Microbial Diversity and Community Structure of Chinese Fresh Beef during Cold Storage and Their Correlations with Off-Flavors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. DNA Extraction and Sequencing
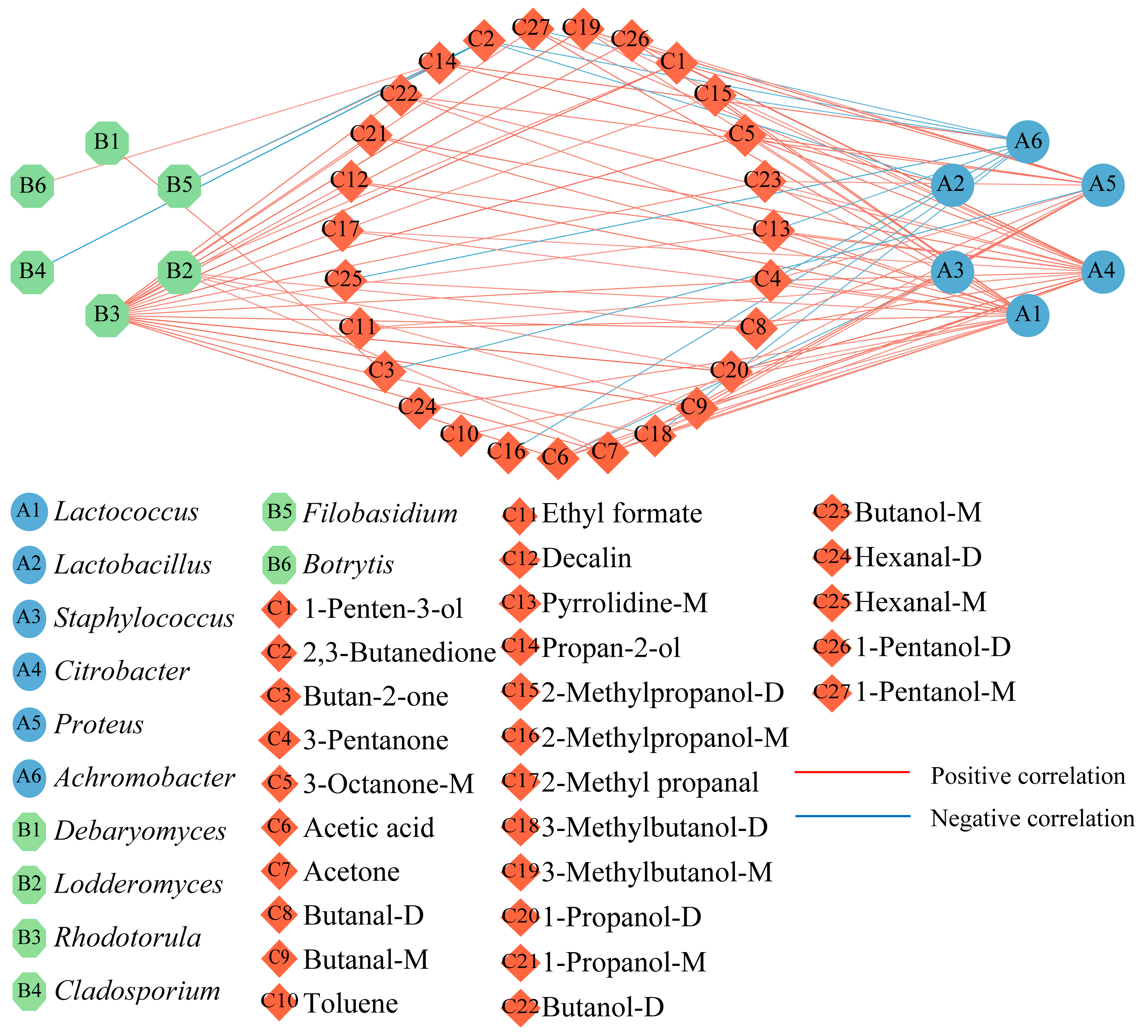
2.3. Correlation between the Core Microbial Genera and Differential Off-Flavor Substances
2.4. Data Processing
3. Results and Discussion
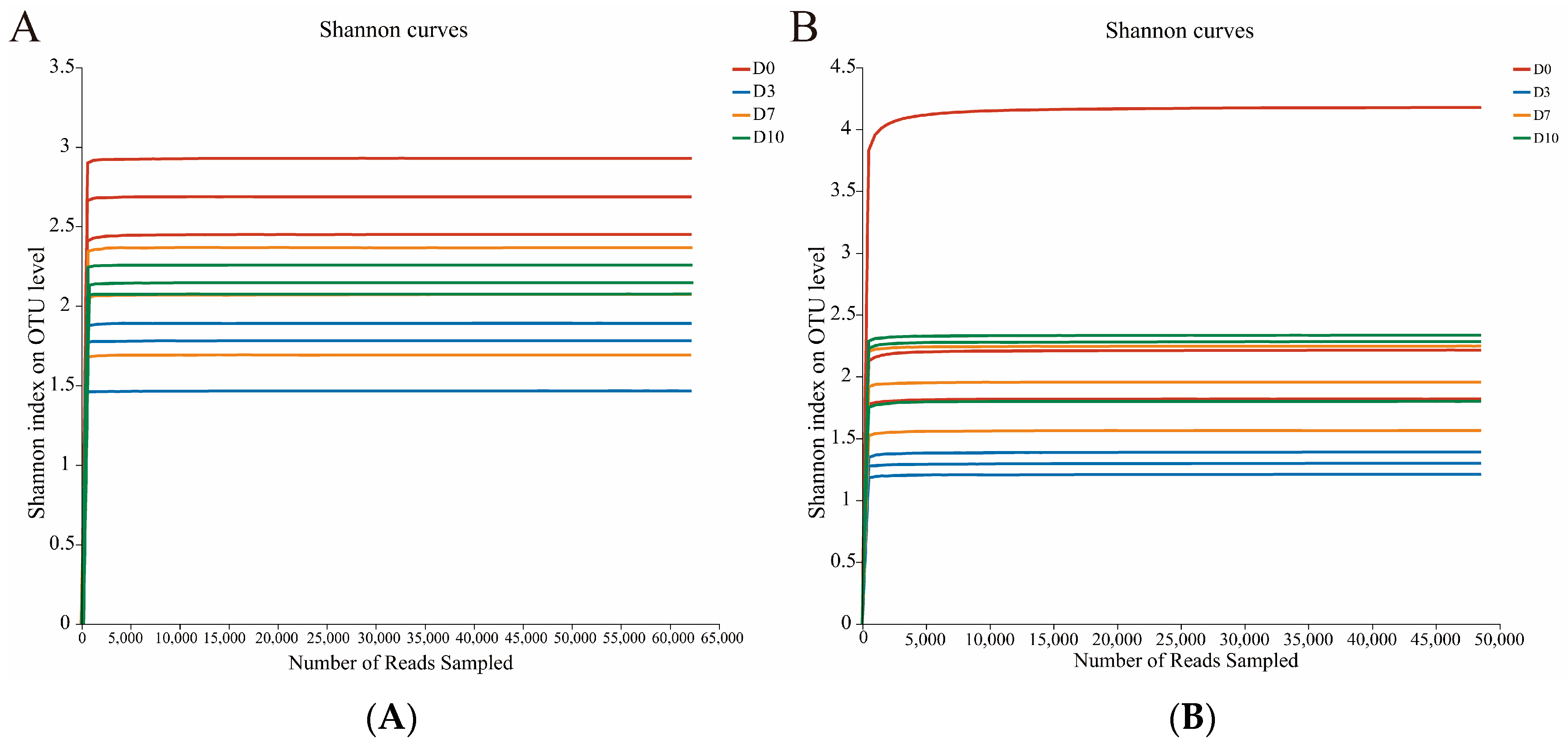
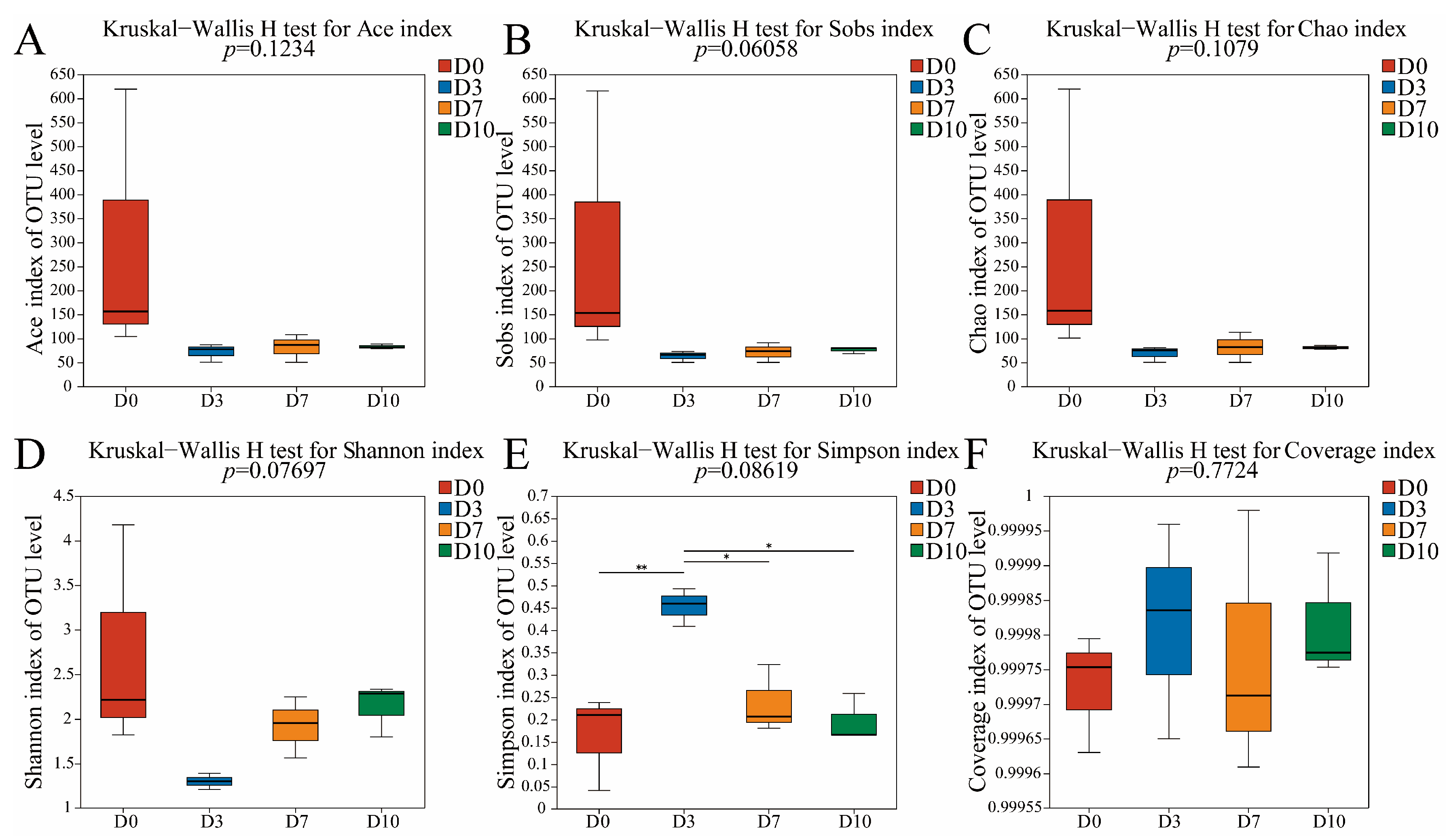
3.1. α-Diversity Analysis
3.2. OTU Venn Analysis
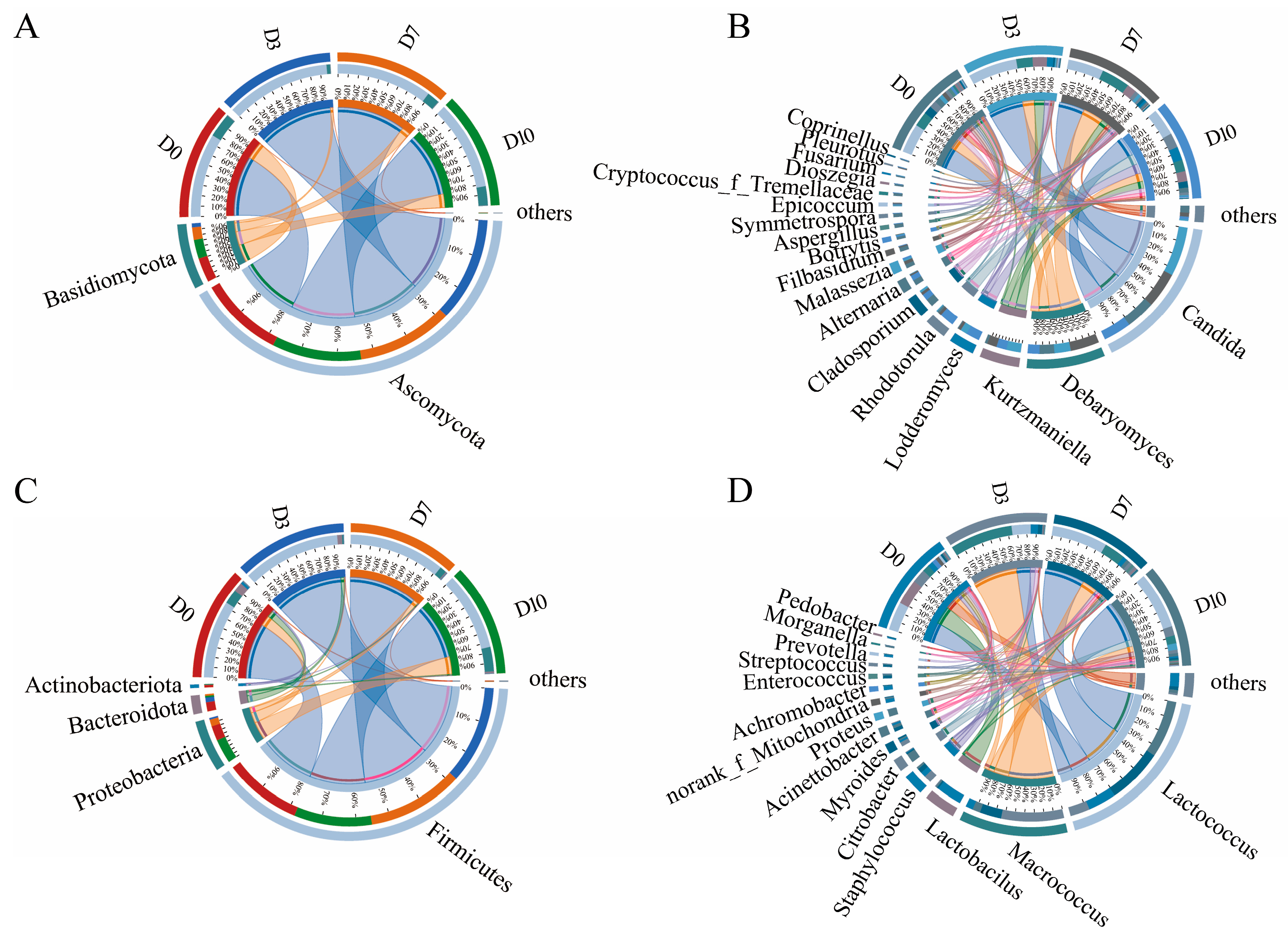
3.3. Community Composition Analysis
3.4. 16S-Based KEGG Functional Pathway Prediction
3.5. Bugbase Phenotype Prediction Analysis
3.6. Correlation between the Core Microbial Genera and Differential Off-Flavor Substances
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greenwood, P.L. Review: An overview of beef production from pasture and feedlot globally, as demand for beef and the need for sustainable practices increase. Animal 2021, 15, 100295. [Google Scholar] [CrossRef]
- Fu, R.; Li, C.; Wang, L.; Gao, Z. Chinese consumer preference for beef with geographical indications and other attributes. Meat Sci. 2024, 212, 109475. [Google Scholar] [CrossRef]
- Zhang, D.; Hou, C. Humble opinion on the quality difference between hot meat and chilled meat. Meat Res. 2020, 34, 83–90. [Google Scholar] [CrossRef]
- Ali, S.; Rezende, V.T.; Ullah, S.; de Paiva, E.L.; Tonin, F.G.; Abdullah; Corassin, C.H.; Oliveira, C.A.F.d. Food processing and challenges in the food production and quality: The foodomics approach. Food Biosci. 2023, 56, 103217. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; Zhang, W.; Dong, P.; Hao, J.; Luo, X. An Overview of Spoilage Microorganisms in Fresh Beef. Food Sci. 2018, 39, 289–296. [Google Scholar] [CrossRef]
