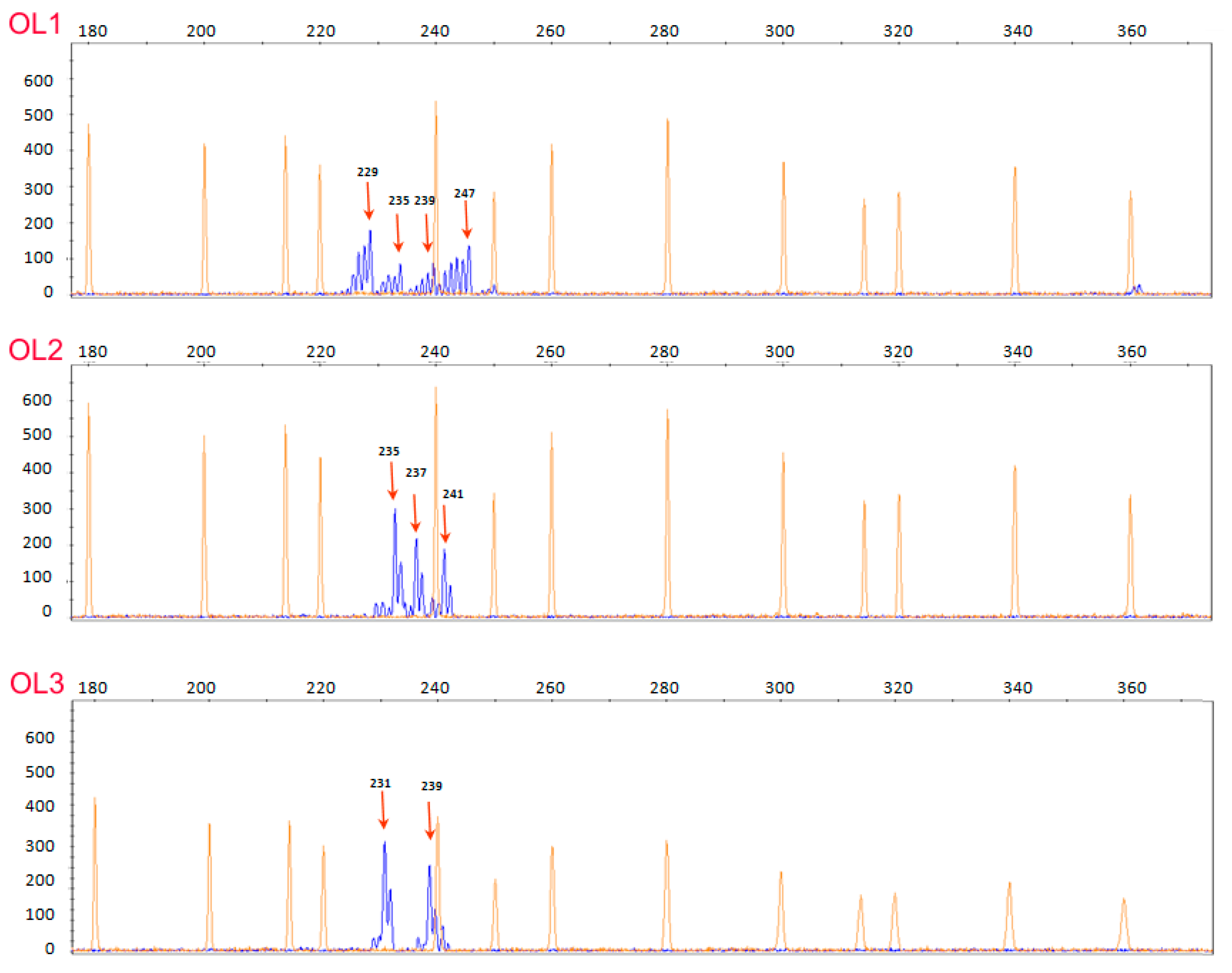
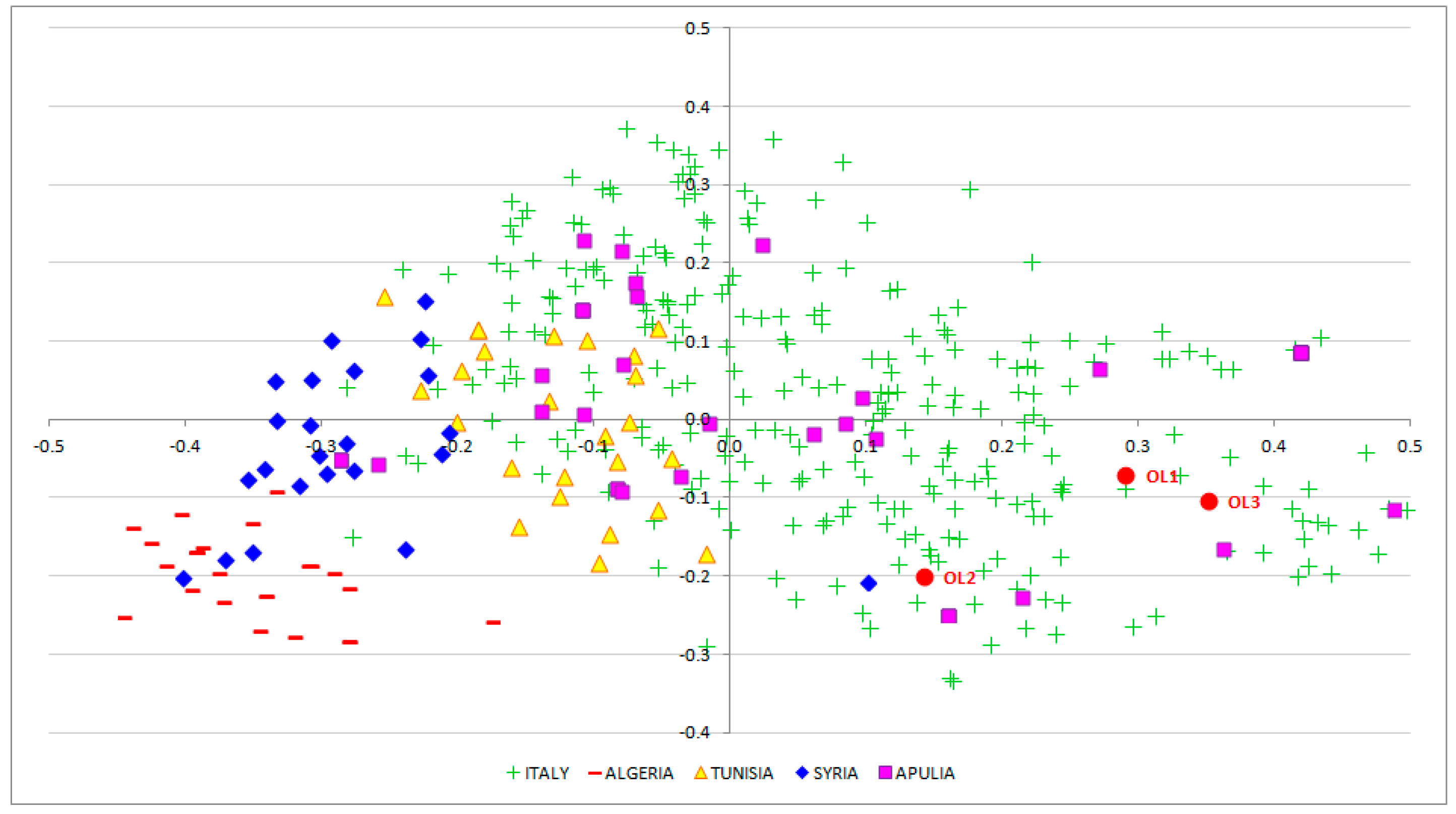
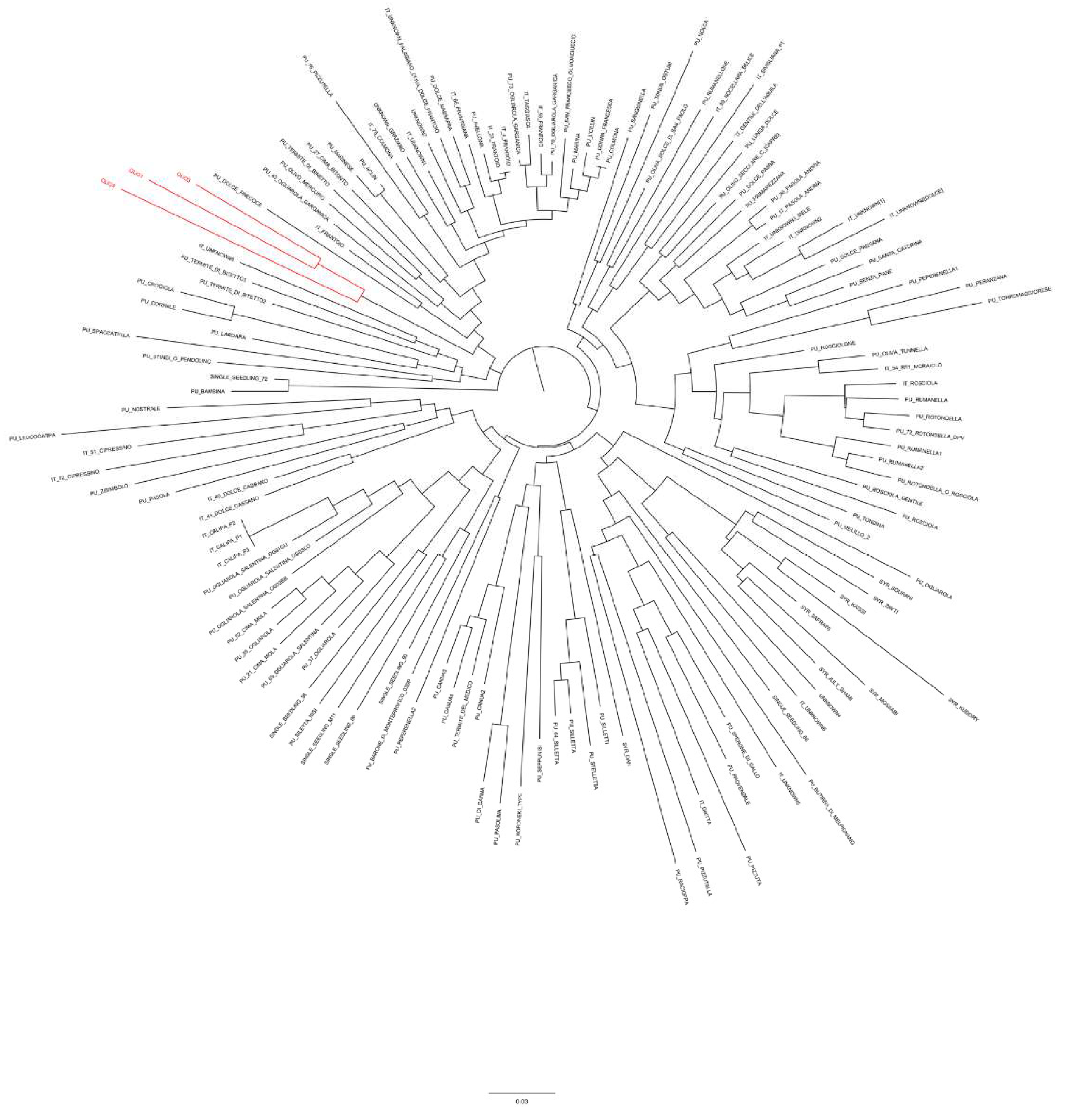
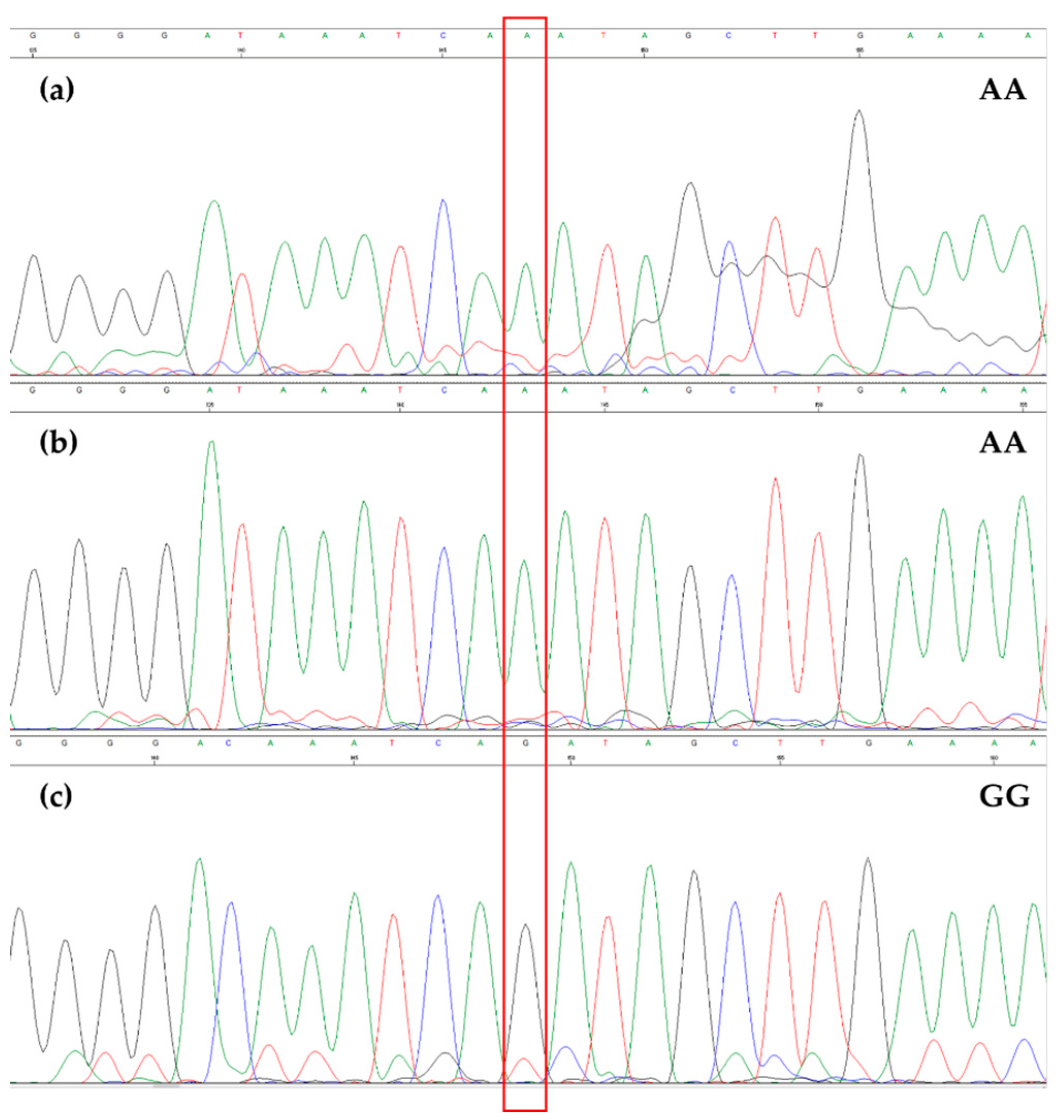
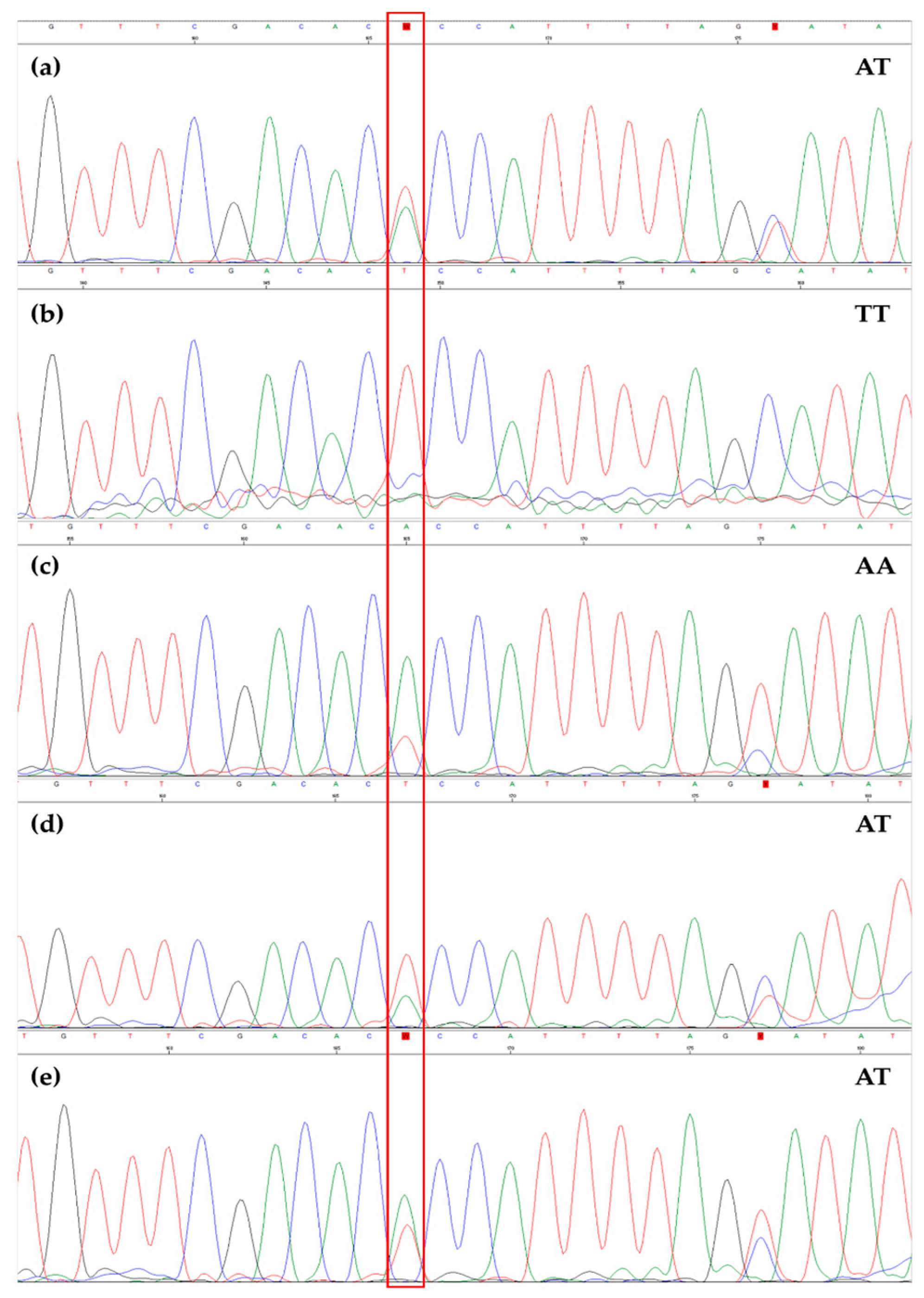
3.2. EVOO Genotyping and Traceability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Besnard, G.; Terral, J.F.; Cornille, A. On the origins and domestication of the olive: A review and perspectives. Ann. Bot. 2018, 121, 385–403. [Google Scholar] [CrossRef]
- Sardaro, R.; Bozzo, F.; Petrontino, A.; Fucilli, V. Community preferences in support of a conservation programme for olive landraces in the Mediterranean area. Acta Hort. 2018, 1199, 183–188. [Google Scholar] [CrossRef]
- International Olive Council. Available online: http://www.internationaloliveoil.org/ (accessed on 26 August 2019).
- Ismea–Istituto di Servizi per il Mercato Agricolo Alimentare. Available online: http://www.ismea.it/istituto-di-servizi-per-il-mercato-agricolo-alimentare (accessed on 26 August 2019).
- Martelli, G.P.; Boscia, D.; Porcelli, F.; Saponari, M. The olive quick decline syndrome in south-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016, 144, 235–243. [Google Scholar] [CrossRef]
- Tanasijevic, L.; Todorovic, M.; Pereira, L.S.; Pizzigalli, C.; Lionello, P. Impacts of climate change on olive crop evapotranspiration and irrigation requirements in the Mediterranean region. Agric. Water Manag. 2014, 144, 54–68. [Google Scholar] [CrossRef]
- Istat. Available online: https://www.istat.it/ (accessed on 8 August 2019).
- Likudis, Z. Olive Oils with Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI). In Products from Olive Tree; Boskou, D., Ed.; IntechOpen Publisher: London, UK, 2016; pp. 175–190. [Google Scholar]
