Screening and In Silico Analyses of the Yeast Saccharomyces cerevisiae Σ1278b Bank Mutants Using Citral as a Natural Antimicrobial
Abstract
:1. Introduction
2. Materials and Methods
2.1. S. cerevisiae Σ1278b Bank of Mutants
2.2. Determination of the MIC of Citral on S. cerevisiae Σ1278b
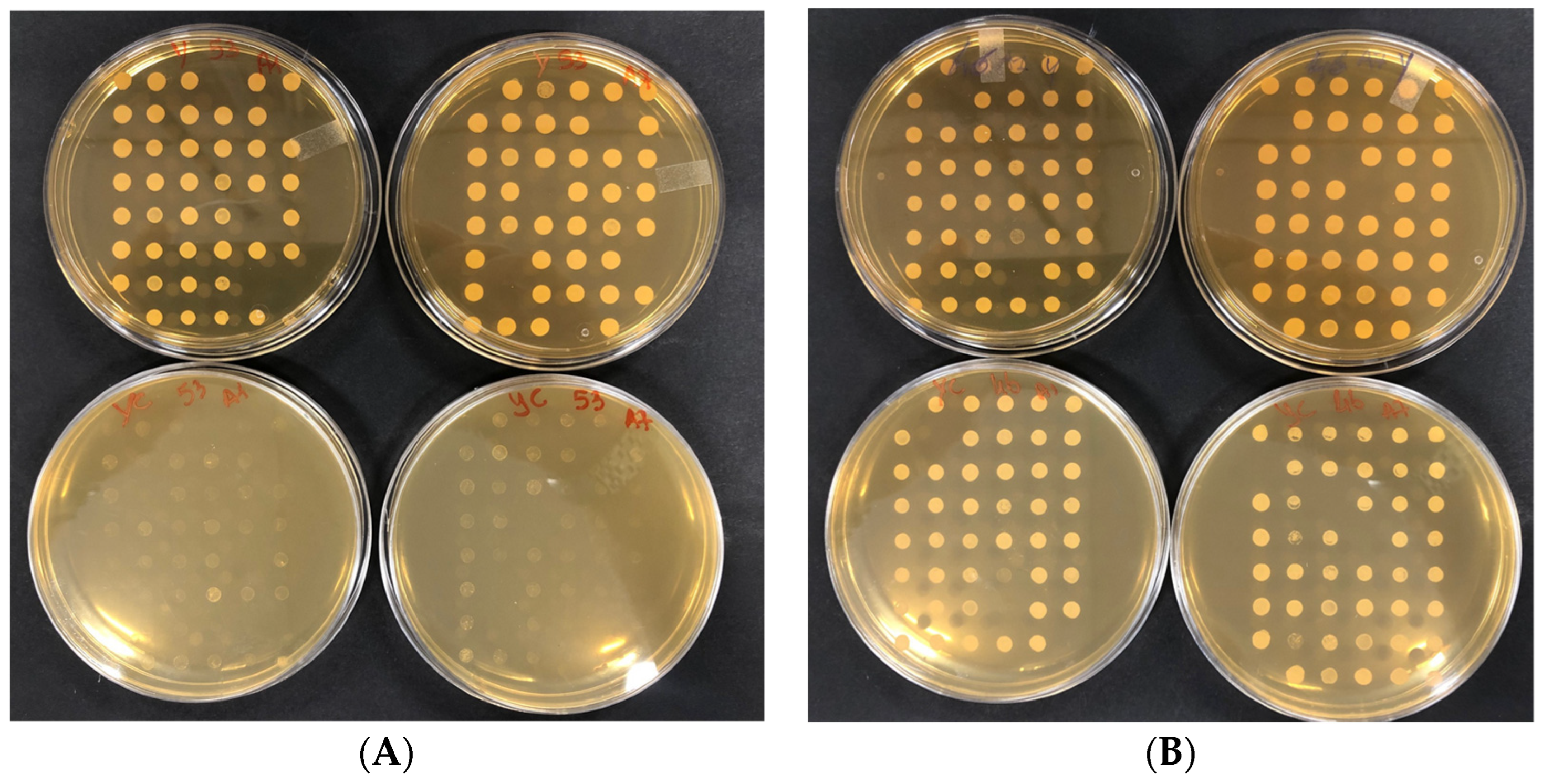
2.3. Screening of the S. cerevisiae Σ1278b Bank of Mutants
2.4. In Silico Analyses
3. Results and Discussion
3.1. Molecular Function of the Identified Genes
3.2. Cellular Component of the Identified Genes
3.3. Biological Process of the Identified Genes
3.4. Genes Related to the Cellular Response to Oxidative Stress
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bastidas, R.J.; Heitman, J.; Cárdenas, M.E. The Protein Kinase Tor1 Regulates Adhesin Gene Expression in Candida albicans. PLoS Pathog. 2009, 5, e1000294. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B.; Fink, G.R. Bakers’ Yeast, a Model for Fungal Biofilm Formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.J.; Sabbagh, W.; Graham, E.; Irick, M.M.; Van Olden, E.K.; Neal, C.L.; Delrow, J.J.; Bardwell, L.; Sprague, G.F. A Signaling Mucin at the Head of the Cdc42- and MAPK-Dependent Filamentous Growth Pathway in Yeast. Genes Dev. 2004, 18, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.J.; Sprague, G.F. Glucose Depletion Causes Haploid Invasive Growth in Yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13619–13624. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, C.J.; Ljungdahl, P.O.; Styles, C.A.; Fink, G.R. Unipolar Cell Divisions in the Yeast S. Cerevisiae Lead to Filamentous Growth: Regulation by Starvation and RAS. Cell 1992, 68, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Zara, G.; Budroni, M.; Mannazzu, I.; Fancello, F.; Zara, S. Yeast biofilm in food realms: Occurrence and control. World J. Microbiol. Biotechnol. 2020, 36, 134. [Google Scholar] [CrossRef] [PubMed]
- Coi, A.; Bigey, F.; Mallet, S.; Marsit, S.; Zara, G.; Gladieux, P.; Galeote, V.; Budroni, M.; Dequin, S.; Legras, J.L. Genomic signatures of adaptation to wine biological ageing conditions in biofilm-forming flor yeasts. Mol. Ecol. 2017, 26, 2150–2166. [Google Scholar] [CrossRef] [PubMed]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.P.; Budroni, M. FLO11 -Based Model for Air-Liquid Interfacial Biofilm Formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef] [PubMed]
- Fidalgo, M.; Barrales, R.R.; Ibeas, J.I.; Jiménez, J. Adaptive Evolution by Mutations in theFLO11gene. Proc. Natl. Acad. Sci. USA 2006, 103, 11228–11233. [Google Scholar] [CrossRef]
- Moreno-García, J.; Martín-García, F.J.; Ogawa, M.; García-Martínez, T.; Moreno, J.; Mauricio, J.C.; Bisson, L.F. FLO1, FLO5 and FLO11 Flocculation Gene Expression Impacts Saccharomyces cerevisiae Attachment to Penicillium chrysogenum in a Co-immobilization Technique. Front. Microbiol. 2018, 9, 2586. [Google Scholar] [CrossRef]
- Martillanes, S.; Rocha-Pimienta, J.; Cabrera-Bañegil, D.M.-V.M.; Delgado-Adámez, J. Application of Phenolic Compounds for Food Preservation: Food Additive and Active Packaging. In Phenolic Compounds—Biological Activity; InTech eBooks; IntechOpen Limited: London, UK, 2017. [Google Scholar]
