Machine Learning Model Stability for Sub-Regional Classification of Barossa Valley Shiraz Wine Using A-TEEM Spectroscopy
Abstract
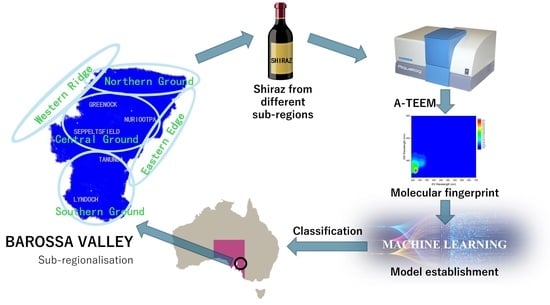
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Wine Samples
2.3. Sample Preparation and A-TEEM Procedure
2.4. Preparation of Wine Blends
2.5. Statistical Analysis
2.5.1. Data Fusion (Multi-Block Modelling)
2.5.2. Unsupervised Data Analysis
2.5.3. Data Analysis with Machine Learning
3. Results and Discussion
3.1. Molecular Fingerprints (EEMs)
3.2. PARAFAC Decomposition of EEMs
3.3. PCA Decomposition of A-TEEM Data
3.4. XGBDA Predictive Modelling
3.4.1. Vintage and Sub-Region Validation
3.4.2. Blended Wine Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castelló, E. The will for terroir: A communicative approach. J. Rural Stud. 2021, 86, 386–397. [Google Scholar] [CrossRef]
- Hill, R.A.D. ‘Le terroir, c’est la vie’: Re-animating a concept among Burgundy’s wine producers. Environ. Plan. E Nat. Space 2022, 5, 447–472. [Google Scholar] [CrossRef]
- Parker, T. Tasting French Terroir: The History of an Idea, 1st ed.; University of California Press: Berkeley, CA, USA, 2015. [Google Scholar]
- Gladstones, J. Wine, Terroir and Climate Change; Wakefield Press: Mile End, SA, Australia, 2011. [Google Scholar]
- Kustos, M.; Gambetta, J.M.; Jeffery, D.W.; Heymann, H.; Goodman, S.; Bastian, S.E.P. A matter of place: Sensory and chemical characterisation of fine Australian Chardonnay and Shiraz wines of provenance. Food Res. Int. 2020, 130, 108903. [Google Scholar] [CrossRef] [PubMed]
- Souza Gonzaga, L.; Capone, D.L.; Bastian, S.E.P.; Jeffery, D.W. Defining wine typicity: Sensory characterisation and consumer perspectives. Aust. J. Grape Wine Res. 2021, 27, 246–256. [Google Scholar] [CrossRef]
- Cramer, G.R.; Cochetel, N.; Ghan, R.; Destrac-Irvine, A.; Delrot, S. A sense of place: Transcriptomics identifies environmental signatures in Cabernet Sauvignon berry skins in the late stages of ripening. BMC Plant Biol. 2020, 20, 41. [Google Scholar] [CrossRef] [PubMed]
- Unwin, T. Terroir: At the heart of geography. In The Geography of Wine; Dougherty, P., Ed.; Springer: Dordrecht, The Netherlands, 2011; pp. 37–48. [Google Scholar]
- Charters, S.; Spielmann, N. Characteristics of strong territorial brands: The case of champagne. J. Bus. Res. 2014, 67, 1461–1467. [Google Scholar] [CrossRef]
- Barham, E. Translating terroir: The global challenge of French AOC labeling. J. Rural Stud. 2003, 19, 127–138. [Google Scholar] [CrossRef]
- Rosen, S. Institutional transformation: Supply or demand? Concluding comment. J. Inst. Theor. Econ. 1996, 152, 275–286. [Google Scholar]
- Meloni, G.; Swinnen, J. Trade and terroir. The political economy of the world’s first geographical indications. Food Policy 2018, 81, 1–20. [Google Scholar] [CrossRef]
- Schamel, G. Geography versus brands in a global wine market. Agribusiness 2006, 22, 363–374. [Google Scholar] [CrossRef]
- Robins, N. The Barossa Grounds—The journey so far. Aust. N. Z. Grapegrow. Winemak. 2014, 601, 42. [Google Scholar]
- Bramley, R.G.V.; Ouzman, J. Underpinning terroir with data: On what grounds might subregionalisation of the Barossa Zone geographical indication be justified? Aust. J. Grape Wine Res. 2022, 28, 196–207. [Google Scholar] [CrossRef]
- Thach, L.; Charters, S.; Cogan-Marie, L. Core tensions in luxury wine marketing: The case of Burgundian wineries. Int. J. Wine Bus. Res. 2018, 30, 343–365. [Google Scholar] [CrossRef]
- Anderson, K.; Nelgen, S.; Pinilla, V. Global Wine Markets: 1860 to 2016: A Statistical Compendium; University of Adelaide Press: Adelaide, SA, Australia, 2017. [Google Scholar]
- Guy, K.M. “Oiling the wheels of social life”: Myths and marketing in Champagne during the Belle Epoque. Fr. Hist. Stud. 1999, 22, 211–239. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, L. Wine fraud. Int. J. Wine Res. 2010, 2, 105–113. [Google Scholar] [CrossRef]
- Gabel, B. Wine origin authentication linked to terroir—Wine fingerprint. BIO Web Conf. 2019, 15, 02033. [Google Scholar] [CrossRef]
- Smith, M. Chinese police seize fake Penfolds haul. Aust. Financ. Rev. 2018. Available online: https://www.afr.com/world/asia/chinese-police-seize-50000-bottles-of-fake-penfolds-20180327-h0y19c (accessed on 17 March 2024).