- Bhandari, M.P.; Carmona, E.N.; Abbatangelo, M.; Sberveglieri, V.; Duina, G.; Malla, R.; Sberveglieri, G. Discrimination of Quality and Geographical Origin of Extra Virgin Olive Oil by S3 Device with Metal Oxides Gas Sensors. Multidiscip. Digit. Publ. Inst. Proc. 2018, 2, 1061. [Google Scholar] [CrossRef]
- Kalaitzis, P.; El-Zein, Z. Olive oil authentication, traceability and adulteration detection using DNA-based approaches. Lipid Technol. 2016, 28, 10–11. [Google Scholar] [CrossRef]
- Meenu, M.; Cai, Q.; Xu, B. A critical review on analytical techniques to detect adulteration of extra virgin olive oil Trends. Food Sci. Technol. 2019, 91, 391–408. [Google Scholar] [CrossRef]
- Xanthopoulou, A.; Ganopoulos, I.; Bosmali, I.; Tsaftaris, A.; Madesis, P. DNA fingerprinting as a novel tool for olive and olive oil authentication, traceability, and detection of functional compounds. In Olives and Olive Oil as Functional Foods: Bioactivity, Chemistry and Processing; Shahidi, F., Kiritsakis, A., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; pp. 587–601. [Google Scholar]
- Bazakos, C.; Spaniolas, S.; Kalaitzis, P. DNA-Based Approaches for Traceability and Authentication of Olive Oil. In Products from Olive Tree; Boskou, D., Ed.; IntechOpen Publisher: London, UK, 2016; pp. 115–133. [Google Scholar]
- Espiñeira, M.; Santaclara, F. Advances in Food Traceability Techniques and Technologies; Woodhead Publishing: Cambridge, UK, 2016. [Google Scholar]
- Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Savino, V.; Pollastro, S.; Colucci, F.; Miccolupo, A.; Blanco, A.; Pasqualone, A.; Montemurro, C. An enhanced analytical procedure to discover table grape DNA adulteration in industrial musts. Food Control 2016, 60, 124–130. [Google Scholar] [CrossRef]
