Romanian Bee Product Analysis: Chemical Composition, Antimicrobial Activity, and Molecular Docking Insights
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples of Bee Products
2.2. Individual Polyphenols Content Detected by Liquid Chromatography-Mass Spectrometry (LC-MS)
2.3. Determination of the Antimicrobial Activity
2.3.1. Bacterial Culture
2.3.2. Fungal Culture
2.4. Molecular Docking Study
3. Results
3.1. Individual Polyphenols Content Detected by LC-MS
3.2. Antimicrobial Activity
3.3. Inhibition of Haemolysis Values and Protein Denaturation
- Inhibition of Haemolysis Values
- Inhibition of Protein Denaturation
3.4. Docking Analysis
4. Discussion
4.1. Individual Polyphenols Content Detected by LC-MS
4.2. Antimicrobial Activity
4.3. Inhibition of Haemolysis Values and Protein Denaturation
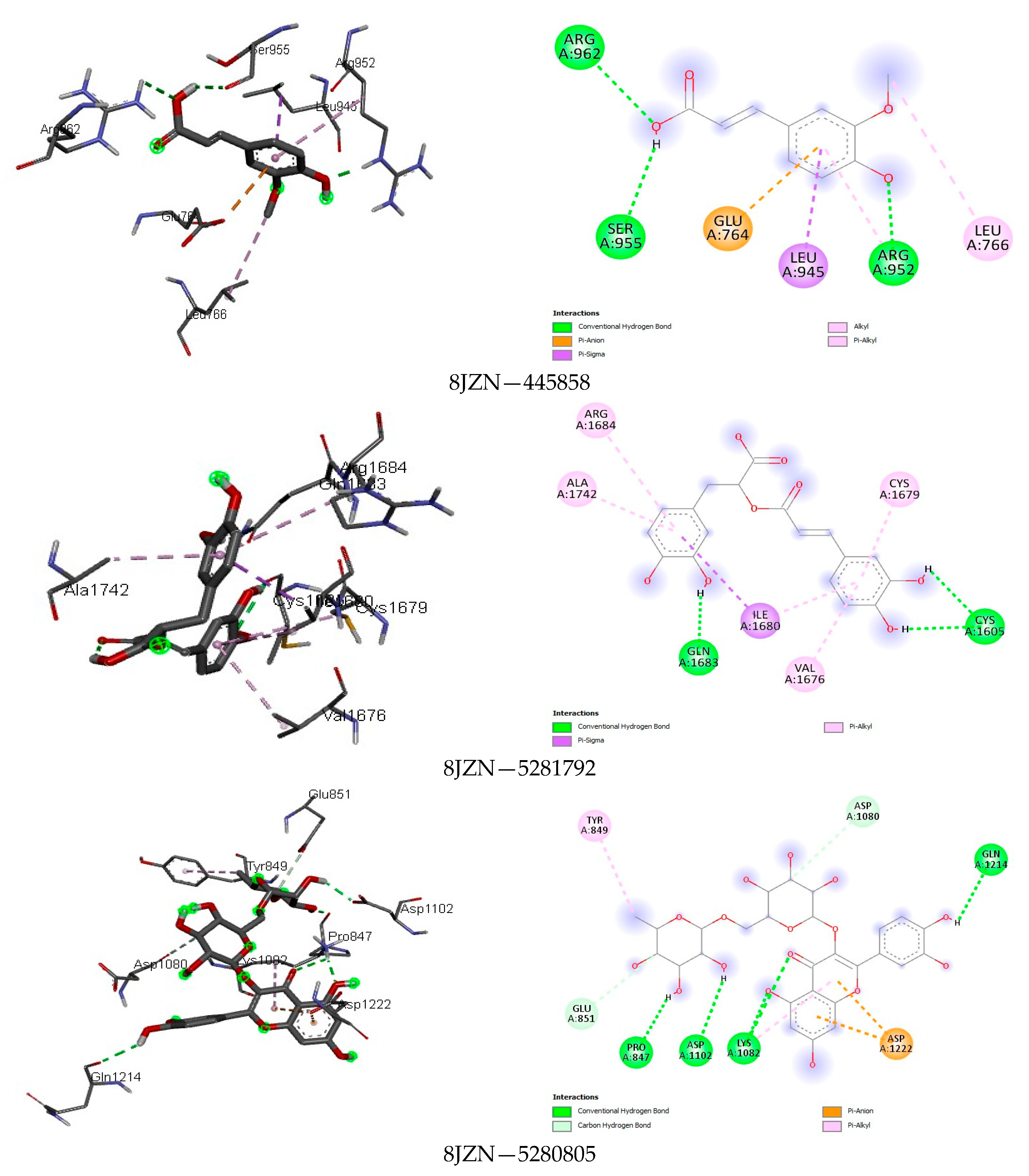
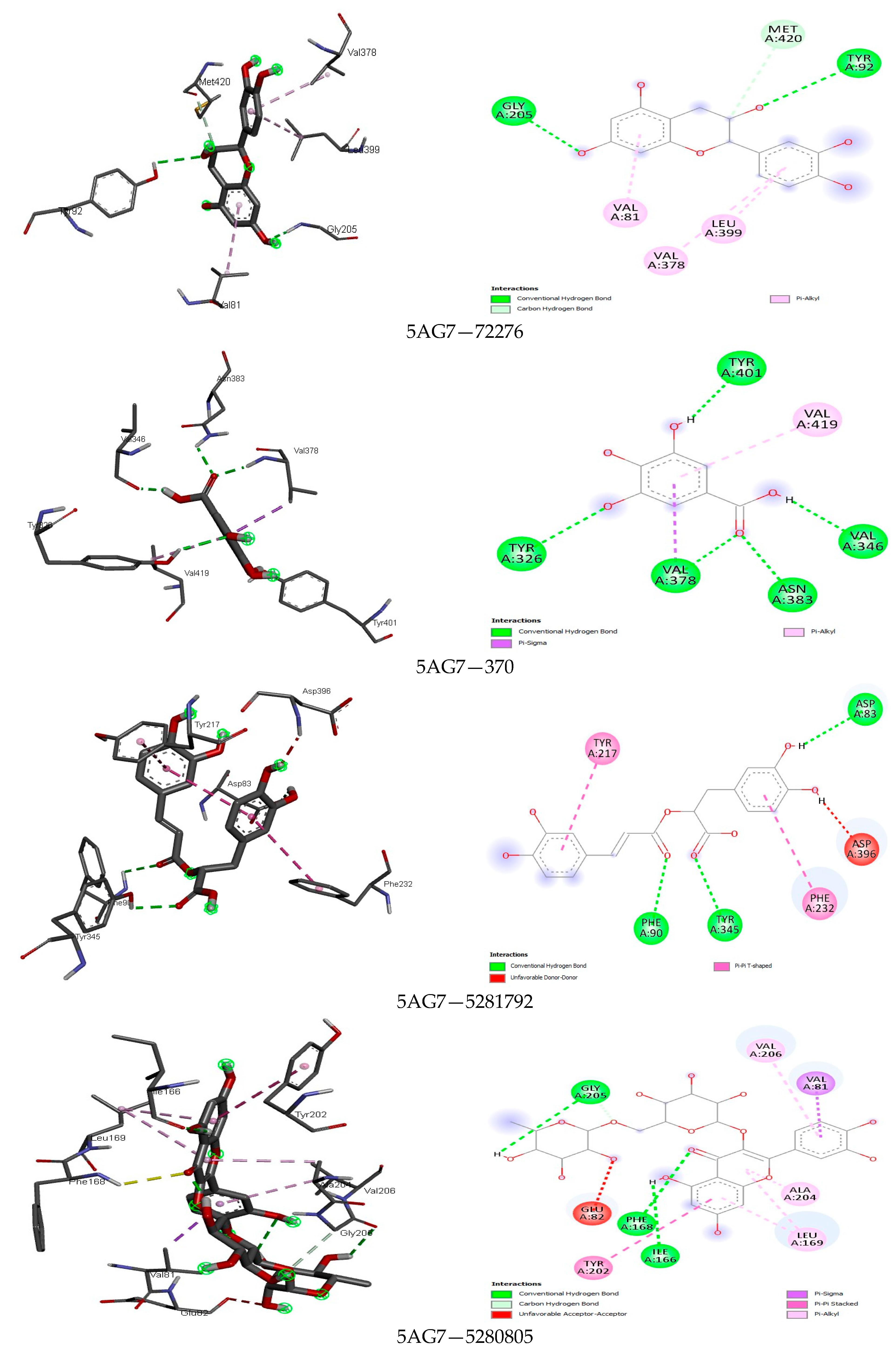
4.4. Docking Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Scumaci, D.; Giuzio, F.; Saturnino, C.; Aquaro, S.; Rosano, C.; Sinicropi, M.S. Multidrug Resistance (MDR): A Widespread Phenomenon in Pharmacological Therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef]
- Beicu, R.; Alexa, E.; Obiștioiu, D.; Cocan, I.; Imbrea, F.; Pop, G.; Circioban, D.; Moisa, C.; Lupitu, A.; Copolovici, L.; et al. Antimicrobial Potential and Phytochemical Profile of Wild and Cultivated Populations of Thyme (Thymus sp.) Growing in Western Romania. Plants 2021, 10, 1833. [Google Scholar] [CrossRef] [PubMed]
- Hulea, A.; Obiștioiu, D.; Cocan, I.; Alexa, E.; Negrea, M.; Neacșu, A.-G.; Hulea, C.; Pascu, C.; Costinar, L.; Iancu, I.; et al. Diversity of Monofloral Honey Based on the Antimicrobial and Antioxidant Potential. Antibiotics 2022, 11, 595. [Google Scholar] [CrossRef]
- Alexa, V.T.; Szuhanek, C.; Cozma, A.; Galuscan, A.; Borcan, F.; Obistioiu, D.; Dehelean, C.A.; Jumanca, D. Natural Preparations Based on Orange, Bergamot and Clove Essential Oils and Their Chemical Compounds as Antimicrobial Agents. Molecules 2020, 25, 5502. [Google Scholar] [CrossRef]
- Bota, V.; Sumalan, R.M.; Obistioiu, D.; Negrea, M.; Cocan, I.; Popescu, I.; Alexa, E. Study on the Sustainability Potential of Thyme, Oregano, and Coriander Essential Oils Used as Vapours for Antifungal Protection of Wheat and Wheat Products. Sustainability 2022, 14, 4298. [Google Scholar] [CrossRef]
- Siheri, W.; Alenezi, S.; Tusiimire, J.; Watson, D.G. The Chemical and Biological Properties of Propolis. In Bee Products—Chemical and Biological Properties; Alvarez-Suarez, J., Ed.; Springer: Cham, Switzerland, 2017; Volume 16, pp. 137–178. [Google Scholar] [CrossRef]
- Pătruică, S.; Alexa, E.; Obiștioiu, D.; Cocan, I.; Radulov, I.; Berbecea, A.; Lazăr, R.N.; Simiz, E.; Vicar, N.M.; Hulea, A.; et al. Chemical Composition, Antioxidant and Antimicrobial Activity of Some Types of Honey from Banat Region, Romania. Molecules 2022, 27, 4179. [Google Scholar] [CrossRef] [PubMed]
- Moraru, D.; Alexa, E.; Cocan, I.; Obiștioiu, D.; Radulov, I.; Simiz, E.; Berbecea, A.; Grozea, A.; Dragomirescu, M.; Vintilă, T.; et al. Chemical Characterization and Antioxidant Activity of Apilarnil, Royal Jelly, and Propolis Collected in Banat Region, Romania. Appl. Sci. 2024, 14, 1242. [Google Scholar] [CrossRef]
- De Figueiredo, S.M.; Binda, N.S.; Almeida, B.D.M.; Lemos Abreu, S.R.; De Abreu, J.A.S.; Pastore, G.M.; Sato, H.H.; Toreti, V.C.; Tapia, E.V.; Park, Y.K.; et al. Green propolis: Thirteen constituents of polar extract and total flavonoids evaluated during six years through RP-HPLC. Curr. Drug Discov. Technol. 2015, 12, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Al-Ani, I.; Zimmermann, S.; Reichling, J.; Wink, M. Antimicrobial Activities of European Propolis Collected from Various Geographic Origins Alone and in Combination with Antibiotics. Medicines 2018, 5, 2. [Google Scholar] [CrossRef]
- Devequi-Nunes, D.; Machado, B.A.S.; Barreto, G.A.; Rebouças Silva, J.; Da Silva, D.F.; Da Rocha, J.L.C.; Brandão, H.N.; Borges, V.M.; Umsza-Guez, M.A. Chemical characterisation and biological activity of six different extracts of propolis through conventional methods and supercritical extraction. PLoS ONE 2018, 13, e0207676. [Google Scholar] [CrossRef] [PubMed]
- Wieczyńska, A.; Weżgowiec, J.; Więckiewicz, W.; Czarny, A.; Kulbacka, J.; Nowakowska, D.; Gancarz, R.; Wilk, K.A. Antimicrobial activity, cytotoxicity and total phenolic content of different extracts of propolis from the west Pomeranian region in Poland. Acta Pol. Pharm. 2017, 74, 715–722. [Google Scholar]
- Przybyłek, I.; Karpiński, T.M. Antibacterial Properties of Propolis. Molecules 2019, 24, 2047. [Google Scholar] [CrossRef]
- Graikou, K.; Popova, M.; Gortzi, O.; Bankova, V.; Chinou, I. Characterization and biological evaluation of selected Mediterranean propolis samples. Is it a new type? LWT Food Sci. Technol. 2016, 65, 261–267. [Google Scholar] [CrossRef]
- Das Neves, M.V.M.; Da Silva, T.M.S.; Lima, E.D.O.; Da Cunha, E.V.L.; Oliveira, E.D.J. Isoflavone formononetin from red propolis acts as a fungicide against Candida sp. Braz. J. Microbiol. 2016, 47, 159–166. [Google Scholar] [CrossRef]
- Grecka, K.; Kuś, P.M.; Okińczyc, P.; Worobo, R.W.; Walkusz, J.; Szweda, P. The Anti-Staphylococcal potential of ethanolic Polish propolis extracts. Molecules 2019, 24, 1732. [Google Scholar] [CrossRef] [PubMed]
- Fratini, F.; Cilia, G.; Mancini, S.; Felicioli, A. Royal Jelly: An ancient remedy with remarkable antibacterial properties. Microbiol. Res. 2016, 192, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Pasupuleti, V.R.; Sammugam, L.; Ramesh, N.; Gan, S.H. Honey, propolis, and royal jelly: A comprehensive review of their biological actions and health benefits. Oxid. Med. Cell. Longev. 2017, 2017, 1259510. [Google Scholar] [CrossRef]
- Melliou, E.; Chinou, I. Chemistry and Bioactivity of Royal Jelly from Greece. J. Agric. Food Chem. 2005, 53, 8987–8992. [Google Scholar] [CrossRef]