- Van Vuuren, S.F.; Suliman, S.; Viljoen, A.M. The antimicrobial activity of four commercial essential oils in combination with conventional antimicrobials. Lett. Appl. Microbiol. 2009, 48, 440–446. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, L.L.; de Andrade, S.C.; Athayde, A.J.; de Oliveira, C.E.; de Sales, C.V.; Madruga, M.S.; de Souza, E.L. Efficacy of Origanum vulgare L. and Rosmarinus officinalis L. essential oils in combination to control postharvest pathogenic Aspergilli and autochthonous mycoflora in Vitis labrusca L. (table grapes). Int. J. Food Microbiol. 2013, 165, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Białoń, M.; Krzyśko–Łupicka, T.; Koszałkowska, M.; Wieczorek, P. The influence of chemical composition of commercial lemon essential oils on the growth of Candida strains. Mycopathologia 2014, 177, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Rajkowska, K.; Kunicka-Styczyńska, A.; Maroszyńska, M.; Dąbrowska, M. The Effect of Thyme and Tea Tree Oils on Morphology and Metabolism of Candida albicans. Acta Biochim. Pol. 2014, 61, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Boire, N.; Zhang, S.X.; Khuvis, J.; Lee, R.T.; Rivers, J.L.; Crandall, P.G.; Keel, M.K.; Parrish, N. Potent Inhibition of Pseudogymnoascus destructans, the Causative Agent of White-Nose Syndrome in Bats, by Cold-Pressed, Terpeneless, Valencia Orange Oil. PLoS ONE 2016, 11, e0148473. [Google Scholar] [CrossRef]
- Fancello, F.; Petretto, G.L.; Zara, S.; Sanna, M.L.; Addis, R.; Maldini, M.; Foddai, M.; Rourke, J.P.; Chessa, M.; Pintore, G.A.M. Chemical Characterization, Antioxidant Capacity and Antimicrobial Activity against Food Related Microorganisms of Citrus limon var. pompia Leaf Essential Oil. LWT 2016, 69, 579–585. [Google Scholar] [CrossRef]
- Zheng, S.; Jing, G.; Wang, X.; Ouyang, Q.; Jia, L.; Tao, N. Citral exerts its antifungal activity against Penicillium digitatum by affecting the mitochondrial morphology and function. Food Chem. 2015, 178, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Scariot, F.J.; Pansera, M.S.; Delamare, A.P.L.; Echeverrigaray, S. Citral and Geraniol Induce Necrotic and Apoptotic Cell Death on Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2021, 37, 42. [Google Scholar] [CrossRef]
- Shi, C.; Song, K.; Zhang, X.; Sun, Y.; Sui, Y.; Chen, Y.; Jia, Z.; Sun, H.; Sun, Z.; Xia, X. Antimicrobial Activity and Possible Mechanism of Action of Citral against Cronobacter sakazakii. PLoS ONE 2016, 11, e0159006. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Chen, Y.; Hou, C. Antioxidant and Antibacterial Activity of Seven Predominant Terpenoids. Int. J. Food Prop. 2019, 22, 230–238. [Google Scholar] [CrossRef]
- Ye, C.J.; Li, S.A.; Zhang, Y.; Lee, W.H. Geraniol targets KV1.3 ion channel and exhibits anti-inflammatory activity in vitro and in vivo. Fitoterapia 2019, 139, 104394. [Google Scholar] [CrossRef] [PubMed]
- Lima, I.O.; de Medeiros Nóbrega, F.; de Oliveira, W.A.; de Oliveira Lima, E.; Albuquerque Menezes, E.; Afrânio Cunha, F.; de Fátima Formiga Melo Diniz, M. Anti-Candida albicans effectiveness of citral and investigation of mode of action. Pharm. Biol. 2012, 50, 1536–1541. [Google Scholar] [CrossRef] [PubMed]