- Pasqualone, A.; di Rienzo, V.; Nasti, R.; Blanco, A.; Gomes, T.; Montemurro, C. Traceability of Italian Protected Designation of Origin (PDO) Table Olives by Means of Microsatellite Molecular Markers. J. Agr. Food Chem. 2013, 61, 3068–3073. [Google Scholar] [CrossRef] [PubMed]
- Uncu, A.T.; Frary, A.; Doganlar, S. Cultivar Origin and Admixture Detection in Turkish Olive Oils by SNP-Based CAPS Assays. J. Agric. Food Chem. 2015, 63, 2284–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualone, A.; Montemurro, C.; di Rienzo, V.; Summo, C.; Paradiso, V.M.; Caponio, F. Evolution and perspectives of cultivar identification and traceability from tree to oil and table olives by means of DNA markers. J. Sci. Food Agric. 2016, 96, 3642–3657. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, C.; Miazzi, M.M.; Pasqualone, A.; Fanelli, V.; Sabetta, W.; di Rienzo, V. Traceability of PDO Olive Oil “Terra di Bari” Using High Resolution Melting. J. Chem. 2015, 205, 1–7. [Google Scholar] [CrossRef]
- Raieta, K.; Muccillo, L.; Colantuoni, V. A novel reliable method of DNA extraction from olive oil suitable for molecular traceability. Food Chem. 2015, 172, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ayed, R.; Grati-Kamoun, N.; Sans-Grout, C.; Moreau, F.; Rebai, A. Characterization and authenticity of virgin olive oil (Olea europaea L.) cultivars by microsatellite markers. Eur. Food Res. Technol. 2012, 234, 263–271. [Google Scholar] [CrossRef]
- Hrnčírová, Z.; Bergerová, E.; Siekel, P. Effects of technological treatment on DNA degradation in selected food matrices of plant origin. J. Food Nutr. Res. 2008, 47, 23–28. [Google Scholar]
- De la Torre, F.; Bautista, R.; Cánovas, F.M.; Claros, G. Isolation of DNA from olive oil and oil sediments: Application in oil fingerprinting. Food Agric. Environ. 2004, 2, 84–89. [Google Scholar]
