Hyperspectral Imaging and Machine Learning as a Nondestructive Method for Proso Millet Seed Detection and Classification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Imaging Data Acquisition Procedure
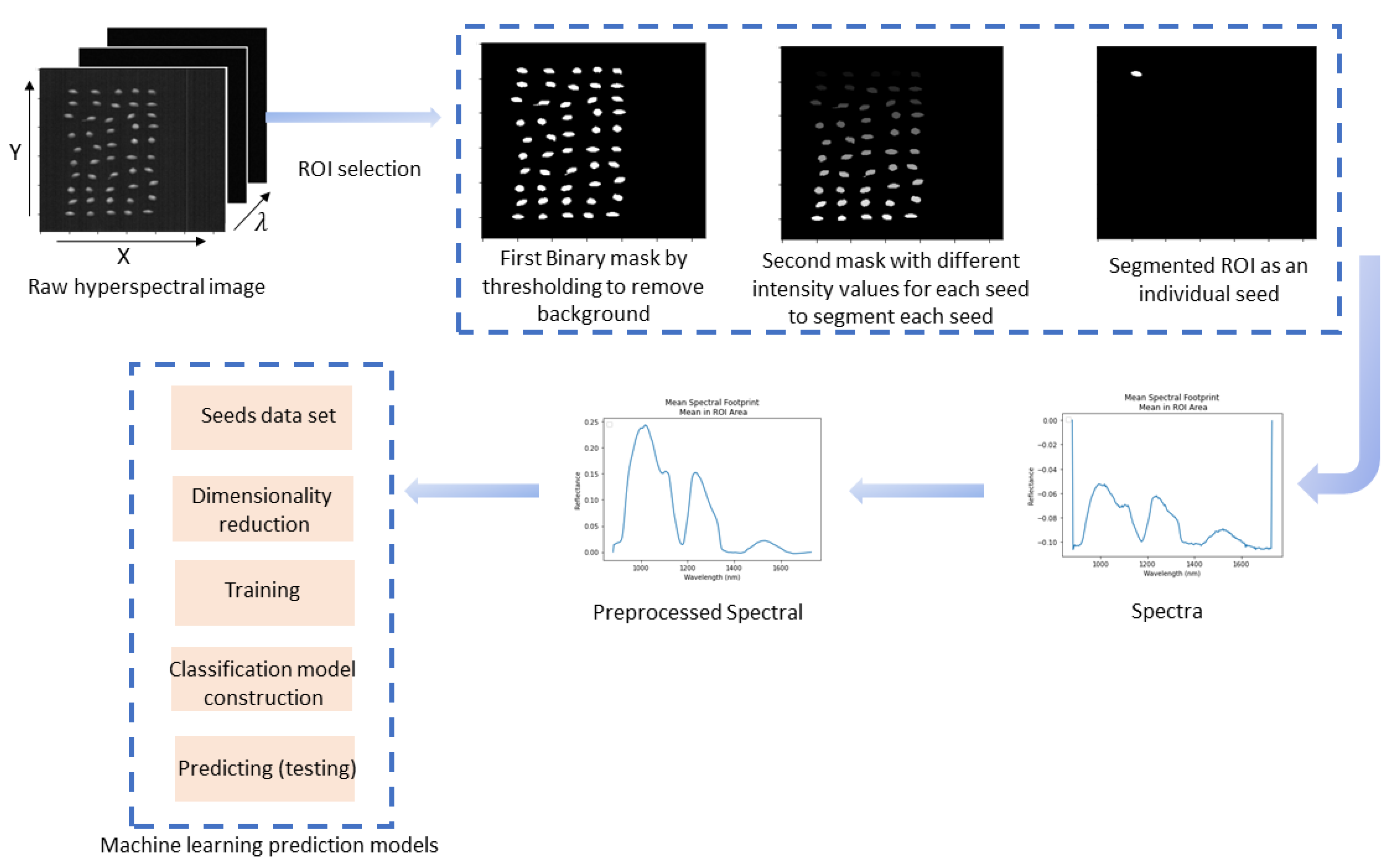
2.3. HSI Data Preprocessing
2.4. Data Analysis
2.5. Spectral Feature Extraction and Preprocessing
2.6. Classification Models
3. Results and Discussion
3.1. Spectral Characteristics of Proso Millet Seeds
3.2. PCA and Preprocessing
3.3. Machine Learning Classifiers for Millet Cultivars
3.4. Optimal Wavelength Selection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Wan, Z.; Perry, L.; Lu, H.; Wang, Q.; Zhao, C.; Li, J.; Xie, F.; Yu, J.; Cui, T.; et al. Early millet use in northern China. Proc. Natl. Acad. Sci. USA 2012, 109, 3726–3730. [Google Scholar] [CrossRef]
- Singh, P.; Boote, K.; Kadiyala, M.; Nedumaran, S.; Gupta, S.; Srinivas, K.; Bantilan, M. An assessment of yield gains under climate change due to genetic modification of pearl millet. Sci. Total Environ. 2017, 601, 1226–1237. [Google Scholar] [CrossRef]
- Woomer, J.; Singh, M.; Vijayakumar, P.P.; Adedeji, A. Physical properties and organoleptic evaluation of gluten-free bread from proso millet. Br. Food J. 2020, 122, 547–560. [Google Scholar] [CrossRef]
- Baltensperger, D. Progress with proso, pearl and other millets. Trends new Crop and new uses. In Proceedings of the fifth National Symposium, Atlanta, GA, USA, 10–13 November 2001. [Google Scholar]
- Kalinova, J.; Moudry, J. Content and quality of protein in proso millet (Panicum miliaceum L.) varieties. Plant Foods Hum. Nutr. 2006, 61, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, H.; Wu, M.; Yi, H. Enhanced drought tolerance of foxtail millet seedlings by sulfur dioxide fumigation. Ecotoxicol. Environ. Saf. 2019, 178, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Niranjan, B.N.; Prakasha, H.C.; Meena, H.M.; Kiran, S.K.; Yadawad, B. Effect of biochar on growth, yield and nutrient uptake by finger millet in acidic soil. J. Pharmacogn. Phytochem. 2020, 9, 1061–1067. [Google Scholar]
- Amadou, I.; Gounga, M.E.; Le, G.-W. Millets: Nutritional composition, some health benefits and processing—A review. Emir. J. Food Agric. 2013, 25, 501–508. [Google Scholar] [CrossRef]
- Fuller, D. A Millet Atlas—Some Identification Guidance; Institute of Archaeology, University College London: London, UK, 2008. [Google Scholar]
- USDA-NASS, National Agricultural Statistics Service. Acreage—1 June 2023. Available online: https://www.nass.usda.gov/Statistics_by_State/Colorado/Publications/News_Releases/2023/CO-June-Acreage-06302023.pdf (accessed on 12 April 2024).
- Zhang, X.; Liu, F.; He, Y.; Li, X. Application of hyperspectral imaging and chemometric calibrations for variety discrimination of maize seeds. Sensors 2012, 12, 17234–17246. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Adedeji, A.; Santra, D. Physico-Chemical and Functional Properties of Nine Proso Millet Cultivars. Trans. ASABE 2018, 61, 1165–1174. [Google Scholar] [CrossRef]