- Leite, M.S.A.; De Brito Bezerra, A.P.; De Sousa, J.P.; Guerra, F.Q.S.; De Oliveira Lima, E. Evaluation of Antifungal Activity and Mechanism of Action of Citral against Candida albicans. Evid.-Based Complement. Altern. Med. 2014, 2014, 378280. [Google Scholar] [CrossRef]
- Li, J.; Lei, Z.; Li, L.; Xie, R.; Xi, W.; Guan, Y.; Sumner, L.W.; Zhou, Z. Antifungal Activity of Citrus Essential Oils. J. Agric. Food Chem. 2014, 62, 3011–3033. [Google Scholar]
- Tang, X.; Shao, Y.; Tang, Y.; Zhou, W.-W. Antifungal Activity of Essential Oil Compounds (Geraniol and Citral) and Inhibitory Mechanisms on Grain Pathogens (Aspergillus flavus and Aspergillus ochraceus). Molecules 2018, 23, 2108. [Google Scholar] [CrossRef] [PubMed]
- Mitropoulou, G.; Fitsiou, E.; Spyridopoulou, K.; Tiptiri-Kourpeti, A.; Bardouki, H.; Vamvakias, M.; Panas, P.; Chlichlia, K.; Pappa, A.; Kourkoutas, Y. Citrus Medica Essential Oil Exhibits Significant Antimicrobial and Antiproliferative Activity. LWT 2017, 84, 344–352. [Google Scholar] [CrossRef]
- Bard, M.; Albrecht, M.R.; Gupta, N.; Guynn, C.J.; Stillwell, W. Geraniol Interferes with Membrane Functions in Strains of Candida and Saccharomyces. Lipids 1988, 23, 534–538. [Google Scholar] [CrossRef]
- Lin, N.-X.; Xu, Y.; Yu, X. Overview of Yeast Environmental Stress Response Pathways and the Development of Tolerant Yeasts. Syst. Microbiol. Biomanufacturing 2021, 2, 232–245. [Google Scholar] [CrossRef]
- Farrugia, G.; Balzan, R. Oxidative Stress and Programmed Cell Death in Yeast. Front. Oncol. 2012, 2, 64. [Google Scholar] [CrossRef]
- Gaonkar, R.; Avti, P.; Hegde, G. Differential Antifungal Efficiency of Geraniol and Citral. Nat. Prod. Commun. 2018, 13, 1934578X1801301. [Google Scholar] [CrossRef]
- Gutiérrez-Pacheco, M.M.; Torres-Moreno, H.; Flores-Lopez, M.L.; Velázquez Guadarrama, N.; Ayala-Zavala, J.F.; Ortega-Ramírez, L.A.; López-Romero, J.C. Mechanisms and Applications of Citral’s Antimicrobial Properties in Food Preservation and Pharmaceuticals Formulations. Antibiotics 2023, 12, 1608. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.M.; André, B.; et al. Functional Profiling of the Saccharomyces cerevisiae Genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Page, N.F.; Robinson, M.D.; Raghibizadeh, S.; Hogue, C.W.V.; Bussey, H.; et al. Systematic Genetic Analysis with Ordered Arrays of Yeast Deletion Mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef] [PubMed]
- Baryshnikova, A.; Costanzo, M.; Dixon, S.J.; Vizeacoumar, F.J.; Myers, C.L.; Andrews, B.; Boone, C. Synthetic Genetic Array (SGA) Analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2010; pp. 145–179. [Google Scholar]
- Ryan, O.; Shapiro, R.; Kurat, C.F.; Mayhew, D.; Baryshnikova, A.; Chin, B.; Lin, Z.; Cox, M.J.; Vizeacoumar, F.S.; Cheung, D.; et al. Global Gene Deletion Analysis Exploring Yeast Filamentous Growth. Science 2012, 337, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Dowell, R.D.; Ryan, O.; Jansen, A.; Cheung, D.; Agarwala, S.D.; Danford, T.; Bernstein, D.A.; Rolfe, P.A.; Heisler, L.E.; Chin, B.; et al. Genotype to Phenotype: A Complex Problem. Science 2010, 328, 469. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.a.L.; Sherlock, G.; Myers, C.L.; Burrows, N.M.; Deng, C.; Wu, H.; McCann, K.; Troyanskaya, O.G.; Brown, J.M. Global Analysis of Gene Function in Yeast by Quantitative Phenotypic Profiling. Mol. Syst. Biol. 2006, 2, 2006.0001. [Google Scholar] [CrossRef] [PubMed]