- Montemurro, C.; Sabetta, W.; di Rienzo, V.; Pasqualone, A. Advances in DNA Extraction for Detecting Extra-Virgin Olive Oil Adulteration. In Authentication and Detection of Adulteration of Olive Oil; Kontominas, M., Ed.; Nova Science: New York, NY, USA, 2018; pp. 315–328. [Google Scholar]
- Ramos-Gómez, S.; Busto, M.D.; Perez-Mateos, M.; Ortega, N. Development of a method to recovery and amplification DNA by real-time PCR from commercial vegetable oils. Food Chem. 2014, 158, 374–383. [Google Scholar] [CrossRef]
- Martins-Lopes, P.; Gomes, S.; Santos, E.; Guedes-Pinto, H. DNA Markers for Portuguese Olive Oil Fingerprinting. J. Agr. Food Chem. 2008, 56, 11786–11791. [Google Scholar] [CrossRef]
- Giménez, M.J.; Pistón, F.; Martín, A.; Atienza, S.G. Application of real-time PCR on the development of molecular markers and to evaluate critical aspects for olive oil authenticatio. Food Chem. 2010, 118, 482–487. [Google Scholar]
- Gomes, S.; Breia, R.; Carvalho, T.; Carnide, V.; Martins-Lopes, P. Microsatellite High-Resolution Melting (SSR-HRM) to Track Olive Genotypes: From Field to Olive Oil. J. Food Sci. 2018, 8, 2415–2423. [Google Scholar] [CrossRef]
- Consolandi, C.; Palmieri, L.; Severgnini, M.; Maestri, E.; Marmiroli, N.; Agrimonti, C.; Baldoni, L.; Donini, P.; De Bellis, G.; Castiglioni, B. A procedure for olive oil traceability and authenticity: DNA extraction, multiplex PCR and LDR-universal array analysis. Eur. Food Res. Technol. 2008, 227, 1429–1438. [Google Scholar] [CrossRef]
- Muzzalupo, I.; Perri, E. Recovery and characterization of DNA from virgin olive oil. Eur. Food Res. Technol. 2002, 214, 528–531. [Google Scholar] [CrossRef]
