Nondestructive and Rapid Screening of Aflatoxin-Contaminated Single Peanut Kernels Using Field-Portable Spectroscopy Instruments (FT-IR and Raman)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
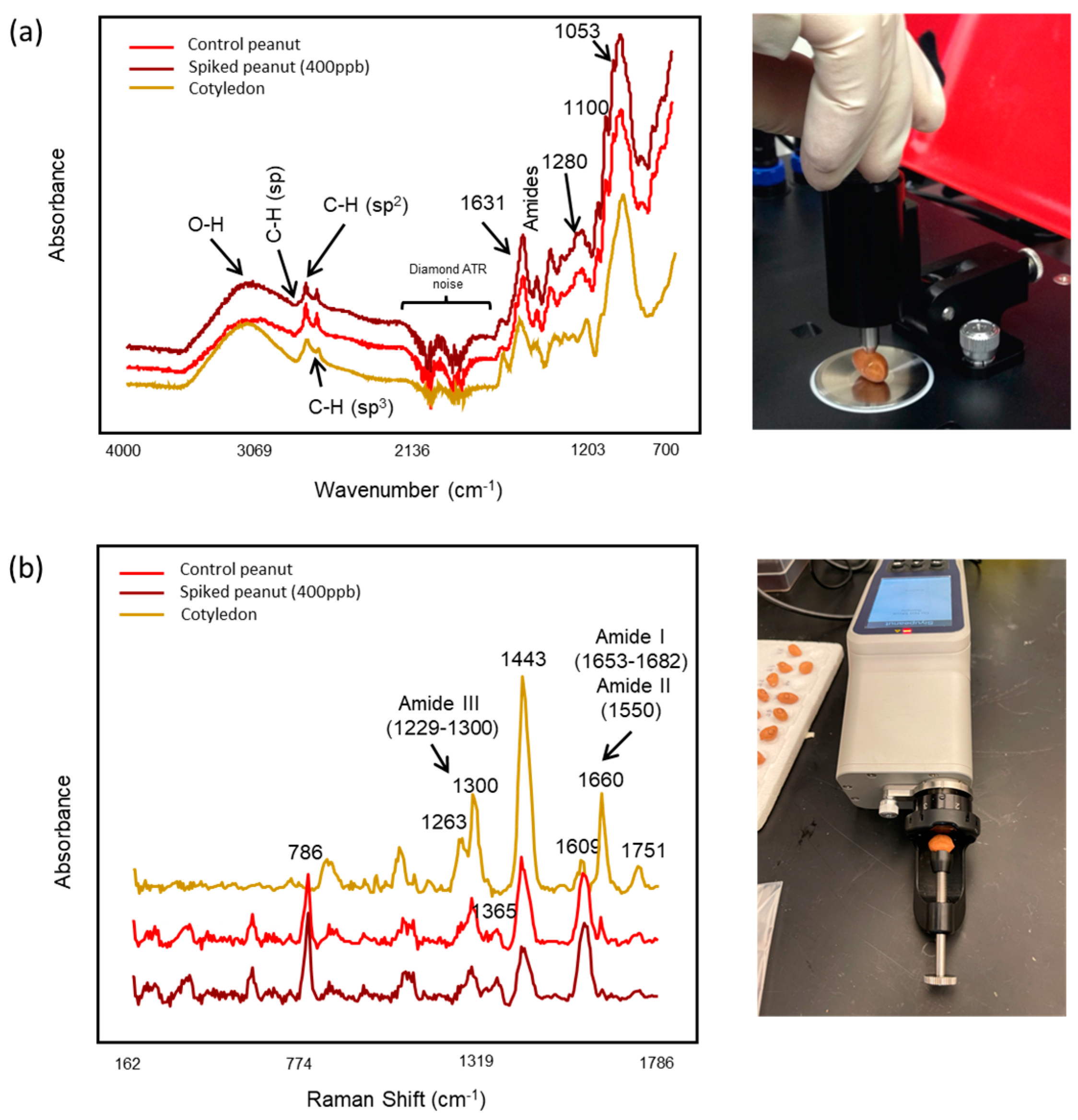