- Germany, E.M.; Zahayko, N.; Huebsch, M.L.; Fox, J.L.; Prahlad, V.; Khalimonchuk, O. The AAA ATPase Afg1 preserves mitochondrial fidelity and cellular health by maintaining mitochondrial matrix proteostasis. J. Cell Sci. 2018, 131, jcs219956. [Google Scholar] [CrossRef]
- Le, D.T.; Lee, B.C.; Marino, S.M.; Zhang, Y.; Fomenko, D.E.; Kaya, A.; Hacıoğlu, E.; Kwak, G.H.; Koç, A.; Kim, H.Y.; et al. Functional Analysis of Free Methionine-R-Sulfoxide Reductase from Saccharomyces Cerevisiae. J. Biol. Chem. 2009, 284, 4354–4364. [Google Scholar] [CrossRef]
- Demasi, A.P.; Pereira, G.A.; Netto, L.E. Yeast oxidative stress response. Influences of cytosolic thioredoxin peroxidase I and of the mitochondrial functional state. FEBS J. 2006, 273, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Helsen, J.; Voordeckers, K.; Vanderwaeren, L.; Santermans, T.; Tsontaki, M.; Verstrepen, K.J.; Jelier, R. Gene Loss Predictably Drives Evolutionary Adaptation. Mol. Biol. Evol. 2020, 37, 2989–3002. [Google Scholar] [CrossRef] [PubMed]
- Skoneczna, A.; Miciałkiewicz, A.; Skoneczny, M. Saccharomyces Cerevisiae Hsp31p, a Stress Response Protein Conferring Protection against Reactive Oxygen Species. Free. Radic. Biol. Med. 2007, 42, 1409–1420. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Antas, P.; Pais, T.F.; Smalley, J.L.; Giorgini, F.; Outeiro, T.F. Yeast DJ-1 Superfamily Members Are Required for Diauxic-Shift Reprogramming and Cell Survival in Stationary Phase. Proc. Natl. Acad. Sci. USA 2014, 111, 7012–7017. [Google Scholar] [CrossRef]
- Park, S.G.; Kyung, M.; Jeong, W.; Kim, I.H. Distinct Physiological Functions of Thiol Peroxidase Isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 5723–5732. [Google Scholar] [CrossRef]
- Okada, N.; Ogawa, J.; Shima, J. Comprehensive Analysis of Genes Involved in the Oxidative Stress Tolerance Using Yeast Heterozygous Deletion Collection. FEMS Yeast Res. 2014, 14, 425–434. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Taggart, J.; Kerdsomboon, K.; Kubisiak, M.; Panascharoen, S.; Schelble, K.; Eide, D.J. Peroxiredoxin Chaperone Activity Is Critical for Protein Homeostasis in Zinc-Deficient Yeast*. J. Biol. Chem. 2013, 288, 31313–31327. [Google Scholar] [CrossRef]
- Brombacher, K.; Fischer, B.B.; Rüfenacht, K.; Eggen, R.I.L. The Role of Yap1p and Skn7p-Mediated Oxidative Stress Response in the Defence of Saccharomyces Cerevisiae against Singlet Oxygen. Yeast 2006, 23, 741–750. [Google Scholar] [CrossRef]
- Du, L.; Yu, Y.; Li, Z.; Chen, J.; Liu, Y.; Xia, Y.; Liu, X. Tim18, a Component of the Mitochondrial Translocator, Mediates Yeast Cell Death Induced by Arsenic. Biochemistry 2007, 72, 843–847. [Google Scholar] [CrossRef]
- Nakamura, T.; Andõ, A.; Takagi, H.; Shima, J. EOS1, Whose Deletion Confers Sensitivity to Oxidative Stress, Is Involved in N-Glycosylation in Saccharomyces Cerevisiae. Biochem. Biophys. Res. Commun. 2007, 353, 293–298. [Google Scholar] [CrossRef]