- Caporaso, N.; Whitworth, M.B.; Fisk, I.D. Near-Infrared spectroscopy and hyperspectral imaging for non-destructive quality assessment of cereal grains. Appl. Spectrosc. Rev. 2018, 53, 667–687. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Zheng, D.; Wang, W. Nondestructive identification of millet varieties using hyperspectral imaging technology. J. Appl. Spectrosc. 2020, 87, 54–61. [Google Scholar] [CrossRef]
- Zhu, S.; Chao, M.; Zhang, J.; Xu, X.; Song, P.; Zhang, J.; Huang, Z. Identification of soybean seed varieties based on hyperspectral imaging technology. Sensors 2019, 19, 5225. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ren, X.; Zhang, Q.; Diao, X.; Shen, Q. Determination of protein, total carbohydrates and crude fat contents of foxtail millet using effective wavelengths in NIR spectroscopy. J. Cereal Sci. 2013, 58, 241–247. [Google Scholar] [CrossRef]
- Wu, C.; Kong, L.; Wang, S.; Guo, Y. Near infrared spectroscopy (NIRS) technology applied in millet feature extraction and variety identification. Afr. J. Agric. Res. 2017, 12, 2223–2231. [Google Scholar] [CrossRef]
- Baath, G.S.; Flynn, K.C.; Gowda, P.H.; Kakani, V.G.; Northup, B.K. Detecting biophysical characteristics and nitrogen status of finger millet at hyperspectral and multispectral resolutions. Front. Agron. 2021, 2, 38. [Google Scholar] [CrossRef]
- Venkatesan, S.; Sujatha, K. Characterization of barnyard millet cultivars using seed image analysis. Seed Res. 2018, 45, 1–3. [Google Scholar]
- ElMasry, G.; Sun, D.-W. Principles of hyperspectral imaging technology. In Hyperspectral Imaging for Food Quality Analysis and Control; Elsevier: Amsterdam, The Netherlands, 2010; pp. 3–43. [Google Scholar]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, Y.; Wang, D.; Wu, Q.; Zhang, D.; Wang, C. The identification of single soybean seed variety by laser light backscattering imaging. Sens. Lett. 2012, 10, 399–404. [Google Scholar] [CrossRef]
- Dong, G.; Guo, J.; Wang, C.; Chen, Z.-L.; Zheng, L.; Zhu, D.-Z. The classification of wheat varieties based on near infrared hyperspectral imaging and information fusion. Guang Pu Xue Yu Guang Pu Fen Xi = Guang Pu 2015, 35, 3369–3374. [Google Scholar]
- Wu, N.; Zhang, Y.; Na, R.; Mi, C.; Zhu, S.; He, Y.; Zhang, C. Variety identification of oat seeds using hyperspectral imaging: Investigating the representation ability of deep convolutional neural network. RSC Adv. 2019, 9, 12635–12644. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, W.-Z.; Lu, J.-F.; Qiu, Z.-J.; He, Y. Study on visual identification of corn seeds based on hyperspectral imaging technology. Guang Pu Xue Yu Guang Pu Fen Xi = Guang Pu 2016, 36, 511–514. [Google Scholar]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety identification of single rice seed using hyperspectral imaging combined with convolutional neural network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Song, S. Origin identification of foxtail millet (Setaria italica) by using green spectral imaging coupled with chemometrics. Infrared Phys. Technol. 2022, 123, 104179. [Google Scholar] [CrossRef]