- Busconi, M.; Foroni, C.; Corradi, M.; Bongiorni, C.; Cattapan, F.; Fogher, C. DNA extraction from olive oil and its use in the identification of the production cultivar. Food Chem. 2003, 83, 127–134. [Google Scholar] [CrossRef]
- Spadoni, A.; Sion, S.; Gadaleta, S.; Savoia, M.A.; Piarulli, L.; Fanelli, V.; di Rienzo, V.; Taranto, F.; Miazzi, M.M.; Montemurro, C.; et al. A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agr. Sci. Technol. 2019, 21, 1215–1226. [Google Scholar]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors–Occurrence, properties and removal. J. App. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, R.; James, C.M.; Tobutt, K.R. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol. Ecol. Notes 2002, 1, 346–348. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lopes, M.S.; Mendonça, D.; Rodrigues Dos Santos, M.; Laimer Da Câmara Machado, M.; Da Câmara Machado, A. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojeda, M.A.; et al. A consensus list of microsatellite markers for olive genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Alba, V.; Montemurro, C.; Sabetta, W.; Pasqualone, A.; Blanco, A. SSR-based identification key of cultivars of Olea europaea L. diffused in Southern-Italy. Sci. Hortic. 2009, 123, 11–16. [Google Scholar] [CrossRef]
- Sion, S.; Taranto, F.; Montemurro, C.; Mangini, G.; Camposeo, S.; Falco, V.; Gallo, A.; Mita, G.; Debbabi, O.S.; Amar, F.B.; et al. Genetic Characterization of Apulian Olive Germplasm as Potential Source in New Breeding Programs. Plants 2019, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, V.; Sion, S.; Taranto, F.; D’Agostino, N.; Montemurro, C.; Fanelli, V.; Sabetta, W.; Boucheffa, S.; Tamendjari, A.; Pasqualone, A.; et al. Genetic flow among olive populations within the Mediterranean basin. PeerJ 2018, 6, e5260. [Google Scholar] [CrossRef]