2.2. FT-IR and Raman Spectral Acquisition
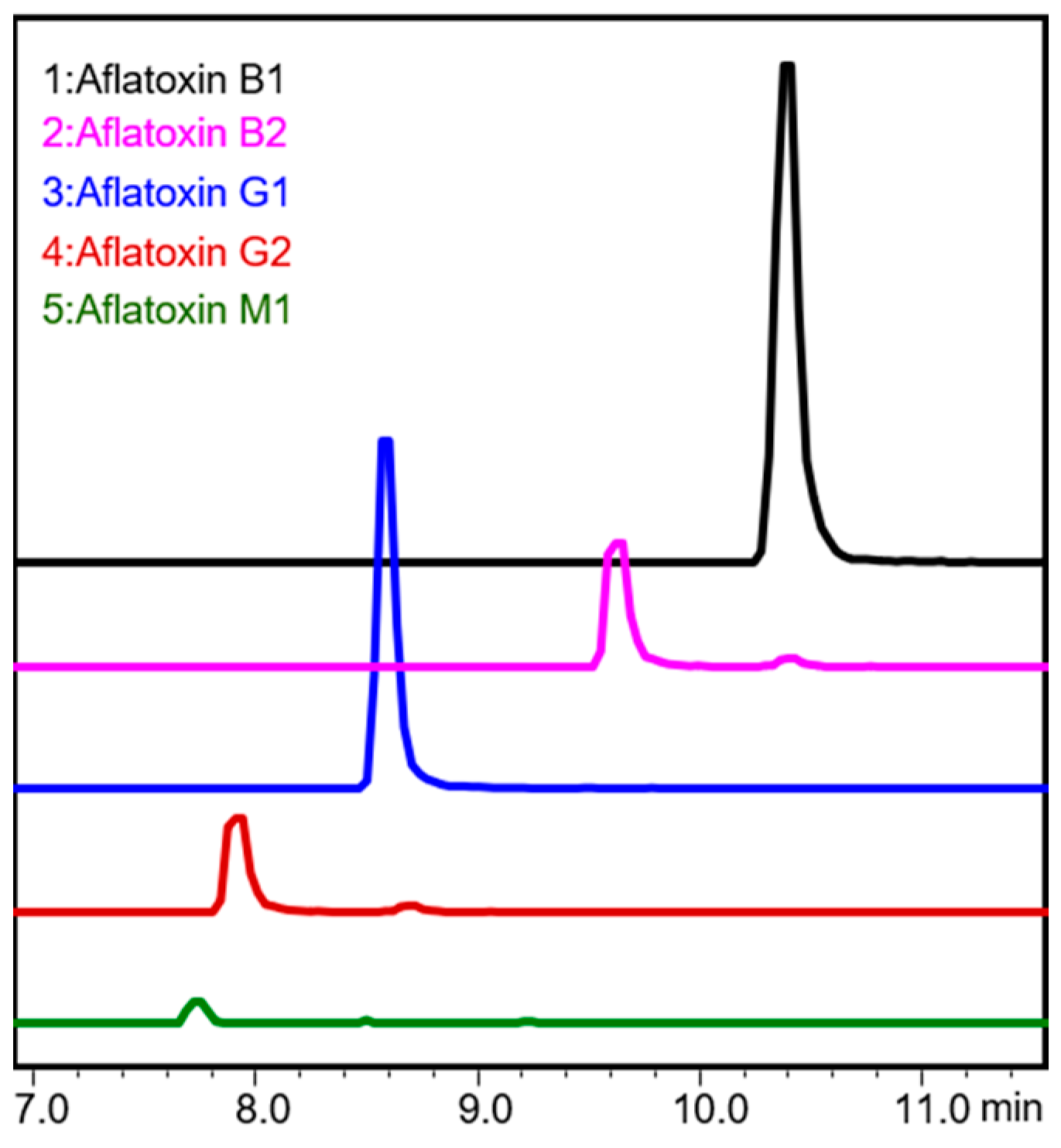
2.3. Aflatoxin Spiking Verification Analysis (Ultra-High-Performance Liquid Chromatography–Tandem Mass Spectrometry (uHPLC-MS/MS) Analysis) of Peanuts