- Ando, A.; Nakamura, T.; Murata, Y.; Takagi, H.; Shima, J. Identification and classification of genes required for tolerance to freeze-thaw stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. FEMS Yeast Res. 2007, 7, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Higgins, V.J.; Alic, N.; Thorpe, G.W.; Breitenbach, M.; Larsson, V.; Dawes, I.W. Phenotypic Analysis of Gene Deletant Strains for Sensitivity to Oxidative Stress. Yeast 2001, 19, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Outten, C.E.; Falk, R.; Culotta, V.C. Cellular Factors Required for Protection from Hyperoxia Toxicity in Saccharomyces Cerevisiae. Biochem. J. 2005, 388, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Begley, T.J.; Rosenbach, A.; Ideker, T.; Samson, L.D. Hot Spots for Modulating Toxicity Identified by Genomic Phenotyping and Localization Mapping. Mol. Cell 2004, 16, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Goehring, A.; Rivers, D.M.; Sprague, G.F. Urmylation: A Ubiquitin-like Pathway That Functions during Invasive Growth and Budding in Yeast. Mol. Biol. Cell 2003, 14, 4329–4341. [Google Scholar] [CrossRef] [PubMed]
- Greetham, D.; Grant, C.M. Antioxidant Activity of the Yeast Mitochondrial One-Cys Peroxiredoxin Is Dependent on Thioredoxin Reductase and Glutathione in Vivo. Mol. Cell. Biol. 2009, 29, 3229–3240. [Google Scholar] [CrossRef] [PubMed]
- Pedrajas, J.R.; Kosmidou, E.; Miranda-Vizuete, A.; Gustafsson, J.; Wright, A.P.H.; Spyrou, G. Identification and Functional Characterization of a Novel Mitochondrial Thioredoxin System in Saccharomyces Cerevisiae. J. Biol. Chem. 1999, 274, 6366–6373. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Tsang, C.K.; Zheng, X.F.S. Mechanisms of Regulation of RNA Polymerase III-Dependent Transcription by TORC1. EMBO J. 2009, 28, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Takagi, H. N-Acetyltransferase Mpr1 Confers Ethanol Tolerance on Saccharomyces Cerevisiae by Reducing Reactive Oxygen Species. Appl. Microbiol. Biotechnol. 2007, 75, 1343–1351. [Google Scholar] [CrossRef]
- Lee, J.S.; Huh, W.-K.; Lee, B.; Baek, Y.-U.; Hwang, C.-S.; Kim, S.-T.; Kim, Y.-R.; Kang, S.-O. Mitochondrial NADH-Cytochrome B5 Reductase Plays a Crucial Role in the Reduction of d-Erythroascorbyl Free Radical in Saccharomyces Cerevisiae. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2001, 1527, 31–38. [Google Scholar] [CrossRef]
- Kaya, A.; Koç, A.; Lee, B.C.; Fomenko, D.E.; Rederstorff, M.; Krol, A.; Lescure, A.; Gladyshev, V.N. Compartmentalization and Regulation of Mitochondrial Function by Methionine Sulfoxide Reductases in Yeast. Biochemistry 2010, 49, 8618–8625. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.; Kershaw, C.J.; Castelli, L.M.; Costello, J.L.; Ashe, M.P.; Grant, C.M.; Sims, P.F.G.; Pavitt, G.D.; Hubbard, S.J. Puf3p Induces Translational Repression of Genes Linked to Oxidative Stress. Nucleic Acids Res. 2013, 42, 1026–1041. [Google Scholar] [CrossRef] [PubMed]
- Gebert, N.; Gebert, M.; Oeljeklaus, S.; Von Der Malsburg, K.; Stroud, D.A.; Kulawiak, B.; Wirth, C.; Zahedi, R.P.; Doležal, P.; Wiese, S.; et al. Dual Function of SDH3 in the Respiratory Chain and TIM22 Protein Translocase of the Mitochondrial Inner Membrane. Mol. Cell 2011, 44, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.S.; Amorim, M.A.; Reis, E.M.; Quintanilha, A.; Moradas-Ferreira, P. Mitochondrial Superoxide Dismutase Is Essential for Ethanol Tolerance of Saccharomyces Cerevisiae in the Post-Diauxic Phase. Microbiology 1997, 143, 1649–1656. [Google Scholar] [CrossRef]