- Altuntaș, S.; Cinar, A.; Altuntaș, V. Modelling of Listeria Monocytogenes Growth and Survival in Presence of Royal Jelly: A Promising Anti-Biofilm Agent. J. Food Nutr. Res. 2020, 59, 7–15. [Google Scholar]
- Bílikova, K.; Huang, S.C.; Linb, I.P.; Simuth, J.; Peng, C.C. Structure and Antimicrobial Activity Relationship of Royalisin, an Antimicrobial Peptide from Royal Jelly of Apis mellifera. Peptides 2015, 68, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Khosla, A.; Gupta, S.J.; Jain, A.; Shetty, D.C.; Sharma, N. Evaluation and Comparison of the Antimicrobial Activity of Royal Jelly—A Holistic Healer against Periodontopathic Bacteria. J. Indian Soc. Periodontol. 2020, 24, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Z.; Chen, Y.; Cao, J.; Tian, W.; Ma, B.; Dong, Y. Active components and biological functions of royal jelly. J. Funct. Foods. 2021, 82, 104514. [Google Scholar] [CrossRef]
- Nainu, F.; Masyita, A.; Bahar, M.A.; Raihan, M.; Prova, S.R.; Mitra, S.; Emran, T.B.; Simal-Gandara, J. Pharmaceutical Prospects of Bee Products: Special Focus on Anticancer, Antibacterial, Antiviral, and Antiparasitic Properties. Antibiotics 2021, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Sidor, E.; Dżugan, M. Drone Brood Homogenate as Natural Remedy for Treating Health Care Problem: A Scientific and Practical Approach. Molecules 2020, 25, 5699. [Google Scholar] [CrossRef] [PubMed]
- Isopescu, R.D.; Josceanu, A.M.; Cota, T.; Spulber, R. Romanian honey: Characterization and classification. Honey Anal. 2017, 27, 27–62. [Google Scholar]
- Popescu, A. Bee honey production in Romania, 2007–2015 and 2016–2020 forecast, Scientific Papers Series Management. Econ. Eng. Agric. Rural. Dev. 2023, 17, 339–349. [Google Scholar]
- Gatea, F.; Matei, A.O.; Teodor, E.D.; Radu, G.L. Antioxidant properties and polyphenols composition of some Romanian propolis samples. Rev. Roum. Chim. 2014, 60, 65–74. [Google Scholar]
- Mărghitaş, L.A.; Dezmirean, D.; Drâglă, F.; Bobiş, O. Caffeic acid phenethyl ester (CAPE) in Romanian propolis. Bull. Univ. Agric. Sci. Veter-Med. Cluj-Napoca. Anim. Sci. Biotechnol. 2014, 71, 111–112. [Google Scholar]
- Dezmirean, D.S.; Mărghitaş, L.A.; Chirilă, F.; Copaciu, F.; Simonca, V.; Bobiş, O.; Erler, S. Influence of geographic origin, plant source and polyphenolic substances on antimicrobial properties of propolis against human and honey bee pathogens. J. Apic. Res. 2017, 56, 588–597. [Google Scholar] [CrossRef]
- Paşca, C.; Dezmirean, D.S.; Bobiş, O.; Mărghitaș, L.A.; Bonta, V. Biotechnological potential of apilarnil and royal jelly used in obtaining some functional foods. Sci. Pap.-Anim. Sci. Ser. Lucr. Ştiinţifice Ser. Zooteh. 2019, 75, 81–86. [Google Scholar]
- Obistioiu, D.; Hulea, A.; Cocan, I.; Alexa, E.; Negrea, M.; Popescu, I.; Herman, V.; Imbrea, I.M.; Heghedus-Mindru, G.; Suleiman, M.A.; et al. Boswellia Essential Oil: Natural Antioxidant as an Effective Antimicrobial and Anti-Inflammatory Agent. Antioxidants 2023, 12, 1807. [Google Scholar] [CrossRef] [PubMed]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; Hassane Anouar, E.; Aouadi, K.; Snoussi, M.; Kadri, A. New substituted pyrazolones and dipyrazolotriazines as promising tyrosyltRNA synthe-tase and peroxiredoxin-5 inhibitors: Design, synthesis, molecular docking and struc-ture-activity relationship (SAR) analysis. Bioorg. Chem. 2021, 109, 104704. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Hackbarth, C.; Ni, Z.J.; Wu, C.; Wang, W.; Jain, R.; He, Y.; Bracken, K.; Weidmann, B.; Patel, D.V.; et al. Peptide Deformylase Inhibitors as Antibacterial Agents: Identification of VRC3375, a Proline-3-Alkylsuccinyl Hydroxamate Derivative, by Using an Integrated Combinatorial and Medicinal Chemistry Approach. Antimicrob. Agents Chemother. 2004, 48, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Nollmann, M.; Crisona, N.J.; Arimondo, P.B. Thirty years of Escherichia coli DNA gyrase: From in vivo function to single molecule mechanism. Biochimie 2007, 89, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by an-ticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef]
- Nowosielski, M.; Hoffmann, M.; Wyrwicz, L.S.; Stepniak, P.; Plewczynski, D.M.; Lazniewski, M.; Ginalski, K.; Rychlewski, L. Detailed mechanism of squalene epoxidase inhibition by terbinafine. J. Chem. Inf. Model. 2011, 51, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Shematek, E.M.; Braatz, J.A.; Cabib, E.J. Preparation and properties of beta-(1 leads to 3)glucan synthetase. Biol. Chem. 1980, 255, 888–894. [Google Scholar] [CrossRef]
- Zhu, J.; Sheng, C.Q.; Zhang, W.N. N-myristoyltransferase: A novel antifungal target. Yao Xue Xue Bao 2005, 40, 775–781. [Google Scholar] [PubMed]
- Farazi, T.A.; Waksman, G.; Gordon, J.I. Structures of Saccharomyces cerevisiae N myristoyltransferase with bound myristoylCoA and peptide provide insights about sub-strate recognition and catalysis. Biochemistry 2001, 40, 6335–6343. [Google Scholar] [CrossRef]
- Gohlk, H.; Klebe, G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew. Chem. Int. Ed. Engl. 2002, 41, 2644–2676. [Google Scholar] [CrossRef]
- Cadariu, A.I.; Cocan, I.; Negrea, M.; Alexa, E.; Obistioiu, D.; Hotea, I.; Radulov, I.; Poiana, M. Exploring the potential of tomato processing by-product as a natural antioxidant in nitrite-free reformulated sausages. Sustainability 2022, 14, 11802. [Google Scholar] [CrossRef]
- Dégi, J.; Herman, V.; Igna, V.; Dégi, D.M.; Hulea, A.; Muselin, F.; Cristina, R.T. Antibacterial Activity of Romanian Propolis against Staphylococcus aureus Isolated from Dogs with Superficial Pyoderma: In Vitro Test. Vet. Sci. 2022, 9, 299. [Google Scholar] [CrossRef]
- Obistioiu, D.; Cocan, I.; Tîrziu, E.; Herman, V.; Negrea, M.; Cucerzan, A.; Neacsu, A.-G.; Cozma, A.L.; Nichita, I.; Hulea, A.; et al. Phytochemical Profile and Microbiological Activity of Some Plants Belonging to the Fabaceae Family. Antibiotics 2021, 10, 662. [Google Scholar] [CrossRef]
- ISO 20776-1:2019. Available online: https://www.iso.org/standard/70464 (accessed on 4 May 2024).
- Available online: http://www.pdb.org (accessed on 18 December 2023).