- Xia, C.; Yang, S.; Huang, M.; Zhu, Q.; Guo, Y.; Qin, J. Maize seed classification using hyperspectral image coupled with multi-linear discriminant analysis. Infrared Phys. Technol. 2019, 103, 103077. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Liu, F.; Nie, P.; He, Y. Rice seed cultivar identification using near-infrared hyperspectral imaging and multivariate data analysis. Sensors 2013, 13, 8916–8927. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Huang, W.; Fan, S.; Zhao, F.; Liang, D.; Tian, X. Non-destructive discrimination of the variety of sweet maize seeds based on hyperspectral image coupled with wavelength selection algorithm. Infrared Phys. Technol. 2020, 109, 103418. [Google Scholar] [CrossRef]
- Ekramirad, N.; Khaled, A.Y.; Doyle, L.E.; Loeb, J.R.; Donohue, K.D.; Villanueva, R.T.; Adedeji, A.A. Nondestructive Detection of Codling Moth Infestation in Apples Using Pixel-Based NIR Hyperspectral Imaging with Machine Learning and Feature Selection. Foods 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, J.; Wang, Q.; Fan, S.; Huang, W.; Zhao, C. A multi-region combined model for non-destructive prediction of soluble solids content in apple, based on brightness grade segmentation of hyperspectral imaging. Biosyst. Eng. 2019, 183, 110–120. [Google Scholar] [CrossRef]
- An, D.; Zhang, L.; Liu, Z.; Liu, J.; Wei, Y. Advances in infrared spectroscopy and hyperspectral imaging combined with artificial intelligence for the detection of cereals quality. Crit. Rev. Food Sci. Nutr. 2023, 63, 9766–9796. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Liu, Y.; Cui, Q.; Ning, J.; Zhang, Z. Enhanced quality monitoring during black tea processing by the fusion of NIRS and computer vision. J. Food Eng. 2021, 304, 110599. [Google Scholar] [CrossRef]
- Esteban-Dıez, I.; González-Sáiz, J.; Pizarro, C. An evaluation of orthogonal signal correction methods for the characterisation of arabica and robusta coffee varieties by NIRS. Anal. Chim. Acta 2004, 514, 57–67. [Google Scholar] [CrossRef]
- Workman, J., Jr.; Weyer, L. Practical Guide to Interpretive Near-Infrared Spectroscopy; Chu, X., Xu, Y., Tian, G., Eds.; Chemical Industry Press: Beijing, China, 2009. [Google Scholar]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Bujang, S.D.A.; Selamat, A.; Ibrahim, R.; Krejcar, O.; Herrera-Viedma, E.; Fujita, H.; Ghani, N.A.M. Multiclass Prediction Model for Student Grade Prediction Using Machine Learning. IEEE Access 2021, 9, 95608–95621. [Google Scholar] [CrossRef]
- Pang, S.; Yu, Z.; Orgun, M.A. A novel end-to-end classifier using domain transferred deep convolutional neural networks for biomedical images. Comput. Methods Programs Biomed. 2017, 140, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Baek, I.; Kim, M.S.; Cho, B.-K.; Mo, C.; Barnaby, J.Y.; McClung, A.M.; Oh, M. Selection of Optimal Hyperspectral Wavebands for Detection of Discolored, Diseased Rice Seeds. Appl. Sci. 2019, 9, 1027. [Google Scholar] [CrossRef]