- Haque, M.A.; Wang, Y.; Shen, Z.; Li, X.; Saleemi, M.K.; He, C. Mycotoxin contamination and control strategy in human, domestic animal and poultry: A review. Microb. Pathog. 2020, 142, 104095. [Google Scholar] [CrossRef]
- Hou, X.; Zhao, H.; Yan, L.; Li, S.; Chen, X.; Fan, J. Effect of CO2 on the preservation effectiveness of chilled fresh boneless beef knuckle in modified atmosphere packaging and microbial diversity analysis. LWT 2023, 187, 115262. [Google Scholar] [CrossRef]
- Yang, X.; Zhu, L.; Zhang, Y.; Liang, R.; Luo, X. Microbial community dynamics analysis by high-throughput sequencing in chilled beef longissimus steaks packaged under modified atmospheres. Meat Sci. 2018, 141, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Dorn-In, S.; Führer, L.; Gareis, M.; Schwaiger, K. Cold-tolerant microorganisms causing spoilage of vacuum-packed beef under time-temperature abuse determined by culture and qPCR. Food Microbiol. 2023, 109, 104147. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, L.; Liang, R.; Mao, Y.; Hopkins, D.L.; Li, K.; Dong, P.; Yang, X.; Niu, L.; Zhang, Y.; et al. Shelf-life and bacterial community dynamics of vacuum packaged beef during long-term super-chilled storage sourced from two Chinese abattoirs. Food Res. Int. 2020, 130, 108937. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Wang, A.; Wang, Q.; Lee, D.-J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, S.; Chen, H.; Ling, Z.; Jia, X.; Liu, D. Changes of Volatile Flavor Substances of Beeves in Spoilage Process based on Gas ChromatographyIon Mobility Spectrometry and Electronic Nose. Sci. Technol. Food Ind. 2024, 45, 1–10. [Google Scholar] [CrossRef]