- Parmenopoulou, V.; Kantsadi, A.L.; Tsirkone, V.G.; Chatzileontiadou, D.S.M.; Manta, S.; Zogra-phos, S.E.; Kollatos, N.; Archontis, G.; Agius, L.; Hayes, J.M.; et al. Bioorg. Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-β-d-glucopyranosylamines. Med. Chem. 2014, 22, 4810–4825. [Google Scholar] [CrossRef]
- Elkolli, M.; Elkolli, H.; Alam, M.; Benguerba, Y. In silico study of antibacterial tyrosyl-tRNA syn-thetase and toxicity of main phytoconstituents from three active essential oils. J. Biomol. Struct. Dyn. 2024, 42, 1404–1416. [Google Scholar] [CrossRef]
- Woźniak, M.; Sip, A.; Mrówczyńska, L.; Broniarczyk, J.; Waśkiewicz, A.; Ratajczak, I. Biological Activity and Chemical Composition of Propolis from Various Regions of Poland. Molecules 2023, 28, 141. [Google Scholar] [CrossRef]
- Galeotti, F.; Maccari, F.; Fachini, A.; Volpi, N. Chemical Composition and Antioxidant Activity of Propolis Prepared in Different Forms and in Different Solvents Useful for Finished Products. Foods 2018, 7, 41. [Google Scholar] [CrossRef]
- Kim, D.H.; Auh, J.-H.; Oh, J.; Hong, S.; Choi, S.; Shin, E.J.; Woo, S.O.; Lim, T.-G.; Byun, S. Propolis Suppresses UV-Induced Photoaging in Human Skin through Directly Targeting Phosphoinositide 3-Kinase. Nutrients 2020, 12, 3790. [Google Scholar] [CrossRef]
- Grassi, G.; Capasso, G.; Gambacorta, E.; Perna, A.M. Chemical and Functional Characterization of Propolis Collected from Different Areas of South Italy. Foods 2023, 12, 3481. [Google Scholar] [CrossRef] [PubMed]
- Kurek-Górecka, A.; Keskin, Ş.; Bobis, O.; Felitti, R.; Górecki, M.; Otręba, M.; Stojko, J.; Olczyk, P.; Kolayli, S.; Rzepecka-Stojko, A. Comparison of the Antioxidant Activity of Propolis Samples from Different Geographical Regions. Plants 2022, 11, 1203. [Google Scholar] [CrossRef]
- Salatino, A.; Salatino, M.L.F.; Negri, G. How diverse is the chemistry and plant origin of Brazilian propolis? Apidologie 2021, 52, 1075–1097. [Google Scholar] [CrossRef]
- Pujirahayu, N.; Suzuki, T.; Katayama, T. Cycloartane-Type Triterpenes and Botanical Origin of Propolis of Stingless Indonesian Bee Tetragonula sapiens. Plants 2019, 8, 57. [Google Scholar] [CrossRef]
- Qiao, J.; Wang, Y.; Zhang, Y.; Kong, L.; Zhang, H. Botanical Origins and Antioxidant Activities of Two Types of Flavonoid-Rich Poplar-Type Propolis. Foods 2023, 12, 2304. [Google Scholar] [CrossRef]
- Touzani, S.; Imtara, H.; Katekhaye, S.; Mechchate, H.; Ouassou, H.; Alqahtani, A.S.; Noman, O.M.; Nasr, F.A.; Fearnley, H.; Fearnley, J.; et al. Determination of Phenolic Compounds in Various Propolis Samples Collected from an African and an Asian Region and Their Impact on Antioxidant and Antibacterial Activities. Molecules 2021, 26, 4589. [Google Scholar] [CrossRef]
- Kasote, D.M.; Pawar, M.V.; Bhatia, R.S.; Nandre, V.S.; Gundu, S.S.; Jagtap, S.D.; Kulkarni, M.V. HPLC, NMR based chemical profiling and biological characterisation of Indian propolis. Fitoterapia 2017, 122, 52–60. [Google Scholar] [CrossRef]
- Lv, L.; Cui, H.; Ma, Z.; Liu, X.; Yang, L. Recent progresses in the pharmacological activities of caffeic acid phenethyl ester. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 394, 1327–1339. [Google Scholar] [CrossRef]
- Osés, S.M.; Marcos, P.; Azofra, P.; de Pablo, A.; Fernández-Muíño, M.Á.; Sancho, M.T. Phenolic Profile, Antioxidant Capacities and Enzymatic Inhibitory Activities of Propolis from Different Geographical Areas: Needs for Analytical Harmonization. Antioxidants 2020, 9, 75. [Google Scholar] [CrossRef]
- Ozturk, G.; Ginis, Z.; Akyol, S.; Erden, G.; Gurel, A.; Akyol, O. The anticancer mechanism of caffeic acid phenethyl ester (CAPE): Review of melanomas, lung and prostate cancers. Eur. Rev. Med. Pharmacol. Sci. 2012, 16, 2064–2068. [Google Scholar]
- Duca, A.; Sturza, A.; Moacă, E.A.; Negrea, M.; Lalescu, V.-D.; Lungeanu, D.; Dehelean, C.-A.; Muntean, D.-M.; Alexa, E. Identification of Resveratrol as Bioactive Compound of Propolis from Western Romania and Characterization of Phenolic Profile and Antioxidant Activity of Ethanolic Extracts. Molecules 2019, 24, 3368. [Google Scholar] [CrossRef]
- Nichitoi, M.M.; Josceanu, A.M.; Isopescu, R.D.; Isopencu, G.O.; Geana, E.-I.; Ciucure, C.T.; Lavric, V. Polyphenolics profile effects upon the antioxidant and antimicrobial activity of propolis extracts. Sci. Rep. 2021, 11, 20113. [Google Scholar] [CrossRef]
- Brenelli de Paiva, L.; Goldbeck, R.; Dantas dos Santos, W.; Squina, F.M. Ferulic acid and derivatives: Molecules with potential application in the pharmaceutical field. Braz. J. Pharm. Sci. 2013, 49, 395–411. [Google Scholar] [CrossRef]
- Kumar, N.; Goel, N. Phenolic acids: Natural versatile molecules with promising therapeutic applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef]
- Prasad, R.; Pradad, S.B. A review on the chemistry and biological properties of Rutin, a promising nutraceutical agent. Asian J. Pharm Pharm. 2019, 5, 1–20. [Google Scholar] [CrossRef]
- Altun, S.K.; Aydemir, M.E. Determination of Antioxidant Capacities with the Phenolic and Flavonoid Contents of Royal Jelly Mixtures. Bee Stud. 2022, 14, 9–15. [Google Scholar] [CrossRef]
- Balkanska, R.; Mărghitaș, L.A.; Pavel, C.I.; Ignatova, M.; Tomoș, L.I. Comparison of physicochemical parameters in royal jelly from Romania and Bulgaria. Bull UASVM Anim. Sci. Biotech. 2013, 70, 117–121. [Google Scholar] [CrossRef]
- Mureşan, C.I.; Mãrghitaş, L.A.; Dezmirean, D.S.; Bobiş, O.; Bonta, V.; Zacharias, I.; Mãrgãoan, R.; Paşca, C. Quality Parameters for commercialized Royal Jelly. Bull. Univ. Agric. Sci. Veter-Med. Cluj-Napoca. Anim. Sci. Biotechnol. 2016, 73, 37–44. [Google Scholar] [CrossRef]
- El-Wahed, A.A.; Khalifa, S.A.M.; Aldahmash, B.; Zhang, H.; du, M.; Zhao, C.; Tahir, H.E.; Saeed, A.; Hussain, H.; Guo, Z.; et al. Exploring the Chemical Constituents and Nutritive Potential of Bee Drone (Apilarnil): Emphasis on Antioxidant Properties. Chem. Biodivers. 2024, e202400085. [Google Scholar] [CrossRef]
- Koşum, N.; Yücel, B.; Kandemir, Ç.; Taşkın, T.; Duru, M.E.; Küçükaydın, S.; Margaoan, R.; Cornea-Cipcigan, M. Chemical Composition and Androgenic Effect of Bee Drone Larvae (Apilarnil) for Goat Male Kids. Chem. Biodivers. 2022, 19, e202200548. [Google Scholar] [CrossRef]
- Inci, H.; Izol, E.; Yilmaz, M.A.; Ilkaya, M.; Bingöl, Z.; Gülçin, I. Comprehensive Phytochemical Content by LC/MS/MS and Anticholinergic, Antiglaucoma, Antiepilepsy, and Antioxidant Activity of Apilarnil (Drone Larvae). Chem. Biodivers. 2023, 20, e202300654. [Google Scholar] [CrossRef]