- Wine Australia. The Blending Rules. Available online: https://www.wineaustralia.com/labelling/further-information/the-blending-rules (accessed on 17 March 2024).
- Ranaweera, R.K.R.; Gilmore, A.M.; Capone, D.L.; Bastian, S.E.P.; Jeffery, D.W. Authentication of the geographical origin of Australian Cabernet Sauvignon wines using spectrofluorometric and multi-element analyses with multivariate statistical modelling. Food Chem. 2021, 335, 127592. [Google Scholar] [CrossRef] [PubMed]
- Ranaweera, R.K.R.; Capone, D.L.; Bastian, S.E.P.; Cozzolino, D.; Jeffery, D.W. A review of wine authentication using spectroscopic approaches in combination with chemometrics. Molecules 2021, 26, 4334. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, A.; Akaji, S.; Csatorday, K. Spectroscopic analysis of red wines with A-TEEM molecular fingerprinting. Readout 2017, E49, 41–48. [Google Scholar]
- Gilmore, A.M.; Sui, Q.; Blair, B.; Pan, B.S. Accurate varietal classification and quantification of key quality compounds of grape extracts using the absorbance-transmittance fluorescence excitation emission matrix (A-TEEM) method and machine learning. OENO One 2022, 56, 107–115. [Google Scholar] [CrossRef]
- Ranaweera, R.K.R.; Gilmore, A.M.; Capone, D.L.; Bastian, S.E.P.; Jeffery, D.W. Spectrofluorometric analysis combined with machine learning for geographical and varietal authentication, and prediction of phenolic compound concentrations in red wine. Food Chem. 2021, 361, 130149. [Google Scholar] [CrossRef] [PubMed]
- Airado-Rodríguez, D.; Durán-Merás, I.; Galeano-Díaz, T.; Wold, J.P. Front-face fluorescence spectroscopy: A new tool for control in the wine industry. J. Food Compos. Anal. 2011, 24, 257–264. [Google Scholar] [CrossRef]
- Cabrera-Bañegil, M.; Hurtado-Sánchez, M.d.C.; Galeano-Díaz, T.; Durán-Merás, I. Front-face fluorescence spectroscopy combined with second-order multivariate algorithms for the quantification of polyphenols in red wine samples. Food Chem. 2017, 220, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Ranaweera, R.K.R.; Gilmore, A.M.; Bastian, S.E.P.; Capone, D.L.; Jeffery, D.W. Spectrofluorometric analysis to trace the molecular fingerprint of wine during the winemaking process and recognise the blending percentage of different varietal wines. OENO One 2022, 56, 189–196. [Google Scholar] [CrossRef]
- Ranaweera, R.K.R.; Bastian, S.E.P.; Gilmore, A.M.; Capone, D.L.; Jeffery, D.W. Absorbance-transmission and fluorescence excitation-emission matrix (A-TEEM) with multi-block data analysis and machine learning for accurate intraregional classification of Barossa Shiraz wine. Food Control 2023, 144, 109335. [Google Scholar] [CrossRef]
- Wu, Q.; Geng, T.; Yan, M.-L.; Peng, Z.-X.; Chen, Y.; Lv, Y.; Yin, X.-L.; Gu, H.-W. Geographical origin traceability and authenticity detection of Chinese red wines based on excitation-emission matrix fluorescence spectroscopy and chemometric methods. J. Food Compos. Anal. 2024, 125, 105763. [Google Scholar] [CrossRef]
- Gomes, S.; Castro, C.; Barrias, S.; Pereira, L.; Jorge, P.; Fernandes, J.R.; Martins-Lopes, P. Alternative SNP detection platforms, HRM and biosensors, for varietal identification in Vitis vinifera L. using F3H and LDOX genes. Sci. Rep. 2018, 8, 5850. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.J.; Guillou, C.; Martin, M.L.; Cabanis, M.T.; Tep, Y.; Aerny, J. Natural factors of isotope fractionation and the characterization of wines. J. Agric. Food Chem. 1988, 36, 316–322. [Google Scholar] [CrossRef]
- Bonello, F.; Cravero, M.; Dell’Oro, V.; Tsolakis, C.; Ciambotti, A. Wine traceability using chemical analysis, isotopic parameters, and sensory profiles. Beverages 2018, 4, 54. [Google Scholar] [CrossRef]
- Collins, C.; Petrie, P.; Bastian, S.; Bindon, K.; Bonada, M.; Boss, P.; Bramley, R.; Cavagnaro, T.; Danner, L.; Francis, L.; et al. Understanding the drivers of terroir in the Barossa. In Final Report to Wine Australia; 2022; pp. 1–279. Available online: https://www.wineaustralia.com/getmedia/07afbc68-8596-466b-95fd-b05899d673c7/UA-1602-Final-Report_Final.pdf (accessed on 17 March 2024).