- Ovuru, K.F.; Izah, S.C.; Ogidi, O.I.; Imarhiagbe, O.; Ogwu, M.C. Slaughterhouse facilities in developing nations: Sanitation and hygiene practices, microbial contaminants and sustainable management system. Food Sci. Biotechnol. 2023, 33, 519–537. [Google Scholar] [CrossRef]
- Guan, N.; Li, J.; Shin, H.-d.; Du, G.; Chen, J.; Liu, L. Microbial response to environmental stresses: From fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 2017, 101, 3991–4008. [Google Scholar] [CrossRef] [PubMed]
- Rangel, D.E.N.; Finlay, R.D.; Hallsworth, J.E.; Dadachova, E.; Gadd, G.M. Fungal strategies for dealing with environment- and agriculture-induced stresses. Fungal Biol. 2018, 122, 602–612. [Google Scholar] [CrossRef]
- Odeyemi, O.A.; Alegbeleye, O.O.; Strateva, M.; Stratev, D. Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Compr. Rev. Food Sci. Food Saf. 2020, 19, 311–331. [Google Scholar] [CrossRef]
- Mazhar, S.; Hill, C.; McAuliffe, O. The Genus Macrococcus: An Insight Into Its Biology, Evolution, and Relationship with Staphylococcus. Adv. Appl. Microbiol. 2018, 105, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, D.; Wen, X.; Li, X.; Chen, L.; Zheng, X.; Fang, F.; Li, J.; Hou, C. Effects of chilling rate on the freshness and microbial community composition of lamb carcasses. LWT 2022, 153, 112559. [Google Scholar] [CrossRef]
- Johansson, P.; Jääskeläinen, E.; Nieminen, T.; Hultman, J.; Auvinen, P.; Björkroth, K.J. Microbiomes in the Context of Refrigerated Raw Meat Spoilage. Meat Muscle Biol. 2020, 4, 9. [Google Scholar] [CrossRef]