- Smiljkovic, M.; Stanisavljevic, D.; Stojkovic, D.; Petrovic, I.; Vicentic, J.M.; Popovic, J.; Golic Grdadolnik, S.; Markovic, D.; Sanković-Babić, S.; Glamoclija, J.; et al. Apigenin-7-O-glucoside versus apigenin: Insight into the modes of anticandidal and cytotoxic actions. EXCLI J. 2017, 16, 795–807. [Google Scholar] [PubMed]
- Ivanov, M.; Kannan, A.; Stojković, D.; Glamočlija, J.; Grdadolnik, S.G.; Sanglard, D.; Soković, M. Revealing the astragalin mode of anticandidal action. EXCLI J. 2020, 19, 1436–1445. [Google Scholar] [PubMed]
- Ivanov, M.; Novović, K.; Malešević, M.; Dinić, M.; Stojković, D.; Jovčić, B.; Soković, M. Polyphenols as Inhibitors of Antibiotic Resistant Bacteria—Mechanisms Underlying Rutin Interference with Bacterial Virulence. Pharmaceuticals 2022, 15, 385. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, M.; Walker, S. Envelope Structures of Gram-Positive Bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Uthaibutra, V.; Kaewkod, T.; Prapawilai, P.; Pandith, H.; Tragoolpua, Y. Inhibition of Skin Pathogenic Bacteria, Antioxidant and Anti-Inflammatory Activity of Royal Jelly from Northern Thailand. Molecules 2023, 28, 996. [Google Scholar] [CrossRef] [PubMed]
- Amly, D.A.; Hajardhini, P.; Jonarta, A.L.; Dedy, H.; Yulianto, K.; Susilowati, H. Enhancement of Pyocyanin Production by Subinhibitory Concentration of Royal Jelly in P. aeruginosa. F1000Research 2021, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.; Finola, M.; Marioli, J. Bioassay Directed Identification of Royal Jelly’s Active Compounds against the Growth of Bacteria Capable of Infecting Cutaneous Wounds. Adv. Microbiol. 2013, 3, 138–144. [Google Scholar] [CrossRef]
- Colomar, A. Apilarnil Therapy: The Healing Power of Bee Larvae. J. Apitherapy 2023, 10, 01–02. [Google Scholar]
- Corrêa, J.L.; Veiga, F.F.; Jarros, I.C.; Costa, M.I.; Castilho, P.F.; de Oliveira, K.M.P.; Rosseto, H.C.; Bruschi, M.L.; Svidzinski, T.I.E.; Negri, M. Propolis extract has bioactivity on the wall and cell membrane of Candida albicans. J. Ethnopharmacol. 2020, 256, 112791. [Google Scholar] [CrossRef]
- Herrera, C.I.; Alvear, M.; Barrientos, I.; Montenegro, G.; Salazar, I.A. The antifungal effect of six commerical extracts of Chilean propolis on Candida spp. Cienc. Investig. Agrar. 2010, 37, 75–84. [Google Scholar]
- Teodoro, G.R.; Ellepola, K.; Seneviratne, C.J.; Koga-Ito, C.Y. Potential Use of Phenolic Acids as Anti-Candida Agents: A Review. Front. Microbiol. 2015, 6, 1420. [Google Scholar] [CrossRef]
- Koç, A.N.; Silici, S.; Kasap, F.; Hörmet-Oz, H.T.; Mavus-Buldu, H.; Ercal, B.D. Antifungal activity of the honeybee products against Candida spp. and Trichosporon spp. J. Med. Food 2011, 14, 128–134. [Google Scholar] [CrossRef]
- Helen, L.B.; Roberts, A.E.L.; Cooper, R.; Jenkins, R.E. A review of selected bee products as potential antibacterial, anti-fungal, and antiviral agents. Med. Res. Arch. 2016, 4, 1–20. [Google Scholar] [CrossRef]
- Moghim, H.; Taghipour, S.; Kheiri, S.; Khabbazi, H.; Baradaran, A. Antifungal Effects of Iranian Propolis Extract and Royal jelly Against Candida albicans In-Vitro. Int. J. Prev. Med. 2021, 12, 163. [Google Scholar] [CrossRef]
- Mendez-Encinas, M.A.; Valencia, D.; Ortega-García, J.; Carvajal-Millan, E.; Díaz-Ríos, J.C.; Mendez-Pfeiffer, P.; Soto-Bracamontes, C.; Garibay-Escobar, A.; Alday, E.; Velazquez, C. Anti-Inflammatory Potential of Seasonal Sonoran Propolis Extracts and Some of Their Main Constituents. Molecules 2023, 28, 4496. [Google Scholar] [CrossRef]
- Pahlavani, N.; Malekahmadi, M.; Firouzi, S.; Rostami, D.; Sedaghat, A.; Moghaddam, A.B.; Ferns, G.A.; Navashenaq, J.G.; Reazvani, R.; Safarian, M.; et al. Molecular and cellular mechanisms of the effects of Propolis in inflammation, oxidative stress and glycemic control in chronic diseases. Nutr. Metab. 2020, 17, 65. [Google Scholar] [CrossRef]
- Bagameri, L.; Botezan, S.; Bobis, O.; Bonta, V.; Dezmirean, D.S. Molecular Insights into Royal Jelly Anti-Inflammatory Properties and Related Diseases. Life 2023, 13, 1573. [Google Scholar] [CrossRef]
- Choi, S.-S.; Park, H.-R.; Lee, K.-A. A Comparative Study of Rutin and Rutin Glycoside: Antioxidant Activity, Anti-Inflammatory Effect, Effect on Platelet Aggregation and Blood Coagulation. Antioxidants 2021, 10, 1696. [Google Scholar] [CrossRef]
- Shanmugasundaram, D.; Roza, J.M. Assessment of Anti-Inflammatory and Antioxidant Activities of a Proprietary Preparation of Quercetin–Rutin Blend (SophorOx™) in Exercised Rats. Sci. World J. 2024, 2024, 9063936. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Qiu, W.; Shi, Y. Ferulic acid exhibits anti-inflammatory effects by inducing autophagy and blocking NLRP3 inflammasome activation. Mol. Cell Toxicol. 2022, 18, 509–519. [Google Scholar] [CrossRef]
- Lopes, R.; Costa, M.; Ferreira, M.; Gameiro, P.; Fernandes, P.; Catarino, C.; Santos-Silva, A.; Paiva-Martins, F. Caffeic acid phenolipids in the protection of cell membranes from oxidative injuries. Interaction with the membrane phospholipid bilayer. Biochim. Biophys. Acta (BBA) Biomembr. 2021, 1863, 183727. [Google Scholar] [CrossRef]
- Gupta, A.; Ganeshpurkar, A.; Bansal, D.; Dubey, N. Protective effect of ferulic acid on erythrocytes against experimental oxidative stress models. Toxicol. Int. 2016, 23, 49–53. [Google Scholar] [CrossRef]
- Dai, F.; Miao, Q.; Zhou, B.; Yang, L.; Liu, Z.-L. Protective effects of flavonols and their glycosides against free radical-induced oxidative hemolysis of red blood cells. Life Sci. 2006, 78, 2488–2493. [Google Scholar] [CrossRef]
- Sun, B.; Gou, Y.; Xue, Z.; Zheng, X.; Ma, Y.; Hu, F.; Zhao, W. Protections of bovine serum albumin protein from damage on functionalized graphene-based electrodes by flavonoids. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 62, 197–205. [Google Scholar] [CrossRef]
- Drăgan, M.; Stan, C.D.; Pânzariu, A.; Profire, L. Evaluation of anti-inflammatory potential of some new ferullic acid derivatives. Farmacia 2016, 64, 194–197. [Google Scholar]
- Drăgan, M.; Tătărîngă, G.; Mircea, C.; Cioancă, O.; Dragostin, O.; Iacob, A.T.; Profire, L.; Stan, C.D. Ferulic acid—A versatile molecule. Acta Biol. Marisiensis J. 2018, 1, 53–60. [Google Scholar] [CrossRef]
- Park, E.H.; Kim, S.H.; Park, S.S. Anti-inflammatory activity of propolis. Arch. Pharmacal Res. 1996, 19, 337–341. [Google Scholar] [CrossRef]
- Yahya, R.; Al-Rajhi, A.M.H.; Alzaid, S.Z.; Al Abboud, M.A.; Almuhayawi, M.S.; Al Jaouni, S.K.; Selim, S.; Ismail, K.S.; Abdelghany, T.M. Molecular Docking and Efficacy of Aloe vera Gel Based on Chitosan Nanoparticles against Helicobacter pylori and Its Antioxidant and AntiInflammatory Activities. Polymers 2022, 14, 2994. [Google Scholar] [CrossRef]