- Perrier, X.; Flori, A.; Bonnot, F. Data Analysis Methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C., Eds.; Science Publishers: Enfield, NH, USA, 2003; pp. 43–76. [Google Scholar]
- D’Agostino, N.; Taranto, F.; Camposeo, S.; Mangini, G.; Fanelli, V.; Gadaleta, S.; Miazzi, M.M.; Pavan, S.; di Rienzo, V.; Sabetta, W.; et al. GBS-derived SNP catalogue unveiled wide genetic variability and geographical relationships of Italian olive cultivars. Sci. Rep. 2018, 8, 15877. [Google Scholar]
- Pasqualone, A.; di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Caponio, F.; Montemurro, C. High resolution melting analysis of DNA microsatellites in olive pastes and virgin olive oils obtained by talc addition. Eur. J. Lipid Sci. Tech. 2015, 117, 2044–2048. [Google Scholar] [CrossRef]
- Pasqualone, A.; Montemurro, C.; Grinn-Gofron, A.; Sonnante, G.; Blanco, A. Detection of soft wheat in semolina and durum wheat bread by analysis of DNA microsatellites. J. Agr. Food Chem. 2007, 55, 3312–3318. [Google Scholar] [CrossRef] [PubMed]
- Pasqualone, A.; Alba, V.; Mangini, G.; Blanco, A.; Montemurro, C. Durum wheat cultivar traceability in PDO Altamura bread by analysis of DNA microsatellites. Eur. Food Res. Technol. 2010, 230, 723–729. [Google Scholar] [CrossRef]
- Agrimonti, C.; Vietina, M.; Pafundo, S.; Marmiroli, N. The use of food genomics to ensure the traceability of olive oil. Trends Food Sci. Technol. 2011, 22, 237–244. [Google Scholar] [CrossRef]
- Breton, C.; Claux, D.; Metton, I.; Skorski, G.; Bervilleä, A. Comparative Study of Methods for DNA Preparation from Olive Oil Samples to Identify Cultivar SSR Alleles in Commercial Oil Samples: Possible Forensic Applications. J. Agr. Food Chem. 2004, 52, 531–537. [Google Scholar] [CrossRef]