- Moghimi, A.; Yang, C.; Marchetto, P.M. Ensemble Feature Selection for Plant Phenotyping: A Journey From Hyperspectral to Multispectral Imaging. IEEE Access 2018, 6, 56870–56884. [Google Scholar] [CrossRef]



| Classifier 1 | Full-Band Data * | PCA | ||
|---|---|---|---|---|
| Training Set | Prediction Set | Training Set | Prediction Set | |
| LDA | 100 ± 0.00 | 99.28 ± 0.20 | 82.20 ± 0.42 | 81.64 ± 1.23 |
| SVM | 69.40 ± 0.38 | 68.62 ± 1.42 | 69.38 ± 0.46 | 68.62 ± 1.42 |
| kNN | 95.88 ± 0.13 | 93.82 ± 0.46 | 95.06 ± 0.12 | 92.74 ± 0.45 |
| RF | 100 ± 0.00 | 98.92 ± 0.33 | 100 ± 0.00 | 98.90 ± 0.17 |
| Gradient tree boosting | 100 ± 0.00 | 99.46 ± 0.10 | 100 ± 0.00 | 99.16 ± 0.30 |
| Classifier | Cultivar | Precision | Recall | F1-Score |
|---|---|---|---|---|
| RF | Cerise | 1.00 | 1.00 | 1.00 |
| Cope | 1.00 | 1.00 | 1.00 | |
| Earlybird | 1.00 | 1.00 | 1.00 | |
| Huntsman | 0.98 | 0.99 | 0.99 | |
| Minco | 1.00 | 1.00 | 1.00 | |
| Plateau | 1.00 | 0.98 | 0.99 | |
| Rise | 0.99 | 0.98 | 0.99 | |
| Snowbird | 1.00 | 0.98 | 0.99 | |
| Sunrise | 0.96 | 0.98 | 0.97 | |
| Sunup | 0.98 | 1.00 | 0.99 | |
| Gradient tree boosting | Cerise | 0.99 | 1.00 | 0.99 |
| Cope | 1.00 | 1.00 | 1.00 | |
| Earlybird | 1.00 | 1.00 | 1.00 | |
| Huntsman | 0.99 | 0.99 | 0.99 | |
| Minco | 1.00 | 1.00 | 1.00 | |
| Plateau | 0.99 | 0.99 | 0.99 | |
| Rise | 1.00 | 0.99 | 1.00 | |
| Snowbird | 1.00 | 0.99 | 1.00 | |
| Sunrise | 1.00 | 0.98 | 0.99 | |
| Sunup | 0.97 | 1.00 | 0.99 |
| Classifier | No. of Features | Wavebands (nm) | Classification Accuracy |
|---|---|---|---|
| Gradient tree boosting | 30 | 900.17, 903.53, 906.88, 910.24, 913.59, 916.95, 920.30, 923.65, 927.01, 930.36, 933.71, 937.07, 940.42, 943.77, 947.13, 950.48, 953.83, 957.18, 960.53, 963.89, 967.24, 970.59, 973.94, 977.29, 980.64, 983.99, 1004.09, 1540.94, 1673.58 | 98.00% |
| 15 | 900.17, 903.53, 906.88, 910.24, 913.59, 916.95, 920.30, 923.65, 927.01, 930.36, 933.71, 1004.09, 1540.94, 1673.58 | 98.14% | |
| 5 | 900.17, 903.53, 1004.09, 1540.94, 1673.58 | 97.60% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekramirad, N.; Doyle, L.; Loeb, J.; Santra, D.; Adedeji, A.A. Hyperspectral Imaging and Machine Learning as a Nondestructive Method for Proso Millet Seed Detection and Classification. Foods 2024, 13, 1330. https://doi.org/10.3390/foods13091330
Ekramirad N, Doyle L, Loeb J, Santra D, Adedeji AA. Hyperspectral Imaging and Machine Learning as a Nondestructive Method for Proso Millet Seed Detection and Classification. Foods. 2024; 13(9):1330. https://doi.org/10.3390/foods13091330
Chicago/Turabian StyleEkramirad, Nader, Lauren Doyle, Julia Loeb, Dipak Santra, and Akinbode A. Adedeji. 2024. "Hyperspectral Imaging and Machine Learning as a Nondestructive Method for Proso Millet Seed Detection and Classification" Foods 13, no. 9: 1330. https://doi.org/10.3390/foods13091330