- Merzoug, A.; Chikhi, A.; Bensegueni, A.; Boucherit, H.; Okay, S. Virtual Screening Approach of Bacterial Peptide Deformylase Inhibitors Result in New Antibiotics. Mol. Inform. 2017, 36, 1700087. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, S.K.; Yoon, H.J.; Lee, H.H.; Kim, K.K.; Lee, B.J.; Lee, B.I.; Suh, S.W. Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors. Acta. Crystallogr. 2012, 68, 784–793. [Google Scholar]
- Sanjay, K.V.; Jat, R.K.; Narendra, N.; Rajkumar, S.; Vineet, S.; Sanjay, P.; Kuldeep, B. A novel antibacterian target: Peptide deformylase. Pharmacophore 2011, 2, 114–123. [Google Scholar]
- Sissi, C.; Palumbo, M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009, 37, 702–711. [Google Scholar] [CrossRef]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef]
- Ganeshpurkar, A.; Saluja, A.K. The pharmacological potential of rutin. Saudi Pharm. J. 2017, 25, 149–164. [Google Scholar] [CrossRef]
- Fang, W.; Robinson, D.A.; Raimi, O.G.; Blair, D.E.; Harrison, J.R.; Lockhart, D.E.A.; Torrie, L.S.; Ruda, G.F.; Wyatt, P.G.; Gilbert, I.H.; et al. N-Myristoyltransferase is a cell wall target in Aspergillus fumigatus. ACS Chem. Biol. 2015, 10, 1425–1434. [Google Scholar] [CrossRef]
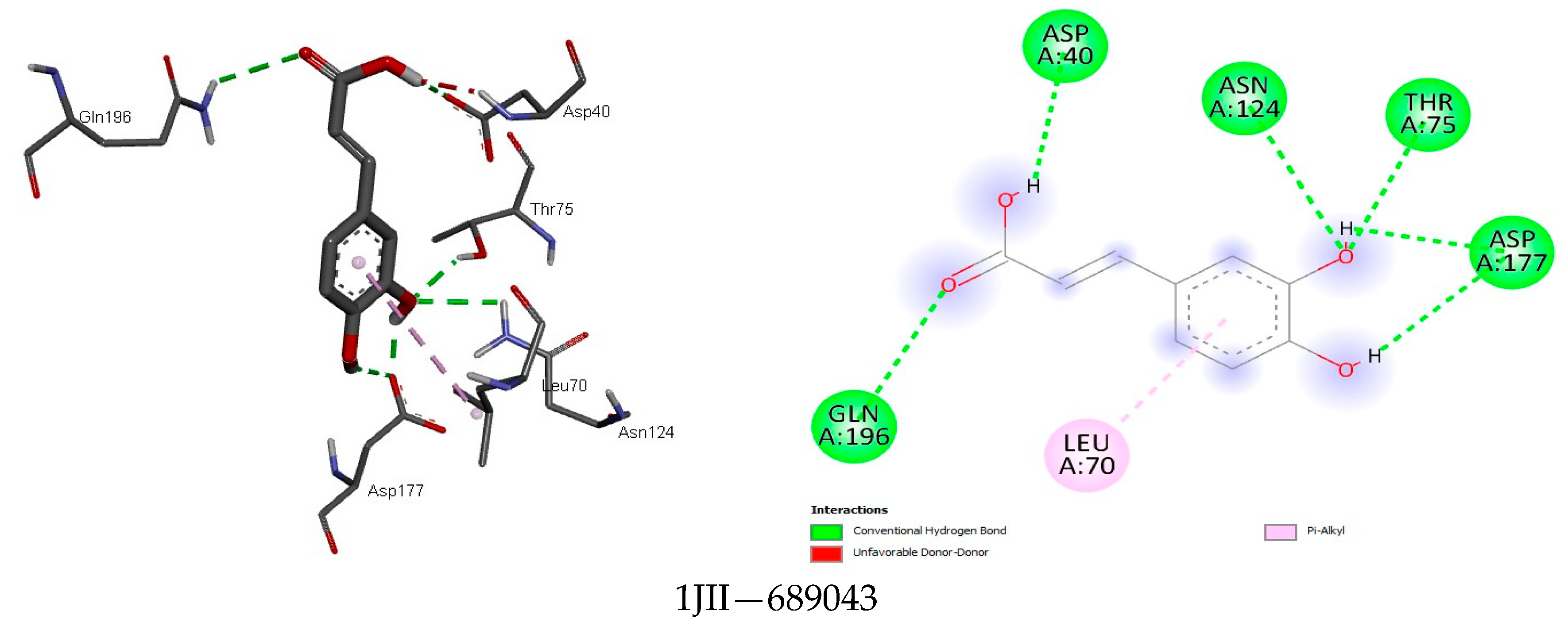
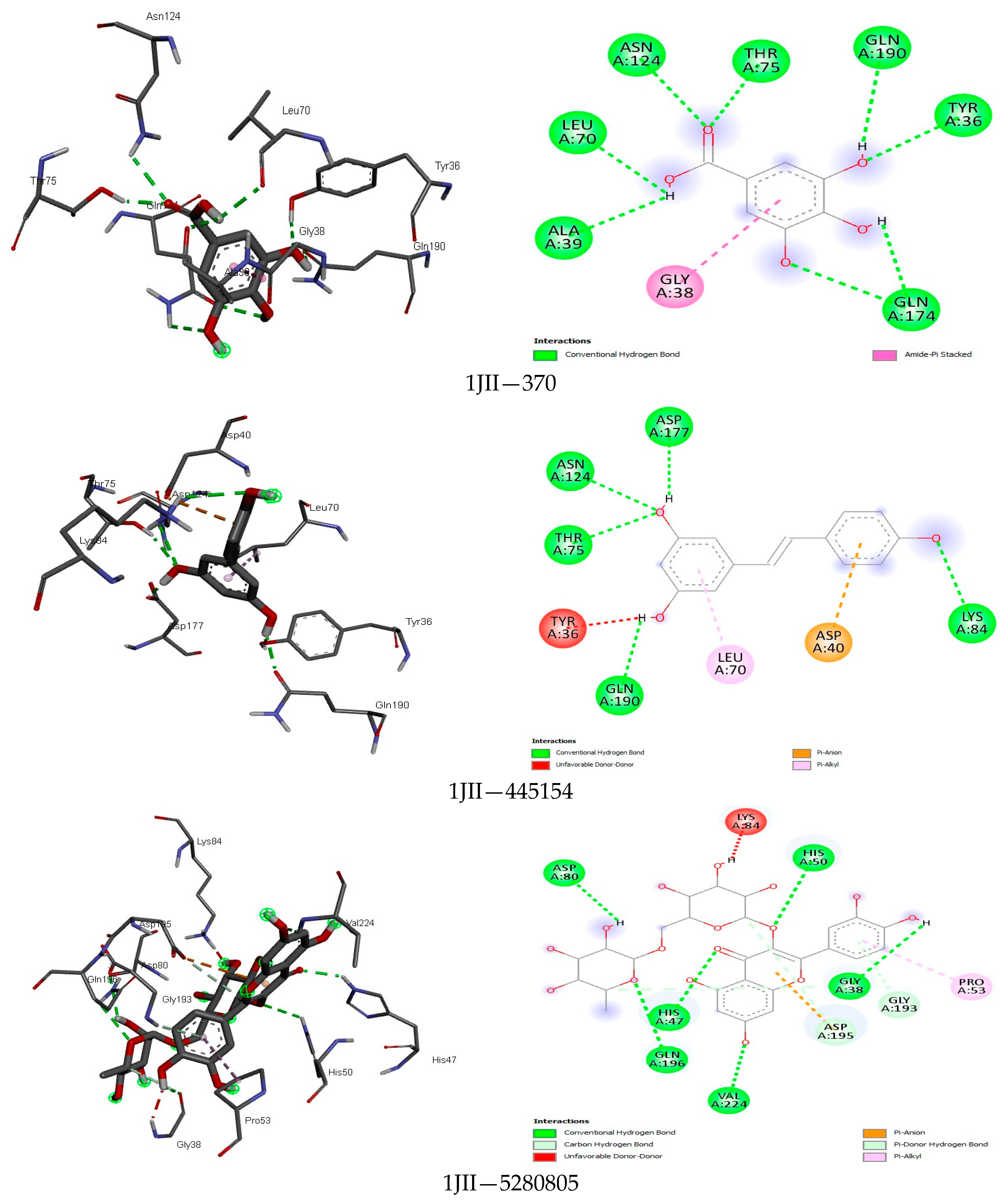




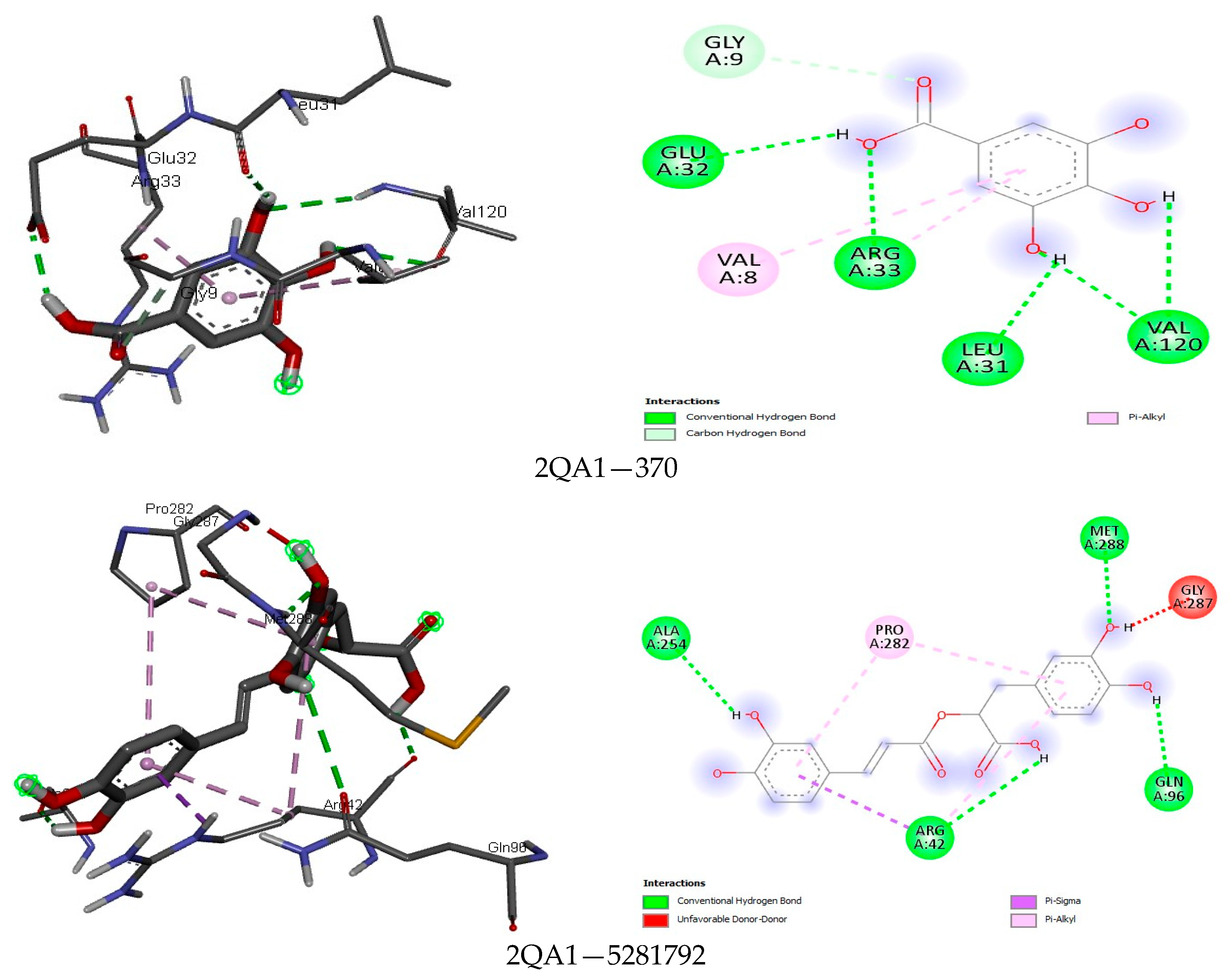
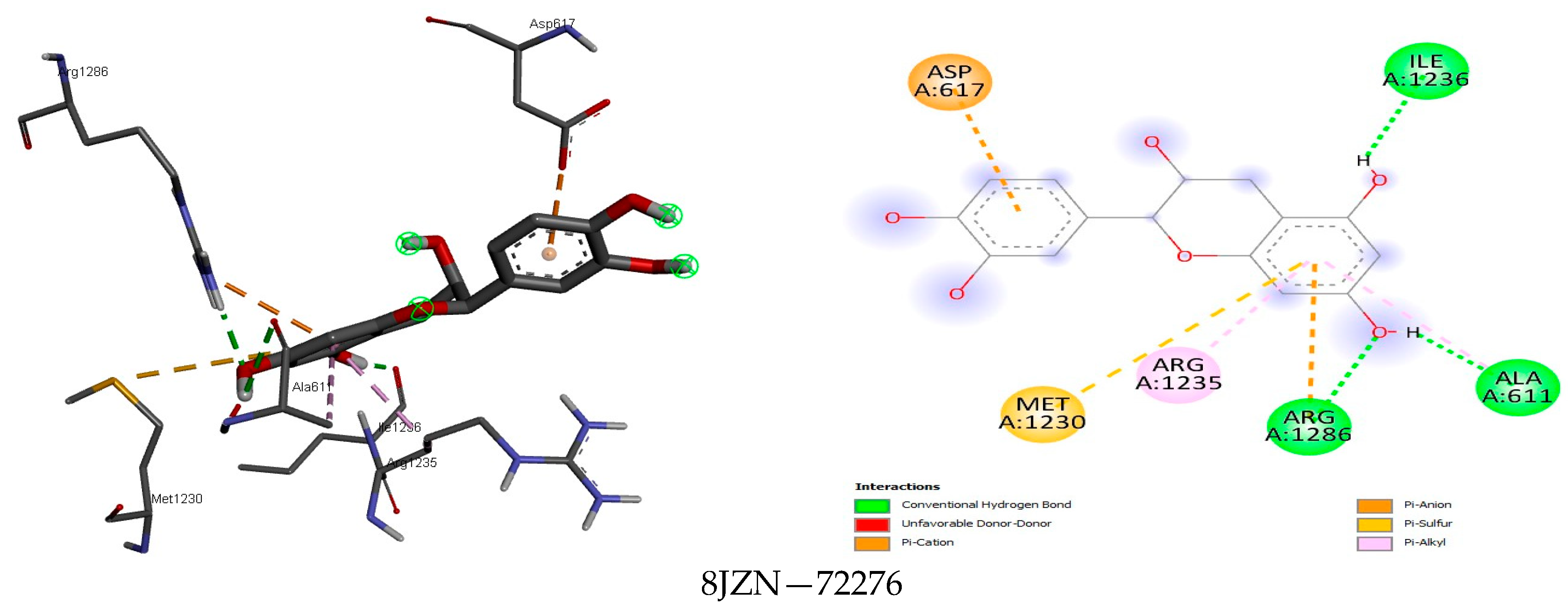
| Class | Compound | MASP | CP | PT | MASRJ | CLRJ | CLJ | MASA | CLA | CA |
|---|---|---|---|---|---|---|---|---|---|---|
| Flavonoids | Epicatechin (µg/g d.w.) | 73.90 ± 1.20 a | 73.90 ± 1.85 a | 74.20 ± 1.86 a | 73.80 ± 1.85 a | 74.20 ± 1.86 a | 74.00 ± 1.85 a | 73.90 ± 1.85 a | 74.20 ± 1.86 a | 74.00 ± 1.85 a |
| Rutin (µg/g d.w.) | 79.30 ± 1.98 a | 79.30 ± 1.98 a | 79.30 ± 1.98 a | nd * | nd | nd | 79.30 ± 1.98 a | 79.30 ± 1.98 a | 79.00 ± 4.38 a | |
| Quercitin (µg/g d.w.) | 15.60 ± 0.39 a | 16.80 ± 1.92 c | 15.70 ± 0.39 a | 15.78 ± 0.39 a | 15.90 ± 0.40 a,b | 15.70 ± 0.39 a | 16.10 ± 0.40 b | 16.50 ± 0.41 b,c | 15.80 ± 0.40 a,b | |
| Total (µg/g d.w.) | 168.8 ± 3.57 | 170 ± 5.75 | 169.2 ± 5.76 | 89.58 ± 2.24 | 90.1 ± 2.26 | 89.7 ± 2.24 | 169.3 ± 4.23 | 170 ± 4.25 | 168.8 ± 6.63 | |
| Phenolic acids | Gallic acid (µ/gd.w.) | 17.70 ± 0.44 b | 16.90 ± 0.42 a | 16.90 ± 0.42 | nd | nd | nd | nd | nd | nd |
| β-rezolcilic acid (µg/g d.w.) | nd | nd | nd | nd | 175.20 ± 4.38 a | 175.00 ± 4.38 a | 175.10 ± 4.38 a | 175.10 ± 4.38 a | 175.00 ± 4.38 a | |
| Caffeic acid (µg/g d.w.) | nd | nd | nd | nd | nd | nd | 18.30 ± 0.46 a | 19.40 ± 0.49 b | 19.30 ± 0.38 a | |
| Cumaric acid (µg/g d.w.) | 41.00 ± 1.03 c | 50.90 ± 5.42 d | 37.00 ± 0.93 a | 39.60 ± 0.99 b | 39.60 ± 0.99 b | 39.10 ± 0.98 b | 39.80 ± 0.02 b | 37.70 ± 0.94 a | 40.80 ± 1.02 c | |
| Ferulic acid (µg/g d.w.) | 126.40 ± 3.16 a | 198.72 ± 12.47 c | 127.00 ± 3.18 a | 129.60 ± 3.24 b | 126.60 ± 3.17 a | 127.80 ± 3.20 a | 126.80 ± 3.17 a | 126.10 ± 3.15 a | 126.20 ± 3.16 a | |
| Rosmarinic acid (µg/g d.w.) | 67.90 ± 1.70 a | 68.60 ± 1.72 a,b | 87.50 ± 2.19 c | 68.10 ± 1.70 a,b | 69.10 ± 1.73 b | 68.20 ± 1.71 a,b | 67.90 ± 1.70 a | 68.00 ± 1.70 a | 68.00 ± 1.70 a | |
| Total (µg/g d.w.) | 253 ± 6.33 | 335.2 ± 20.03 | 268.4 ± 6.72 | 237.3 ± 5.93 | 410.5 ± 10.27 | 410.1 ± 10.27 | 427.9 ± 9.73 | 426.3 ± 10.66 | 429.3 ± 10.64 | |
| Stilbene | Resveratrol (µg/g d.w.) | 137.70 ± 3.44 c | 122.60 ± 3.07 a | 132.70 ± 3.32 b,c | 119.10 ± 2.98 a | 233.30 ± 5.83 e | 226.60 ± 5.67 e | 135.00 ± 4.38 a | 128.20 ± 3.21 b | 160.00 ± 4.00 d |
| Total (µg/g d.w.) | 137.70 ± 3.44 c | 122.60 ± 3.07 a | 132.70 ± 3.32 b,c | 119.10 ± 2.98 a | 233.30 ± 5.83 e | 226.60 ± 5.67 e | 135.00 ± 4.38 a | 128.20 ± 3.21 b | 160.00 ± 4.00 d |
| Sample mg/mL/Strain | S. pyogenes ATCC 19615 | S. aureus ATCC 25923 | B. cereus ATCC 10876 | Cl perfringens ATCC 13124 | L. monocytogenes ATCC 19114 |
|---|---|---|---|---|---|
| MASP 2.5 | −126.65 | 30.25 | −78.55 | −22.46 | −13.61 |
| MASP 5 | −146.08 | 19.61 | −51.04 | −31.69 | 1.29 |
| MASP 7.5 | −147.75 | 13.12 | −19.9 | −42.26 | 7.9 |
| MASP 10 | −149.53 | 4.42 | −19.15 | −43.38 | 13.94 |
| MASP 25 | −163.22 | −0.09 | −16.32 | −137.44 | 17.44 |
| MASP 50 | −168.76 | −11.88 | −12.92 | −113.74 | 28.87 |
| MASP 75 | −196.87 | −20.49 | −11.42 | −176.92 | 34.26 |
| MASP 100 | −210.97 | −47.93 | 18.51 | −224.31 | 36.04 |
| CP 2.5 | −80.36 | 55.71 | 40.02 | 12.82 | 34.95 |
| CP 5 | −90.07 | 49.86 | 32.41 | −1.64 | 37.1 |
| CP 7.5 | −96.03 | 46.87 | 16.32 | −32.62 | 39.84 |
| CP 10 | −102.82 | 45.95 | 11.3 | −37.03 | 41.1 |
| CP 25 | −106.58 | 27.49 | 11.13 | −68.31 | 41.46 |
| CP 50 | −111.6 | 18.92 | 10.03 | −70.46 | 43.87 |
| CP 75 | −135.21 | 17.13 | 9.46 | −73.74 | 46.68 |
| CP 100 | −154.02 | 13.95 | 4.61 | −91.28 | 51.04 |
| PT 2.5 | −86.52 | −30.76 | −68.22 | −58.87 | −94.35 |
| PT 5 | −115.78 | −32 | −88 | −61.03 | −76.71 |
| PT 7.5 | −163.11 | −37.71 | −91.41 | −104.41 | −69.08 |
| PT 10 | −172.73 | −48.76 | −94 | −131.49 | −14.97 |
| PT 25 | −327.17 | −118.19 | −104.33 | −264.92 | −12.39 |
| PT 50 | −409.2 | −134.48 | −106.75 | −353.95 | −2.48 |
| PT 75 | −441.8 | −139.18 | −107.84 | −472.21 | 2.84 |
| PT 100 | −482.65 | −143.42 | −150.52 | −534.97 | 14.17 |
| MASRJ 2.5 | −33.75 | 22.79 | 7.15 | 29.64 | 39.18 |
| MASRJ 5 | −23.3 | 38.86 | 16.44 | 39.08 | 45.13 |
| MASRJ 7.5 | −16.82 | 49.68 | 18.28 | 43.59 | 48.5 |
| MASRJ 10 | −14.42 | 59.39 | 36.79 | 47.79 | 57.05 |
| MASRJ 25 | 12.54 | 57.46 | 47.69 | 47.9 | 62.14 |
| MASRJ 50 | 38.35 | 68.78 | 73.47 | 54.77 | 80.64 |
| MASRJ 75 | 42.11 | 70.3 | 76.93 | 55.59 | 84.01 |
| MASRJ 100 | 45.98 | 72.1 | 78.03 | 60.62 | 84.18 |
| CRJ 2.5 | −253.5 | −23.02 | −5.88 | −95.38 | 24.88 |
| CRJ 5 | −209.2 | −11.23 | 3.86 | −50.05 | 33.27 |
| CRJ 7.5 | −163.85 | −7.46 | 15.74 | −4.41 | 43.84 |
| CRJ 10 | −145.35 | 1.2 | 30.1 | 23.08 | 56.16 |
| CRJ 25 | −127.59 | 12.29 | 37.37 | 51.9 | 60.92 |
| CRJ 50 | 21.32 | 53.18 | 73.36 | 61.95 | 82.72 |
| CRJ 75 | 44.51 | 78.36 | 74.74 | 61.74 | 83.98 |
| CRJ 100 | 48.69 | 78.91 | 75.78 | 62.77 | 84.41 |
| CLRJ 2.5 | −167.71 | 41.3 | −39.45 | −172.72 | 11.63 |
| CLRJ 5 | −138.24 | 50.83 | −30.57 | −150.77 | 17.91 |
| CLRJ 7.5 | −107.63 | 57.83 | −17.07 | −133.85 | 21.18 |
| CLRJ 10 | −33.33 | 64.55 | −8.3 | −105.13 | 28.15 |
| CLRJ 25 | −15.88 | 76.93 | −0.63 | −61.95 | 34.75 |
| CLRJ 50 | 38.24 | 77.81 | 51.5 | 32.92 | 66.3 |
| CLRJ 75 | 41.69 | 78.31 | 71.97 | 56.41 | 84.64 |
| CLRJ 100 | 47.65 | 79.01 | 73.93 | 55.9 | 84.7 |
| MASA 2.5 | −64.79 | 53.22 | 10.9 | 29.54 | 67.16 |
| MASA 5 | −65.41 | 51.1 | 6.11 | 25.85 | 64.95 |
| MASA 7.5 | −89.24 | 14.83 | −16.03 | 15.18 | 47.08 |
| MASA 10 | −92.37 | −5.48 | −21.28 | 11.08 | 11.3 |
| MASA 25 | −204.28 | −9.25 | −72.84 | −128.62 | 9.32 |
| MASA 50 | −345.66 | −93.69 | −163.55 | −332.31 | −45.66 |