- Bracci, T.; Busconi, M.; Fogher, C.; Sebastiani, L. Molecular studies in olive (Olea europaea L.): Overview on DNA markers applications and recent advances in genome analysis. Plant Cell Rep. 2011, 30, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Vietina, M.; Agrimonti, C.; Marmiroli, N. Detection of plant oil DNA using high resolution melting (HRM) post PCR analysis: A tool for disclosure of olive oil adulteration. Food Chem. 2013, 14, 3820–3826. [Google Scholar] [CrossRef] [PubMed]
- Ganopoulos, I.; Bazakos, C.; Madesis, P.; Kalaitzis, P.; Tsaftaris, A. Barcode DNA high-resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J. Sci. Food Agric. 2013, 93, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Gomes, S.; Barrias, S.; Fernandes, J.R.; Martins-Lopes, P. Applying high-resolution melting (HRM) technology to olive oil and wine authenticity. Food Res. Int. 2018, 103, 170–181. [Google Scholar] [CrossRef] [PubMed]

| P1 | P2 | P3 | P4 | P5 | |
|---|---|---|---|---|---|
| Spadoni et al., 2019 [32] | Muzzalupo et al., 2002 [30] | Busconi et al., 2003 [31] | Consolandi et al., 2008 [29] | Consolandi et al., 2008 modified [29] | |
| Extraction time (hours) | ~6 h | ~30 h | ~6 h | ~30 h | ~4 h |
| Sample/tissue and amount * | 2 g leaves | 10 mL not filtered, clear oil | 50 mL not filtered, clear oil | 2 mL not filtered, clear oil | 1 mL filtered, clear oil |
| Starting centrifugation step | No | Yes | Yes | No | No |
| Main solvent | CTAB, phenol, chloroform | CTAB, dichloromethane, chloroform | CTAB, octanol, chloroform | Hexane, chloroform | Hexane, chloroform |
| Liquid nitrogen | No | No | Yes | No | No |
| Proteinase K | No | No | No | Yes | No |
| Pronase | No | Yes | No | No | No |