- Lee, A.; Xu, X.; Blachly-Dyson, E.; Forte, M.; Colombini, M. The Role of Yeast VDAC Genes on the Permeability of the Mitochondrial Outer Membrane. J. Membr. Biol. 1998, 161, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Manfredini, V.; Martins, V.D.; Peralba, M.D.C.R.; Da Silveira Benfato, M. Adaptative Response to Enhanced Basal Oxidative Damage in Sod Mutants from Saccharomyces Cerevisiae. Mol. Cell. Biochem. 2005, 276, 175–181. [Google Scholar] [CrossRef]
- Munhoz, D.C.; Netto, L.E.S. Cytosolic Thioredoxin Peroxidase I and II Are Important Defenses of Yeast against Organic Hydroperoxide Insult. J. Biol. Chem. 2004, 279, 35219–35227. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, A. Thioredoxin and Glutaredoxin Systems. J. Biol. Chem. 1989, 264, 13963–13966. [Google Scholar] [CrossRef]
- Morgan, B.A.; Banks, G.R.; Toone, W.M.; Raitt, D.C.; Kuge, S.; Johnston, L.H. The Skn7 Response Regulator Controls Gene Expression in the Oxidative Stress Response of the Budding Yeast Saccharomyces Cerevisiae. EMBO J. 1997, 16, 1035–1044. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Jamieson, D.J. Oxidative Stress Responses of the yeastSaccharomyces Cerevisiae. Yeast 1998, 14, 1511–1527. [Google Scholar] [CrossRef]
- Temple, M.D.; Perrone, G.G.; Dawes, I.W. Complex Cellular Responses to Reactive Oxygen Species. Trends Cell Biol. 2005, 15, 319–326. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Fassler, J.S. Identification of Novel Yap1p and Skn7p Binding Sites Involved in the Oxidative Stress Response of Saccharomyces Cerevisiae. Mol. Microbiol. 2005, 58, 1454–1467. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, K.; Rovinsky, S.A.; Coleman, S.T.; Moye-Rowley, W.S. Oxidant-Specific Folding of YAP1P Regulates Both Transcriptional Activation and Nuclear Localization. J. Biol. Chem. 2005, 280, 40524–40533. [Google Scholar] [CrossRef] [PubMed]


| ID | Source | Term ID | Term Name | Padj (Query_1) |
|---|---|---|---|---|
| 1 | GO:CC | GO:0031907 | Microbody lumen | 4.029 × 10−2 |
| 2 | GO:BP | GO:0009987 | Cellular process | 2.188 × 10−18 |
| 3 | GO:CC | GO:0005622 | Intracellular anatomical structure | 8.594 × 10−17 |
| 4 | GO:CC | GO:0005737 | Cytoplasm | 5.573 × 10−13 |
| 5 | GO:CC | GO:0043226 | Organelle | 1.212 × 10−11 |
| 6 | GO:CC | GO:0110165 | Cellular anatomical entity | 1.248 × 10−11 |
| 7 | GO:MF | GO:0003824 | Catalytic activity | 4.577 × 10−3 |
| 8 | GO:MF | GO:0005488 | Binding | 2.577 × 10−3 |
| 9 | GO:BP | GO:0008152 | Metabolic process | 5.582 × 10−5 |
| 10 | GO:BP | GO:0007124 | Pseudohyphal growth | 8.798 × 10−3 |
| 11 | GO:BP | GO:0044237 | Cellular metabolic process | 2.940 × 10−3 |
| 12 | GO:BP | GO:0044238 | Primary metabolic process | 1.856 × 10−3 |
| 13 | GO:BP | GO:0044182 | filamentous growth of a population of unicellular organisms. | 3.599 × 10−2 |
| 14 | GO:BP | GO:0070783 | growth of unicellular organism as a thread of attached cells | 2.302 × 10−2 |