- Mansur, A.R.; Song, E.-J.; Cho, Y.-S.; Nam, Y.-D.; Choi, Y.-S.; Kim, D.-O.; Seo, D.-H.; Nam, T.G. Comparative evaluation of spoilage-related bacterial diversity and metabolite profiles in chilled beef stored under air and vacuum packaging. Food Microbiol. 2019, 77, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Duran, P.; Barra, P.J.; Jorquera, M.A.; Viscardi, S.; Fernandez, C.; Paz, C.; de la Luz Mora, M.; Bol, R. Occurrence of Soil Fungi in Antarctic Pristine Environments. Front. Bioeng. Biotechnol. 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Branco, D.; Lockhart, S.R.; Chen, Y.C.; Santos, D.A.; Hagen, F.; Hawkins, N.J.; Lavergne, R.A.; Meis, J.F.; Le Pape, P.; Rocha, M.F.G.; et al. Collateral consequences of agricultural fungicides on pathogenic yeasts: A One Health perspective to tackle azole resistance. Mycoses 2022, 65, 303–311. [Google Scholar] [CrossRef]
- Hwang, B.K.; Choi, H.; Choi, S.H.; Kim, B.-S. Analysis of Microbiota Structure and Potential Functions Influencing Spoilage of Fresh Beef Meat. Front. Microbiol. 2020, 11, 1657. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, D.; Li, X.; Ding, T.; Liang, C.; Zheng, X.; Yang, W.; Hou, C. Dynamic changes of bacteria and screening of potential spoilage markers of lamb in aerobic and vacuum packaging. Food Microbiol. 2022, 104, 103996. [Google Scholar] [CrossRef]
- Mao, J.; Wang, X.; Chen, H.; Zhao, Z.; Liu, D.; Zhang, Y.; Nie, X. The Contribution of Microorganisms to the Quality and Flavor Formation of Chinese Traditional Fermented Meat and Fish Products. Foods 2024, 13, 608. [Google Scholar] [CrossRef] [PubMed]