| MASA 75 | −465.31 | −151.93 | −220.76 | −469.85 | −90.98 |
| MASA 100 | −665.41 | −241.8 | −323.7 | −630.97 | −144.33 |
| CLA 2.5 | −24.03 | 56.81 | 20.82 | 57.23 | 51.17 |
| CLA 5 | −17.55 | 63.81 | 21.63 | 56.31 | 66.9 |
| CLA 7.5 | −17.14 | 64.5 | 44 | 56.21 | 66.93 |
| CLA 10 | −16.93 | 65.84 | 44.41 | 50.15 | 65.94 |
| CLA 25 | −11.39 | 66.3 | 61.36 | 46.36 | 72.91 |
| CLA 50 | 21.84 | 67.91 | 60.73 | 42.36 | 78.72 |
| CLA 75 | 20.48 | 70.17 | 68.74 | 40.62 | 79.19 |
| CLA 100 | 15.36 | 76.29 | 68.8 | 35.9 | 80.38 |
| CA 2.5 | −36.68 | 77.95 | 19.26 | 40.92 | 66.73 |
| CA 5 | −42.84 | 76.43 | 18.34 | 40.62 | 66.07 |
| CA 7.5 | −56.11 | 74.82 | −0.81 | 19.18 | 58.57 |
| CA 10 | −88.09 | 54.33 | −20.07 | 5.85 | 56.56 |
| CA 25 | −115.46 | 37.57 | −23.76 | −36.51 | 24.61 |
| CA 50 | −142.22 | −10.82 | −44.29 | −150.87 | 24.25 |
| CA 75 | −251.72 | −62.02 | −110.32 | −282.26 | −7.83 |
| CA 100 | −360.5 | −120.26 | −176.99 | −393.85 | −56.92 |
| Sample/Strain | S. flexneri | P. aeruginosa | E. coli | S. typhimurium | H. influenzae | C. parapsilopsis | C. albicans |
|---|---|---|---|---|---|---|---|
| MASP 2.5 | 131F.06 | −14.03 | −41.01 | 50.17 | 24.49 | −45.08 | −128.66 |
| MASP 5 | 123.78 | −18.21 | −34.07 | 50.07 | 16.95 | −22.53 | −232.11 |
| MASP 7.5 | 112.64 | −34.15 | −9.28 | 41.61 | 15.02 | −19.56 | −360.16 |
| MASP 10 | 107.68 | −62.57 | 0.9 | 34.08 | 11.95 | −7.51 | −360.57 |
| MASP 25 | 106.26 | −89.25 | 6.37 | 32.18 | 5.91 | 0.88 | −397.56 |
| MASP 50 | 106.22 | −138.43 | 8.11 | 29.64 | −4.24 | 14.51 | −456.71 |
| MASP 75 | 96.79 | −138.89 | 17.39 | 25.78 | −6.27 | 14.81 | −525.2 |
| MASP 100 | 84.51 | −171.95 | 24.19 | 12.96 | −14.41 | 16.16 | −555.08 |
| CP 2.5 | 90.08 | 1.46 | 30.15 | 53.95 | 28.43 | 29.39 | −314.43 |
| CP 5 | 83.21 | −13.11 | 31.69 | 56.09 | 28.66 | 29.97 | −249.8 |
| CP 7.5 | 82.56 | −21.4 | 35.41 | 56.12 | 28.63 | 31.28 | −228.66 |
| CP 10 | 79.31 | −32.79 | 36.68 | 56.33 | 32.8 | 31.92 | −225.61 |
| CP 25 | 79.02 | −43.9 | 38.79 | 56.52 | 37.67 | 33 | −203.66 |
| CP 50 | 70.16 | −52.46 | 41.17 | 57.7 | 40.87 | 33.23 | −200.61 |
| CP 75 | 60.49 | −58.47 | 48.78 | 57.94 | 53.22 | 39.87 | −188.62 |
| CP 100 | 58.25 | −64.94 | 52.86 | 60 | 62.66 | 42.22 | −156.3 |
| PT 2.5 | 105.16 | −41.62 | 12.53 | 41.02 | −15.05 | −76.73 | −213.01 |
| PT 5 | 109.23 | −49.64 | 10.22 | 37.8 | −15.62 | −72.96 | −329.07 |
| PT 7.5 | 126.1 | −76.23 | 9.18 | 13.9 | −15.75 | −68.72 | −354.27 |
| PT 10 | 129.55 | −139.16 | −19.43 | 10.15 | −18.32 | −62.86 | −482.32 |
| PT 25 | 133.17 | −244.08 | −80.23 | 8.25 | −35.84 | −16.16 | −567.07 |
| PT 50 | 182.15 | −335.15 | −80.5 | −20.83 | −67.37 | −15.49 | −778.25 |
| PT 75 | 193.29 | −371.31 | −81.47 | −29.93 | −69.47 | −13.77 | −845.33 |
| PT 100 | 200.69 | −394.9 | −89.45 | −30.44 | −78.68 | −2.73 | −871.54 |
| MASRJ 2.5 | 79.19 | 21.86 | 30.28 | 51.49 | 25.66 | 25.35 | −133.74 |
| MASRJ 5 | 70.08 | 19.4 | 35.08 | 56.92 | 25.99 | 30.27 | −116.46 |
| MASRJ 7.5 | 64.72 | 15.76 | 50.25 | 61.23 | 28.03 | 33.64 | −110.98 |
| MASRJ 10 | 60.81 | 12.57 | 62.75 | 61.5 | 29.5 | 33.74 | −108.74 |
| MASRJ 25 | 57.6 | 8.74 | 67.47 | 86.02 | 53.82 | 54.81 | −60.77 |
| MASRJ 50 | 40.73 | 2.82 | 76.92 | 87.66 | 79.05 | 82.42 | −11.59 |
| MASRJ 75 | 23.86 | −5.65 | 80.7 | 88.35 | 81.65 | 83.43 | 1.42 |
| MASRJ 100 | 20.98 | −4.83 | 85.66 | 89.05 | 83.38 | 85.02 | 14.63 |
| CRJ 2.5 | 81.1 | −33.06 | 49.82 | 50.15 | 19.95 | 14.92 | −188.21 |
| CRJ 5 | 66.5 | −9.84 | 57.39 | 52.9 | 28.5 | 20.74 | −142.68 |
| CRJ 7.5 | 59.72 | 8.65 | 61.98 | 58.8 | 37.54 | 29.09 | −86.18 |
| CRJ 10 | 50.39 | 20.49 | 69.48 | 71.11 | 43.64 | 63.7 | −60.77 |
| CRJ 25 | 34.27 | 39.71 | 77.52 | 85.25 | 51.15 | 46.67 | 6.1 |
| CRJ 50 | 18.29 | 60.29 | 84.69 | 88.49 | 69.7 | 85.89 | 18.7 |
| CRJ 75 | 16.99 | 62.02 | 85.73 | 88.92 | 82.08 | 86.9 | 33.94 |
| CRJ 100 | 16.38 | 62.3 | 86.93 | 88.81 | 84.25 | 87.91 | 23.98 |
| CLRJ 2.5 | 104.11 | −116.67 | 13.94 | 48.03 | 11.34 | 10.57 | −381.5 |
| CLRJ 5 | 96.95 | −92.81 | 21.11 | 53.95 | 14.61 | 19.97 | −326.83 |
| CLRJ 7.5 | 88.27 | −70.31 | 28.31 | 64.79 | 20.25 | 27.74 | −291.67 |
| CLRJ 10 | 77.44 | −41.35 | 37.29 | 72.96 | 27.19 | 35.82 | −239.84 |
| CLRJ 25 | 62.8 | −18.12 | 44.72 | 81.07 | 38.61 | 40.37 | −178.25 |
| CLRJ 50 | 33.46 | 56.65 | 83.32 | 87.6 | 63.56 | 83.74 | −76.02 |
| CLRJ 75 | 18.86 | 57.83 | 84.25 | 87.87 | 82.68 | 84.61 | 17.89 |
| CLRJ 100 | 18.9 | 58.74 | 85.13 | 88.35 | 84.15 | 90.51 | 23.37 |
| MASA 2.5 | 46.59 | 28.69 | 58.93 | 60.29 | 59.29 | 55.39 | −104.67 |
| MASA 5 | 47.03 | 13.3 | 58.36 | 56.97 | 58.99 | 44.28 | −125.81 |
| MASA 7.5 | 49.23 | 7.92 | 42.35 | 52.29 | 52.49 | 29.46 | −183.74 |
| MASA 10 | 54.31 | −9.84 | 14 | 43.43 | 36.54 | 12.15 | −225.61 |
| MASA 25 | 93.17 | −103.92 | 6.4 | 40.11 | 9.28 | 11.85 | −394.51 |
| MASA 50 | 170.28 | −293.08 | −43.85 | 1.31 | −50.12 | −26.63 | −663.21 |
| MASA 75 | 231.5 | −426.14 | −99.43 | −35.26 | −92.69 | −79.12 | −1016.06 |
| MASA 100 | 289.72 | −600.55 | −148.44 | −85.38 | −147.35 | −142.79 | −1415.04 |
| CLA 2.5 | 56.54 | 57.83 | 83.65 | 61.39 | 46.28 | 34.18 | −204.88 |
| CLA 5 | 42.89 | 51.91 | 81.44 | 63.59 | 49.45 | 43.7 | −165.04 |
| CLA 7.5 | 39.11 | 51.46 | 77.82 | 68.94 | 50.15 | 59.49 | −128.46 |
| CLA 10 | 35.65 | 48.54 | 71.29 | 69.75 | 52.45 | 63.94 | −85.16 |
| CLA 25 | 31.91 | 44.35 | 67.74 | 83.56 | 62.06 | 76.6 | −55.28 |
| CLA 50 | 31.42 | 43.99 | 61.07 | 86.13 | 70.64 | 80.1 | −21.34 |
| CLA 75 | 28.46 | 38.52 | 60.44 | 87.1 | 73.61 | 82.79 | 6.1 |
| CLA 100 | 24.63 | 20.49 | 57.39 | 89.37 | 78.28 | 84.21 | 16.06 |
| CA 2.5 | 30.53 | 29.87 | 65.09 | 66.72 | 44.51 | 58.28 | −189.23 |
| CA 5 | 43.21 | 16.12 | 58.22 | 61.93 | 41.14 | 51.85 | −195.53 |
| CA 7.5 | 44.88 | −0.27 | 51.29 | 61.69 | 41.11 | 38.59 | −207.93 |
| CA 10 | 48.37 | −8.65 | 32.43 | 54.08 | 35.1 | 28.59 | −225.2 |
| CA 25 | 62.24 | −60.66 | 24.09 | 40.27 | 23.49 | 22.96 | −307.32 |
| CA 50 | 92.4 | −132.79 | 15.14 | 34.56 | 22.09 | 15.89 | −354.88 |
| CA 75 | 144.92 | −233.06 | −16.05 | 7.6 | −17.08 | −18.65 | −603.25 |
| CA 100 | 175.85 | −341.62 | −51.19 | −22.78 | −51.48 | −54.71 | −818.09 |
| Sample mg/mL/Strain | S. aureus (MLIRP 052022) | S. aureus (MLIRP 072022) | S. aureus (MLIRP 092020) | P. aeruginosa (MLIRP 122021) | P. aeruginosa (MLIRP 092022) | P. aeruginosa (MLIRP 042019) | E. coli (MLIRP 062019) | E. coli (MLIRP 022020) | E. coli (MLIRP 112020) |
|---|---|---|---|---|---|---|---|---|---|
| MASP 0.25 | −17.26 | −4.18 | −14.97 | 4.69 | 0.05 | 2.05 | 3.97 | −0.52 | −3.20 |
| MASP 0.5 | −13.71 | −5.50 | −15.29 | 1.24 | −1.67 | 1.05 | 2.94 | −1.63 | −3.75 |
| MASP 0.75 | −11.07 | −6.32 | −15.08 | −5.00 | −3.52 | −0.08 | 2.60 | −3.47 | −4.30 |