| SDS | Yes | No | No | No | No |
| RNAse treatment | No | Yes | Yes | No | No |
| β-mercaptoethanol | Yes | Yes | Yes | No | No |
| Extraction time (hours) | ~6 h | ~30 h | ~6 h | ~30 h | ~4 h |
| Variety | # SNP | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Frantoio | CC | GG | TC | GG | AT | GG | GG | AT |
| Taggiasca | CC | GT | TC | GG | AT | GC | GG | AT |
| Pendolino | CC | GG | TT | AG | TT | GC | AA | AA |
| Crastu | TT | TT | TT | AG | TT | GG | AA | AA |
| Leccino | CC | GG | TC | GG | TT | GG | AA | AT |
| Ogliarola barese | CC | GT | n.a. | GG | AT | CC | GG | AT |
| OL1 | CC | GT | TT | GG | TT | GG | AA | AT |
| OL2 | CC | GT | TT | n.a. | TT | GG | AA | AT |
| OL3 | CC | GT | TT | GG | TT | GG | GG | AT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piarulli, L.; Savoia, M.A.; Taranto, F.; D’Agostino, N.; Sardaro, R.; Girone, S.; Gadaleta, S.; Fucili, V.; De Giovanni, C.; Montemurro, C.; et al. A Robust DNA Isolation Protocol from Filtered Commercial Olive Oil for PCR-Based Fingerprinting. Foods 2019, 8, 462. https://doi.org/10.3390/foods8100462
Piarulli L, Savoia MA, Taranto F, D’Agostino N, Sardaro R, Girone S, Gadaleta S, Fucili V, De Giovanni C, Montemurro C, et al. A Robust DNA Isolation Protocol from Filtered Commercial Olive Oil for PCR-Based Fingerprinting. Foods. 2019; 8(10):462. https://doi.org/10.3390/foods8100462
Chicago/Turabian StylePiarulli, Luciana, Michele Antonio Savoia, Francesca Taranto, Nunzio D’Agostino, Ruggiero Sardaro, Stefania Girone, Susanna Gadaleta, Vincenzo Fucili, Claudio De Giovanni, Cinzia Montemurro, and et al. 2019. "A Robust DNA Isolation Protocol from Filtered Commercial Olive Oil for PCR-Based Fingerprinting" Foods 8, no. 10: 462. https://doi.org/10.3390/foods8100462