- Doelle, H.W. 5—Carbohydrate Metabolism. In Bacterial Metabolism, 2nd ed.; Doelle, H.W., Ed.; Academic Press: Cambridge, MA, USA, 1975; pp. 208–311. [Google Scholar]
- Illikoud, N.; Gohier, R.; Werner, D.; Barrachina, C.; Roche, D.; Jaffrès, E.; Zagorec, M. Transcriptome and Volatilome Analysis During Growth of Brochothrix thermosphacta in Food: Role of Food Substrate and Strain Specificity for the Expression of Spoilage Functions. Front. Microbiol. 2019, 10, 2527. [Google Scholar] [CrossRef] [PubMed]
- Pellissery, A.J.; Vinayamohan, P.G.; Amalaradjou, M.A.R.; Venkitanarayanan, K. Chapter 17—Spoilage bacteria and meat quality. In Meat Quality Analysis; Biswas, A.K., Mandal, P.K., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 307–334. [Google Scholar]
- Jeckelmann, J.-M.; Erni, B. Transporters of glucose and other carbohydrates in bacteria. Pflügers Arch. Eur. J. Physiol. 2020, 472, 1129–1153. [Google Scholar] [CrossRef] [PubMed]
- Fath, M.J.; Kolter, R. ABC transporters: Bacterial exporters. Microbiol. Rev. 1993, 57, 995–1017. [Google Scholar] [CrossRef]
- Davies, J.S.; Currie, M.J.; Wright, J.D.; Newton-Vesty, M.C.; North, R.A.; Mace, P.D.; Allison, J.R.; Dobson, R.C.J. Selective Nutrient Transport in Bacteria: Multicomponent Transporter Systems Reign Supreme. Front. Mol. Biosci. 2021, 8, 699222. [Google Scholar] [CrossRef]