- Ranaweera, R.K.R.; Gilmore, A.M.; Jeffery, D.W. Fluorescence spectroscopy for red wine authentication. In Wine Analysis and Testing Techniques; Pozo-Bayón, M.Á., Muñoz González, C., Eds.; Springer: New York, NY, USA, 2024; pp. 23–38. [Google Scholar]
- Lenhardt Acković, L.; Zeković, I.; Dramićanin, T.; Bro, R.; Dramićanin, M.D. Modeling food fluorescence with PARAFAC. In Reviews in Fluorescence 2017; Geddes, C.D., Ed.; Springer International Publishing AG: Cham, Switzerland, 2019; pp. 161–197. [Google Scholar]
- Bramley, R.G.V.; Ouzman, J.; Trought, M.C.T. Making sense of a sense of place: Precision viticulture approaches to the analysis of terroir at different scales. OENO One 2020, 54, 903–917. [Google Scholar] [CrossRef]
- Schueuermann, C.; Silcock, P.; Bremer, P. Front-face fluorescence spectroscopy in combination with parallel factor analysis for profiling of clonal and vineyard site differences in commercially produced Pinot Noir grape juices and wines. J. Food Compos. Anal. 2018, 66, 30–38. [Google Scholar] [CrossRef]
- Airado-Rodríguez, D.; Galeano-Díaz, T.; Durán-Merás, I.; Wold, J.P. Usefulness of fluorescence excitation−emission matrices in combination with PARAFAC, as fingerprints of red wines. J. Agric. Food Chem. 2009, 57, 1711–1720. [Google Scholar] [CrossRef] [PubMed]
- Díaz, T.G.; Durán Merás, I.; Airado Rodríguez, D. Determination of resveratrol in wine by photochemically induced second-derivative fluorescence coupled with liquid-liquid extraction. Anal. Bioanal. Chem. 2007, 387, 1999–2007. [Google Scholar] [CrossRef]
- Szukay, B.; Gałęcki, K.; Kowalska-Baron, A.; Przybyt, M.; Saletnik, Ł.; Budzyński, J.; Fisz, J.J. Application of steady-state and time-resolved fluorescence spectroscopy in identification of cold-pressed vegetal oils. Food Anal. Methods 2023, 16, 1571–1582. [Google Scholar] [CrossRef]
- Barossa Australia. Barossa Vintage Reports. Available online: https://barossawine.com/barossa-vintages/barossa-vintage-reports/ (accessed on 17 March 2024).






| Vintage | ||||||
| 2018 | 2021 | |||||
| Sample | Actual | Predicted | Relative Difference | Actual | Predicted | Relative Difference |
| S1 | 95% | 97.30% | 2.4% | 5% | 1.30% | −74% |
| S2 | 95% | 97.80% | 2.9% | 5% | 1.10% | −78% |
| S3 | 90% | 89.10% | −1.0% | 10% | 6.30% | −37% |
| S4 | 90% | 90.80% | 0.9% | 10% | 4.90% | −51% |
| Sub-Region | ||||||
| Southern Grounds (SG) | Western Ridge (WR) | |||||
| Sample | Actual | Predicted | Relative Difference | Actual | Predicted | Relative Difference |
| S1 | 85% | 88.50% | 4.1% | 15% | 6.20% | −59% |
| S2 | 85% | 6.90% | −91.9% | 15% | 1.10% | −93% |
| S3 | 50% | 41.00% | −18.0% | 50% | 32.20% | −36% |
| S4 | 50% | 58.60% | 17.2% | 50% | 34.50% | −31% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Jeffery, D.W. Machine Learning Model Stability for Sub-Regional Classification of Barossa Valley Shiraz Wine Using A-TEEM Spectroscopy. Foods 2024, 13, 1376. https://doi.org/10.3390/foods13091376
Wang H, Jeffery DW. Machine Learning Model Stability for Sub-Regional Classification of Barossa Valley Shiraz Wine Using A-TEEM Spectroscopy. Foods. 2024; 13(9):1376. https://doi.org/10.3390/foods13091376
Chicago/Turabian StyleWang, Han, and David W. Jeffery. 2024. "Machine Learning Model Stability for Sub-Regional Classification of Barossa Valley Shiraz Wine Using A-TEEM Spectroscopy" Foods 13, no. 9: 1376. https://doi.org/10.3390/foods13091376