| MASP 1.0 | −4.97 | −8.87 | −16.34 | −5.67 | −7.81 | −1.84 | 2.26 | −4.58 | −5.04 |
| MASP 2.5 | −2.54 | −10.91 | −18.43 | −7.92 | −9.95 | −2.22 | 1.23 | −4.85 | −6.24 |
| MASP 5.0 | −0.71 | −11.62 | −19.69 | −10.58 | −13.24 | −3.73 | 0.03 | −6.42 | −6.97 |
| MASP 7.5 | −0.10 | −12.84 | −20.63 | −14.17 | −15.95 | −4.86 | −0.74 | −6.97 | −8.26 |
| MASP 10 | 1.93 | −13.25 | −22.30 | −16.82 | −18.95 | −6.88 | −1.69 | −8.08 | −8.91 |
| CP 0.25 | −22.94 | −2.45 | −3.87 | 1.77 | −2.10 | 0.67 | 5.17 | −0.71 | −1.54 |
| CP 0.5 | −22.03 | −3.06 | −5.97 | 0.44 | −2.81 | −0.08 | 4.83 | −1.90 | −2.09 |
| CP 0.75 | −20.61 | −6.32 | −7.33 | −0.62 | −3.24 | −1.97 | 3.89 | −2.18 | −2.83 |
| CP 1.0 | −18.38 | −8.15 | −8.38 | −3.81 | −4.95 | −4.23 | 3.46 | −3.38 | −3.20 |
| CP 2.5 | −15.74 | −9.68 | −9.63 | −5.40 | −7.10 | −4.86 | 2.77 | −4.12 | −3.66 |
| CP 5.0 | −13.50 | −10.40 | −10.37 | −7.66 | −7.81 | −5.62 | 2.17 | −5.50 | −4.12 |
| CP 7.5 | −4.47 | −11.42 | −11.83 | −10.18 | −10.10 | −6.62 | 1.49 | −6.24 | −5.41 |
| CP 10 | −1.22 | −13.05 | −13.09 | −10.71 | −12.38 | −9.01 | 0.03 | −7.34 | −6.05 |
| PT 0.25 | −22.74 | −2.14 | −5.76 | 1.51 | −1.52 | 0.29 | 3.80 | −1.08 | −2.00 |
| PT 0.5 | −20.00 | −3.77 | −7.23 | −0.35 | −2.81 | −0.46 | 2.86 | −2.09 | −2.55 |
| PT 0.75 | −19.29 | −4.49 | −7.64 | −1.28 | −3.24 | −1.84 | 2.17 | −3.75 | −3.29 |
| PT 1.0 | −17.26 | −7.54 | −8.38 | −3.54 | −5.52 | −2.47 | 1.74 | −4.30 | −4.21 |
| PT 2.5 | −14.52 | −10.81 | −9.42 | −4.47 | −7.10 | −4.36 | 0.97 | −5.04 | −4.67 |
| PT 5.0 | −12.59 | −11.93 | −10.79 | −5.27 | −8.24 | −6.88 | 0.46 | −6.42 | −5.50 |
| PT 7.5 | −11.07 | −12.95 | −11.83 | −8.32 | −9.52 | −8.13 | −0.66 | −7.25 | −6.33 |
| PT 10 | −8.12 | −14.78 | −12.88 | −12.04 | −12.67 | −9.01 | −1.17 | −8.17 | −6.88 |
| MASRJ 0.25 | −0.71 | −4.18 | −4.92 | −1.81 | −0.10 | 0.17 | 12.55 | 8.14 | −0.80 |
| MASRJ 0.5 | −1.32 | −2.96 | −4.40 | −2.88 | −1.38 | −2.22 | 11.52 | 7.31 | −1.26 |
| MASRJ 0.75 | −5.18 | −2.04 | −3.87 | −4.07 | −2.24 | −4.36 | 10.66 | 7.13 | −2.37 |
| MASRJ 1.0 | −10.46 | −1.33 | −3.25 | −5.27 | −3.95 | −6.00 | 8.69 | 5.93 | −2.73 |
| MASRJ 2.5 | −12.49 | −0.20 | −2.41 | −5.67 | −4.81 | −8.13 | 8.26 | 5.10 | −3.10 |
| MASRJ 5.0 | −14.52 | 1.53 | −1.47 | −7.53 | −6.81 | −9.39 | 6.98 | 3.90 | −3.84 |
| MASRJ 7.5 | −15.84 | 2.85 | 2.41 | −9.12 | −8.52 | −10.40 | 6.46 | 3.07 | −5.04 |
| MASRJ 10 | −17.16 | 3.36 | 3.87 | −12.17 | −9.52 | −11.03 | 5.69 | 2.43 | −6.33 |
| CLRJ 0.25 | −3.76 | −4.18 | −3.46 | 1.37 | 1.62 | 10.10 | 6.38 | 9.62 | −0.98 |
| CLRJ 0.5 | −7.01 | −3.26 | −2.72 | −0.35 | 0.76 | 9.22 | 6.03 | 9.25 | −1.63 |
| CLRJ 0.75 | −7.61 | −2.34 | −2.09 | −2.08 | 0.05 | 8.22 | 5.60 | 8.79 | −2.18 |
| CLRJ 1.0 | −10.46 | −1.53 | −1.36 | −2.61 | −0.81 | 7.71 | 4.83 | 8.23 | −2.92 |
| CLRJ 2.5 | −12.28 | −0.51 | −0.31 | −3.54 | −2.38 | 6.83 | 3.54 | 7.68 | −3.66 |
| CLRJ 5.0 | −12.99 | 0.31 | 0.31 | −4.47 | −3.52 | 4.57 | 2.86 | 6.67 | −4.12 |
| CLRJ 7.5 | −14.92 | 1.63 | 0.73 | −5.40 | −4.38 | 3.19 | 2.17 | 6.21 | −4.76 |
| CLRJ 10 | −16.55 | 2.96 | 1.26 | −6.33 | −5.95 | 0.67 | 0.97 | 5.47 | −5.78 |
| CRJ 0.25 | −0.20 | −2.34 | −5.55 | −5.40 | 4.48 | 6.08 | 13.58 | 10.08 | 7.31 |
| CRJ 0.5 | −0.81 | −1.73 | −4.40 | −6.20 | 3.48 | 4.44 | 12.46 | 9.43 | 6.67 |
| CRJ 0.75 | −1.93 | −1.12 | −3.87 | −7.13 | 1.62 | 3.19 | 11.86 | 8.88 | 6.11 |
| CRJ 1.0 | −8.22 | −0.41 | −2.93 | −7.79 | 0.90 | 2.18 | 11.18 | 8.23 | 5.19 |
| CRJ 2.5 | −9.95 | 0.00 | −2.09 | −10.18 | −1.10 | 1.05 | 10.75 | 7.40 | 3.90 |
| CRJ 5.0 | −11.37 | 0.51 | −1.36 | −11.51 | −2.67 | −0.08 | 10.06 | 6.85 | 2.80 |
| CRJ 7.5 | −15.23 | 1.33 | −0.31 | −12.17 | −3.95 | −0.96 | 8.86 | 6.02 | 1.78 |
| CRJ 10 | −18.27 | 2.45 | 0.63 | −14.56 | −5.67 | −2.10 | 8.18 | 5.01 | 0.86 |
| MASA 0.25 | −1.62 | −0.41 | −1.05 | −3.54 | 0.76 | 2.43 | −0.31 | 7.86 | −2.92 |
| MASA 0.5 | −1.22 | 0.31 | −0.42 | −4.34 | 0.05 | 0.67 | 0.29 | 8.42 | −2.27 |
| MASA 0.75 | −0.41 | 1.02 | 0.52 | −4.87 | −1.24 | 0.42 | 1.32 | 8.97 | −1.35 |
| MASA 1.0 | 0.51 | 1.83 | 1.26 | −5.53 | −1.67 | −0.96 | 1.83 | 9.16 | −0.89 |
| MASA 2.5 | 1.32 | 2.96 | 2.09 | −6.06 | −3.10 | −1.97 | 3.12 | 9.62 | 0.31 |
| MASA 5.0 | 2.23 | 3.77 | 3.04 | −7.26 | −4.10 | −2.85 | 4.23 | 10.35 | 1.14 |
| MASA 7.5 | 2.44 | 4.59 | 3.77 | −8.46 | −4.95 | −4.11 | 5.00 | 10.63 | 2.33 |
| MASA 10 | 3.05 | 5.30 | 4.40 | −9.25 | −6.67 | −4.86 | 6.38 | 11.46 | 3.35 |
| CLA 0.25 | 1.52 | 0.20 | −1.78 | 2.57 | 4.76 | 9.10 | 5.26 | 9.16 | −1.17 |
| CLA 0.5 | 2.44 | 0.51 | −1.36 | 1.51 | 2.90 | 8.22 | 6.46 | 9.62 | −0.43 |
| CLA 0.75 | 3.35 | 0.92 | −0.63 | 0.58 | 1.48 | 6.96 | 7.32 | 10.17 | 0.40 |
| CLA 1.0 | 4.26 | 1.83 | 0.00 | −0.89 | 0.76 | 6.21 | 7.92 | 10.63 | 1.04 |
| CLA 2.5 | 5.58 | 2.45 | 1.36 | −1.81 | −0.38 | 4.44 | 9.12 | 11.37 | 2.24 |
| CLA 5.0 | 6.50 | 3.67 | 2.30 | −3.14 | −2.24 | 3.31 | 10.06 | 11.83 | 3.07 |
| CLA 7.5 | 6.90 | 4.69 | 3.46 | −4.74 | −2.95 | 1.93 | 11.09 | 12.38 | 4.18 |
| CLA 10 | 7.82 | 5.40 | 4.19 | −5.40 | −4.67 | 0.29 | 12.21 | 13.49 | 5.10 |
| CA 0.25 | −2.34 | 0.71 | −2.93 | −0.89 | −0.81 | 4.82 | −0.40 | 8.42 | −2.09 |
| CA 0.5 | −1.52 | 1.22 | −2.20 | −1.81 | −1.24 | 3.19 | 0.37 | 8.79 | −1.44 |
| CA 0.75 | −0.81 | 1.83 | −1.47 | −1.95 | −2.38 | 2.56 | 1.74 | 9.62 | −0.89 |
| CA 1.0 | 0.20 | 2.85 | −0.73 | −4.34 | −4.52 | 0.67 | 3.12 | 10.08 | 0.03 |
| CA 2.5 | 1.32 | 4.28 | 0.10 | −5.27 | −6.38 | −0.21 | 4.15 | 11.27 | 1.32 |
| CA 5.0 | 2.13 | 5.10 | 0.84 | −6.33 | −7.52 | −1.84 | 5.00 | 11.74 | 2.24 |
| CA 7.5 | 2.84 | 5.61 | 2.41 | −8.59 | −9.24 | −3.61 | 5.52 | 12.47 | 4.36 |
| CA 10 | 3.96 | 6.12 | 3.66 | −9.38 | −11.10 | −6.37 | 6.63 | 13.03 | 5.10 |
| MASP | CP | PT | MASRJ | CLRJ | CLJ | MASA | CLA | AP | |
|---|---|---|---|---|---|---|---|---|---|
| S. pyogenes ATCC 19615 | - | - | - | 2.5 | 5 | 5 | - | 5 | - |
| S. aureus ATCC 25923 | 1 | - | - | 0.25 | 1 | 0.25 | 0.25 | - | 0.25 |
| S. flexneri ATCC 12022 | 0.25 | - | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 |
| P. aeruginosa ATCC 27853 | - | - | - | 0.25 | 0.75 | 5 | 0.25 | - | 0.25 |
| E. coli ATCC 25922 | 1 | - | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 |
| S. typhimurium ATCC 14028 | 0.25 | - | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 |
| H. influenzae ATCC 10211 | 0.25 | - | - | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 |
| B. cereus ATCC 10876 | - | - | - | 0.25 | 0.5 | 5 | 0.25 | - | 0.25 |
| Cl perfringens ATCC 13124 | - | - | - | 0.25 | 1 | 5 | 0.25 | - | 0.25 |
| L. monocytogenes ATCC 19114 | 0.5 | - | 7.5 | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 |
| C. parapsilopsis ATCC 22019 | 2.5 | - | - | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 |