- Coulthurst, S. The Type VI secretion system: A versatile bacterial weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Hernandez, R.E.; Gallegos-Monterrosa, R.; Coulthurst, S.J. Type VI secretion system effector proteins: Effective weapons for bacterial competitiveness. Cell. Microbiol. 2020, 22, e13241. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Ham, H.-I.; Lee, S.-H.; Cho, Y.-J.; Kim, Y.-R.; Yoon, C.-K.; Seok, Y.-J. Functional dissection of the phosphotransferase system provides insight into the prevalence of Faecalibacterium prausnitzii in the host intestinal environment. Environ. Microbiol. 2021, 23, 4726–4740. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Pierneef, R.; Mafuna, T.; Magwedere, K.; Matle, I. Genus-wide genomic characterization of Macrococcus: Insights into evolution, population structure, and functional potential. Front. Microbiol. 2023, 14, 1181376. [Google Scholar] [CrossRef]
- Kerth, C.R.; Legako, J.F.; Woerner, D.R.; Brooks, J.C.; Lancaster, J.M.; O’Quinn, T.G.; Nair, M.; Miller, R.K. A current review of U.S. beef flavor I: Measuring beef flavor. Meat Sci. 2024, 210, 109437. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, S.; Zhu, C.; Sun, L.; Wang, Z.; Wang, X.; Xie, W. Identification of Characteristic Odorants in Duck Meat by Headspace-Gas Chromatography-Ion Mobility Spectrometry. Food Sci. 2023, 44, 247–255. [Google Scholar] [CrossRef]
- Reid, R.; Fanning, S.; Whyte, P.; Kerry, J.; Lindqvist, R.; Yu, Z.; Bolton, D. The microbiology of beef carcasses and primals during chilling and commercial storage. Food Microbiol. 2017, 61, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Zhang, L.; Zhang, D.; Luo, L. Spoilage bacteria in fresh meat products: A review. Qual. Saf. Agro-Prod. 2018, 2018, 21–29. [Google Scholar]