| 15 | GO:BP | GO:0071704 | Organic substance metabolic process | 4.253 × 10−3 |
| 16 | GO:CC | GO:0043231 | Intracellular membrane-bounded organelle | 1.856 × 10−7 |
| 17 | GO:CC | GO:0005739 | mitochondrion | 2.429 × 10−4 |
| 18 | GO:CC | GO:0031090 | Organelle membrane | 1.198 × 10−3 |
| 19 | GO:CC | GO:0005782 | Peroxisomal matrix | 4.029 × 10−2 |
| 20 | GO:CC | GO:0098588 | Bounding membrane of organelle | 6.426 × 10−3 |
| GO Ontology Term | Genes | GO ID |
|---|---|---|
| Oxidoreductase activity | 38 of 590 genes, 6.44% | GO:0016491 |
| Transferase activity | 21 of 590 genes, 3.56% | GO:0016740 |
| Kinase activity | 18 of 590 genes, 3.05% | GO:0016301 |
| Gene Ontology Term | Cluster Frequency | Genome Frequency | Corrected p-Value | FDR | FALSE Positives | Genes Annotated to the Term |
|---|---|---|---|---|---|---|
| pseudohyphal growth | 18 of 590 genes, 3.1% | 73 of 7166 genes, 1.0% | 0.02426 | 0.00% | 0.00 | YOR032C, YKL149C, YNL068C, YHR111W, YNL076W, YKL185W, YOR315W, YNL196C, YLR353W, YER020W, YNL142W, YJR094C, YOR030W, YNL098C, YHR084W, YLR362W, YDR480W, YDR477W |
| regulation of pseudohyphal growth | 9 of 590 genes, 1.5% | 22 of 7166 genes, 0.3% | 0.04274 | 2.00% | 0.04 | YNL068C, YHR111W, YNL076W, YNL098C, YHR084W, YKL185W, YNL196C, YDR477W, YDR480W |
| cell growth | 19 of 590 genes, 3.2% | 85 of 7166 genes, 1.2% | 0.06294 | 1.33% | 0.04 | YOR032C, YKL149C, YNL068C, YHR111W, YNL076W, YKL185W, YOR315W, YNL196C, YJL201W, YLR353W, YER020W, YNL142W, YJR094C, YOR030W, YNL098C, YHR084W, YLR362W, YDR480W, YDR477W |
| growth of unicellular organism as a thread of attached cells | 21 of 590 genes, 3.6% | 100 of 7166 genes, 1.4% | 0.06823 | 2.50% | 0.10 | YOR032C, YKL149C, YNL068C, YHR111W, YNL076W, YKL185W, YOR315W, YNL196C, YOR212W, YLR353W, YER020W, YNL142W, YJR094C, YNL097C, YOR030W, YNL098C, YHR084W, YLR362W, YOR080W, YDR480W, YDR477W |
| lipid metabolic process | 52 of 590 genes, 8.8% | 365 of 7166 genes, 5.1% | 0.07262 | 2.00% | 0.10 | YNL111C, YDR503C, YOR317W, YEL020C, YOR100C, YOR245C, YOR022C, YNL156C, YNL045W, YJL062W, YNL202W, YOR025W, YOR237W, YJR066W, YHR133C, YOR126C, YOR365C, YOR221C, YJR019C, YOR298W, YOR175C, YOR093C, YKL094W, YJL187C, YLR404W, YKL140W, YKL055C, YOR311C, YJL100W, YJL134W, YOR084W, YOR086C, YHR100C, YNL169C, YOR059C, YKL150W, YER024W, YNL123W, YNL054W, YLR386W, YNL231C, YOR180C, YOR081C, YHR004C, YNL128W, YOR196C, YER005W, YKL188C, YJL083W, YLR450W, YER019W, YHL032C |
| regulation of filamentous growth | 12 of 590 genes, 2.0% | 40 of 7166 genes, 0.6% | 0.07885 | 1.67% | 0.10 | YNL068C, YNL142W, YHR111W, YNL097C, YNL076W, YNL098C, YHR084W, YKL185W, YOR315W, YNL196C, YDR477W, YDR480W |
| GO Ontology Term | Genes | GO ID |
|---|---|---|
| mitochondrion organization | 36 of 590 genes, 6.10% | GO:0007005 |
| mitotic cell cycle | 32 of 590 genes, 5.42% | GO:0000278 |
| meiotic cell cycle | 30 of 590 genes, 5.08% | GO:0051321 |
| transmembrane transport | 26 of 590 genes, 4.41% | GO:0055085 |
| organelle fission | 22 of 590 genes, 3.73% | GO:0048285 |
| cell wall organization or biogenesis | 21 of 590 genes, 3.56% | GO:0071554 |
| cellular ion homeostasis | 20 of 590 genes, 3.39% | GO:0006873 |
| nucleobase-containing small molecule metabolic process | 20 of 590 genes, 3.39% | GO:0055086 |