| C. albicans ATCC 10231 | - | - | - | 7.5 | 2.5 | 7.5 | - | 10 | - |
| S. aureus (MLIRP 052022) | - | - | - | 7.5 | 5 | 10 | 7.5 | 2.5 | 5 |
| S. aureus (MLIRP 072022) | - | - | - | 5 | 5 | 2.5 | 2.5 | 0.25 | 5 |
| S. aureus (MLIRP 092020) | 10 | - | - | - | - | - | 1 | 0.25 | 1 |
| P. aeruginosa (MLIRP 122021) | 0.25 | 0.25 | 0.25 | - | 0.25 | - | - | 0.25 | - |
| P. aeruginosa (MLIRP 092022) | - | - | - | - | 0.25 | 0.25 | 0.25 | 0.25 | - |
| P. aeruginosa (MLIRP 042019) | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| E. coli (MLIRP 062019) | - | - | - | - | - | 0.25 | 5 | 0.75 | 1 |
| E. coli (MLIRP 022020) | - | - | - | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| E. coli (MLIRP 112020) | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 | 0.5 |
| Samples | The Concentrations of the Samples | |||||||
|---|---|---|---|---|---|---|---|---|
| 2.5% | 5% | 7.5% | 10% | 2.5% | 5% | 7.5% | 10% | |
| % Inhibition of Haemolysis Values | % Inhibition of Protein Denaturation | |||||||
| MASP | 47.22 | 52.78 | 57.28 | 58.83 | 19.81 | 82.77 | 83.23 | 83.32 |
| CP | 0.75 | 3.41 | 44.02 | 51.46 | 28.40 | 34.30 | 42.29 | 50.61 |
| PT | - | - | 0.84 | 18.71 | 10.98 | 8.64 | 0.77 | - |
| MASRJ | - | - | - | - | 34.43 | 18.75 | 8.22 | 7.40 |
| CLRJ | 41.56 | 47.2 | 55 | 59.58 | 81.17 | 71.18 | 65.82 | 65.03 |
| CLJ | - | - | - | 14.37 | 58.42 | 53.48 | 51.48 | 51.42 |
| MASA | - | - | - | - | 49.03 | 38.13 | 34.69 | 9.81 |
| CLA | - | - | - | - | 45.63 | 30.35 | 14.07 | 13.99 |
| AP | - | - | - | 44.72 | 58.83 | 58.05 | 51.15 | 50.33 |
| CTR-dexamethasone 0.1 mg/mL | 61.28 | 67.18% | ||||||
| S/no | Compounds | Pubchem CID | 1JII | 1G27 | 3LPX | |||
|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) | Number of Bond Interactions | Binding Energy (kcal/mol) | Number of Bond Interactions | Binding Energy (kcal/mol) | Number of Bond Interactions | |||
| 1 | β-resorcylic acid | 1491 | −6.7 | H: 1, π-H: 1 Unfavourable Acceptor: 1 | −5.8 | H: 3, π-π Stacked: 1, π-Sigma/Alkyl: 1/1 | −5.9 | H: 3, π-π T-Shaped: 1, Unfavourable Donor: 1 |
| 2 | Caffeic acid | 689043 | −7.0 | H: 5, π-Alkyl: 1 | −6.3 | H: 3, π-Sigma: 2, Unfavourable Acceptor: 1 | −6.2 | H: 5, π-Sulfur/Alkyl: 1/1 |
| 3 | Epicatechin | 72276 | −8.3 | H: 2, C-H: 1, Unfavourable Donor/π anion: 1 | −7.8 | H: 4, π-π Stacked: 1, π-Alkyl: 1, Unfavourable Acceptor: 1 | −7.5 | H: 4, π-Sulfur/Alkyl: 1/1 |
| 4 | Ferulic acid | 445858 | −6.5 | C-H: 2, Alkyl: 1, Amide-π: 1 | −6.2 | H: 5, π-Sigma: 1 | −6.0 | H: 3, C-H: 1, π-Sulfur/Alkyl: 1/1 |
| 5 | Gallic acid | 370 | −7.2 | H: 7, Amide π-Stacked: 1 | −5.9 | H: 3, π-H/Sigma: 1/1, Unfavourable Acceptor: 1 | −6.1 | H: 2, π-π T-Shaped: 1, π-Alkyl: 1 |
| 6 | p-coumaric | 637542 | −6.2 | H:2, C-H: 1, π-Alkyl:1 | −5.9 | H: 3, C-H: 1, π-Sigma: 1 | −6.0 | H: 2 |
| 7 | Quercetin | 5280343 | −9.6 | H: 3, π-Anion: 1, Unfavourable Donor: 2 | −7.5 | H: 1, C-H: 1, Π-Anion/Alkyl/Sigma: 1/2/1, π-π Stacked: 1, Unfavourable Donor: 1 | −7.9 | H: 4, C-H: 2, π-Sulfur/Sigma/Alkyl: 1/2/1 |
| 8 | Resveratrol | 445154 | −8.2 | H: 5, π-anion/alkyl: 2, Unfavourable Donor: 1 | −6.5 | H: 1, π-Anion:1, π-π-Stacked: 1, π-Sigma/Alkyl: 1/1 | −7.4 | H: 2, π-π T Shaped: 1, π-Sulfur/Alkyl: 1/1, Unfavourable Donor: 1 |
| 9 | Rosmarinic acid | 5281792 | −8.0 | H: 4, π-Anion: 1 | −8.2 | H: 7, π-π Stacked: 1, Π-Sigma/Alkyl: 1/1 | −7.6 | H: 6, C-H: 1, Π-Π T-Shaped: 1, Π-Alkyl: 3 |
| 10 | Rutin | 5280805 | −9.7 | H: 6, C-H: 1, π-H: 3, π-Anion/Alkyl: 1/1, Unfavourable Donor: 1 | −8.3 | van der Waals: 1, H: 6, Amide-π: 1, π -Alkyl: 2, Unfavourable Donor: 1 | −9.0 | H: 6, π-π T Shaped: 1, π-Alkyl: 2, Unfavourable Donor: 1 |
| S/no | Compounds | Pubchem CID | 2QA1 | 8JZN | 5AG7 | |||
|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) | Number of Bond Interactions | Binding Energy (kcal/mol) | Number of Bond Interactions | Binding Energy (kcal/mol) | Number of Bond Interactions | |||
| 1 | β-resorcylic acid | 1491 | −6.1 | H: 4, π-Alkyl: 2, Unfavourable Donor: 1 | −6.6 | H: 2, π-π Stacked: 1 | −6.3 | H: 1, π-π T Shaped: 1, π-Sigma/Alkyl: 1/1 |
| 2 | Caffeic acid | 689043 | −6.1 | H: 3, π-Alkyl/Sigma: 1/1 | −6.5 | H: 2, π-Sigma/Anion/Alkyl: 1/1/1 | −7.5 | H: 1, π-Sigma/Alkyl: 1/3, π-π T Shaped: 2 |
| 3 | Epicatechin | 72276 | −8.0 | H: 5, Amide-π Stacked: 1, π-π T-Shaped: 1, π-Alkyl: 1 | −8.7 | H: 3, π-Anion/Cation/Sulfur/Alkyl: 1/1/1/2 | −8.1 | H: 2, C-H: 1, π-Alkyl: 3 |
| 4 | Ferulic acid | 445858 | −5.9 | H: 3, C-H: 1, Alkyl: 1 | −6.8 | H: 3, π-Anion/Sigma: 1/1, Alkyl/π-Alkyl: 1/1 | −7.0 | H: 1, π-Sigma: 1, π-π T Shaped: 1, Alkyl/π-Alkyl: 4 |
| 5 | Gallic acid | 370 | −6.8 | H: 5, C-H: 1, π-Alkyl: 2 | −6.9 | H: 2, π-Sigma/Alkyl/Anion: 1/1/1, Unfavourable Donor: 1 | −6.5 | H: 5, π-Sigma/Alkyl: 1/1 |
| 6 | p-coumaric | 637542 | −6.1 | H: 2, π-Alkyl: 3 | −6.6 | H: 3, π-Sigma/Anion/Alkyl: 1/1/1 | −7.6 | H: 1, π-Sigma/Alkyl: 2/2, π-π T Shaped: 1 |
| 7 | Quercetin | 5280343 | −7.9 | H: 4, Unfavourable Acceptor: 1 | −8.7 | H: 1, π-π T Shaped: 1, π-Sigma/Alkyl: 2/5 | −9.2 | H: 2, C-H: 1, π-Anion/Sigma/Alkyl: 1/1/3, Unfavourable Donor/Acceptor: 1/1 |
| 8 | Resveratrol | 445154 | −7.8 | H: 1, π-Alkyl: 2 | −7.8 | H: 1, π-Alkyl: 4, π-π T Shaped: 1 | −9.5 | H: 1, π-Sigma: 4, π-π/Stacked/T Shaped: 1/1, π-Alkyl: 3 |
| 9 | Rosmarinic acid | 5281792 | −8.1 | H: 4, π-Sigma/Alkyl: 1/1, Unfavourable Donor: 1 | −8.6 | H: 3, π-Sigma/Alkyl: 1/5 | −8.3 | H: 3, π-π T Shaped: 2, Unfavourable Donor: 1 |
| 10 | Rutin | 5280805 | −9.4 | H: 1, π-Sigma/Alkyl: 1/1, π-π Stacked: 1, Unfavourable Donor/Acceptor: 1/1 | −9.4 | H: 5, C-H: 2, π-Anion/Alkyl: 2/2 | −9.5 | H: 3, C-H: 1, π-Sigma/Alkyl: 1/4, π-π Stacked: 1, Unfavourable Acceptor: 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pătruică, S.; Adeiza, S.M.; Hulea, A.; Alexa, E.; Cocan, I.; Moraru, D.; Imbrea, I.; Floares, D.; Pet, I.; Imbrea, F.; et al. Romanian Bee Product Analysis: Chemical Composition, Antimicrobial Activity, and Molecular Docking Insights. Foods 2024, 13, 1455. https://doi.org/10.3390/foods13101455
Pătruică S, Adeiza SM, Hulea A, Alexa E, Cocan I, Moraru D, Imbrea I, Floares D, Pet I, Imbrea F, et al. Romanian Bee Product Analysis: Chemical Composition, Antimicrobial Activity, and Molecular Docking Insights. Foods. 2024; 13(10):1455. https://doi.org/10.3390/foods13101455
Chicago/Turabian StylePătruică, Silvia, Suleiman Mukhtar Adeiza, Anca Hulea, Ersilia Alexa, Ileana Cocan, Dragos Moraru, Ilinca Imbrea, Doris Floares, Ioan Pet, Florin Imbrea, and et al. 2024. "Romanian Bee Product Analysis: Chemical Composition, Antimicrobial Activity, and Molecular Docking Insights" Foods 13, no. 10: 1455. https://doi.org/10.3390/foods13101455