- Tofalo, R.; Fusco, V.; Böhnlein, C.; Kabisch, J.; Logrieco, A.F.; Habermann, D.; Cho, G.-S.; Benomar, N.; Abriouel, H.; Schmidt-Heydt, M.; et al. The life and times of yeasts in traditional food fermentations. Crit. Rev. Food Sci. Nutr. 2020, 60, 3103–3132. [Google Scholar] [CrossRef]



| KEGG Pathway_level2 | Relative Abundance/% | |||
|---|---|---|---|---|
| D0 | D3 | D7 | D10 | |
| Carbohydrate metabolism | 15.6 | 15.7 | 15.2 | 15.4 |
| Membrane transport | 13.7 | 14.3 | 13.8 | 13.9 |
| Amino acid metabolism | 10.0 | 9.4 | 10.1 | 9.8 |
| Nucleotide metabolism | 7.2 | 8.4 | 7.9 | 8.1 |
| Translation | 6.7 | 7.9 | 7.4 | 7.6 |
| Replication and repair | 6.1 | 6.7 | 6.3 | 6.5 |
| Metabolism of cofactors and vitamins | 5.8 | 5.2 | 5.6 | 5.5 |
| Energy metabolism | 5.1 | 4.3 | 4.7 | 4.5 |
| Signal transduction | 4.9 | 4.1 | 4.4 | 4.2 |
| Lipid metabolism | 3.9 | 3.6 | 3.7 | 3.7 |
| Glycan biosynthesis and metabolism | 3.3 | 4.1 | 3.9 | 4.0 |
| Xenobiotics biodegradation and metabolism | 3.0 | 2.5 | 2.8 | 2.7 |
| Folding, sorting, and degradation | 2.7 | 2.7 | 2.7 | 2.7 |
| Metabolism of other amino acids | 2.7 | 2.7 | 2.7 | 2.7 |
| Metabolism of terpenoids and polyketides | 2.2 | 2.0 | 2.2 | 2.1 |
| Others | 6.9 | 6.4 | 6.6 | 6.5 |
| Number | Compound | CAS | Odor Description |
|---|---|---|---|
| 1 | 1-Penten-3-ol | 616-25-1 | Butter, pungent, green |
| 2 | 2,3-Butanedione | 431-03-8 | Butter |
| 3 | Butan-2-one | 78-93-3 | Ether |
| 4 | 3-Pentanone | 96-22-0 | Ether |
| 5 | 3-Octanone-M | 106-68-3 | Herb, butter, mold |
| 6 | Acetic acid | 64-19-7 | Acid, pungent, sour |
| 7 | Acetone | 67-64-1 | Pungent |
| 8 | Butanal-D | 123-72-8 | Pungent, green |
| 9 | Butyraldehyde-M | 123-72-8 | Pungent, green |