| chromatin organization | 19 of 590 genes, 3.22% | GO:0006325 |
| nucleobase-containing small molecule metabolic process | 20 of 590 genes, 3.39% | GO:0055086 |
| DNA recombination | 19 of 590 genes, 3.22% | GO:0006310 |
| proteolysis involved in protein catabolic process | 8 of 590 genes, 3.05% | GO:0051603 |
| DNA repair | 25 of 590 genes, 4.24% | GO:0006281 |
| Deleted Gene | Chemical | References | Chemical | References |
|---|---|---|---|---|
| GCY1 (YOR120W) | No references available | |||
| AFG1 (YEL052W) | hydrogen peroxide | oxidative stress resistance: decreased [40,41] | ||
| YKL069W fRMsr, YKG9 | Hydrogen peroxide | Oxidative stress resistance increased [40] | Hydrogen peroxide | oxidative stress resistance: decreased [42] |
| SRX1/YKL086W | Paraquat hydrogen peroxide | oxidative stress resistance: decreased [43,44] | ||
| GRE3/YHR104W | ||||
| LTV1/YKL143W (YKL2) | paraquat | oxidative stress resistance: increased. [44] | ||
| HSP31/YDR533C | lipid hydroperoxide hydrogen peroxide tert-butyl hydroperoxide 1,1′-azobis(N,N-dimethylformamide) | oxidative stress resistance decreased [45,46] | ||
| TSA1/YML028W | hydrogen peroxide tert-butyl hydroperoxide 1,1′-azobis (N,N-dimethylformamide Rose Bengal paraquat | oxidative stress resistance decreased [47,48,49,50] | ||
| TIM18/YOR297C | Hydrogen peroxide | oxidative stress resistance increased [51] | ||
| EOS1/YNL080C | paraquat | oxidative stress resistance increased [44] | cumene hydroperoxide hydrogen peroxide linoleic acid hydroperoxide | oxidative stress resistance: decreased [52,53,54] |
| GRR1/YJR090C | Paraquat hydrogen peroxide tert-butyl hydroperoxide dioxygen | oxidative stress resistance: decreased [53,55,56] | ||
| UBA4/YHR111W | Hydrogen peroxide | oxidative stress resistance increased [40] | 1,1′-azobis(N,N-dimethylformamide) | oxidative stress resistance: decreased [57] |
| TRR2/YHR106W | cadmium (2+) hydrogen peroxide | oxidative stress resistance: decreased [48,58,59] | ||
| TOR1/YJR066W | Hydrogen peroxide | oxidative stress resistance increased [60] | ||
| GPX1/YKL026C | Hydrogen peroxide | oxidative stress resistance: decreased [48,61] | ||
| MCR1/YKL150W | Menadione Hydrogen peroxide | oxidative stress resistance increased [40,62] | Hydrogen peroxide menadione | oxidative stress resistance decreased [62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Harati, R.; Fancello, F.; Multineddu, C.; Zara, G.; Zara, S. Screening and In Silico Analyses of the Yeast Saccharomyces cerevisiae Σ1278b Bank Mutants Using Citral as a Natural Antimicrobial. Foods 2024, 13, 1457. https://doi.org/10.3390/foods13101457
El Harati R, Fancello F, Multineddu C, Zara G, Zara S. Screening and In Silico Analyses of the Yeast Saccharomyces cerevisiae Σ1278b Bank Mutants Using Citral as a Natural Antimicrobial. Foods. 2024; 13(10):1457. https://doi.org/10.3390/foods13101457
Chicago/Turabian StyleEl Harati, Rolla, Francesco Fancello, Chiara Multineddu, Giacomo Zara, and Severino Zara. 2024. "Screening and In Silico Analyses of the Yeast Saccharomyces cerevisiae Σ1278b Bank Mutants Using Citral as a Natural Antimicrobial" Foods 13, no. 10: 1457. https://doi.org/10.3390/foods13101457