| 10 | Toluene | 108-88-3 | Paint |
| 11 | Ethyl formate | 109-94-4 | Pungent |
| 12 | Decalin | 91-17-8 | Pungent |
| 13 | Pyrrolidine-M | 123-75-1 | Alkaline |
| 14 | Propan-2-ol | 67-63-0 | Alcohol |
| 15 | 2-Methylpropanol-D | 78-83-1 | Bitter |
| 16 | 2-Methylpropanol-M | 78-83-1 | Bitter |
| 17 | 2-Methyl propanal | 78-84-2 | Pungent, green |
| 18 | 3-Methylbutan-1-ol-D | 123-51-3 | Burnt |
| 19 | 3-Methylbutanol-M | 123-51-3 | Burnt |
| 20 | 1-Propanol-D | 71-23-8 | Alcohol, pungent |
| 21 | 1-Propanol-M | 71-23-8 | Alcohol, pungent |
| 22 | Butanol-D | 71-36-3 | Medicine |
| 23 | Butanol-M | 71-36-3 | Medicine |
| 24 | Hexanal-D | 66-25-1 | Fat, oil, green |
| 25 | Hexanal-M | 66-25-1 | Fat, oil, green |
| 26 | 1-Pentanol-D | 71-41-0 | Balsamic, pungent, green |
| 27 | 1-Pentanol-M | 71-41-0 | Balsamic, pungent, green |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Ling, Z.; Nie, X.; Liu, D.; Chen, H.; Zhang, S. Microbial Diversity and Community Structure of Chinese Fresh Beef during Cold Storage and Their Correlations with Off-Flavors. Foods 2024, 13, 1482. https://doi.org/10.3390/foods13101482
Zhao Z, Ling Z, Nie X, Liu D, Chen H, Zhang S. Microbial Diversity and Community Structure of Chinese Fresh Beef during Cold Storage and Their Correlations with Off-Flavors. Foods. 2024; 13(10):1482. https://doi.org/10.3390/foods13101482
Chicago/Turabian StyleZhao, Zhiping, Ziqing Ling, Xin Nie, Dayu Liu, Hongfan Chen, and Shengyuan Zhang. 2024. "Microbial Diversity and Community Structure of Chinese Fresh Beef during Cold Storage and Their Correlations with Off-Flavors" Foods 13, no. 10: 1482. https://doi.org/10.3390/foods13101482