2.4. Multivariate Data Analysis
3. Results and Discussion
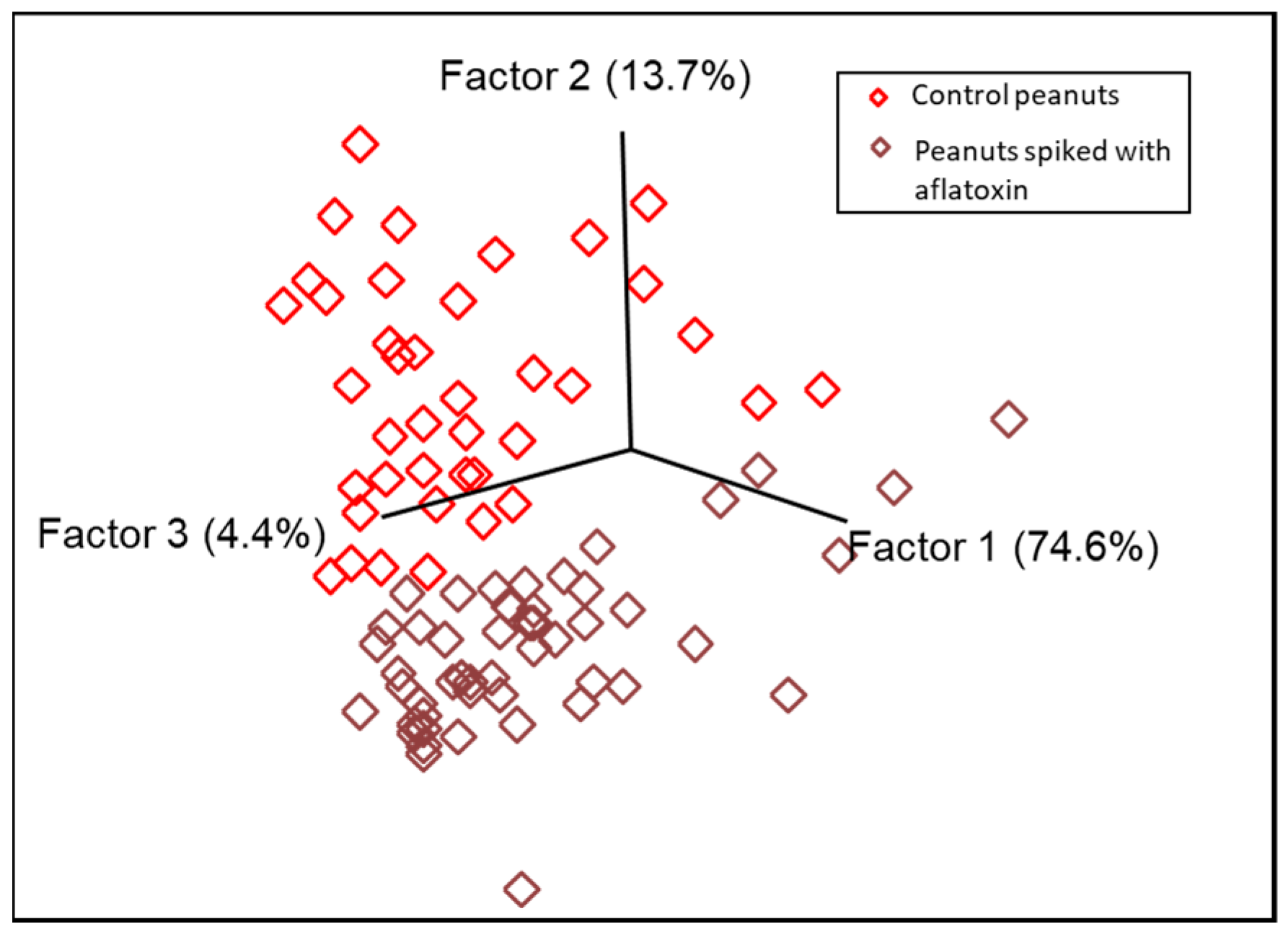
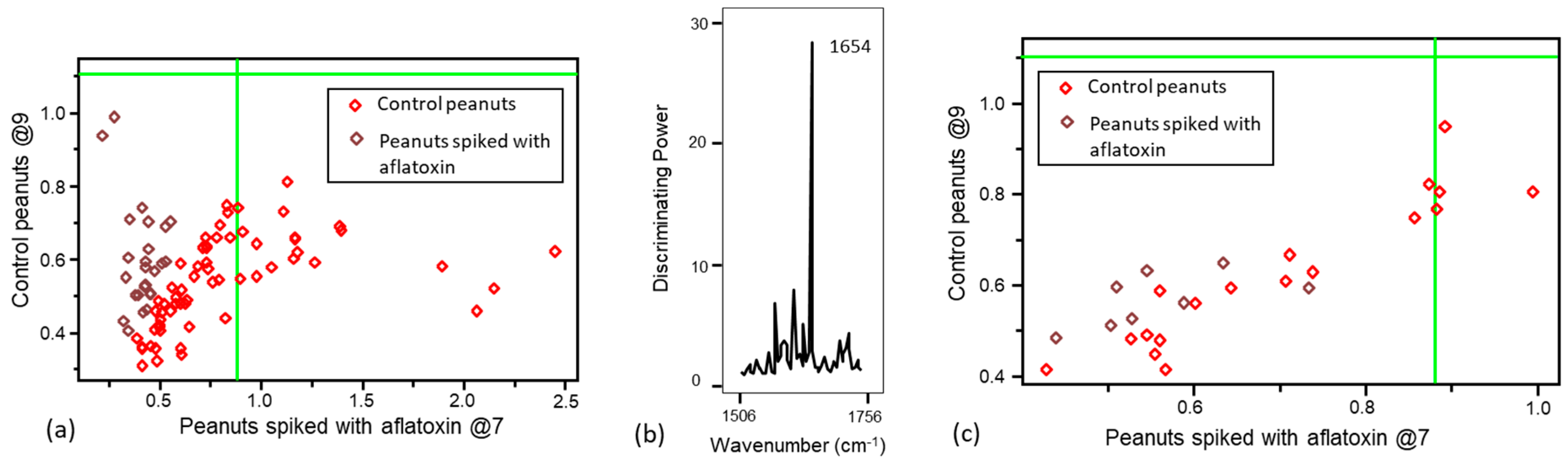
3.1. Pattern Recognition Model Development for Detecting Aflatoxin Contamination
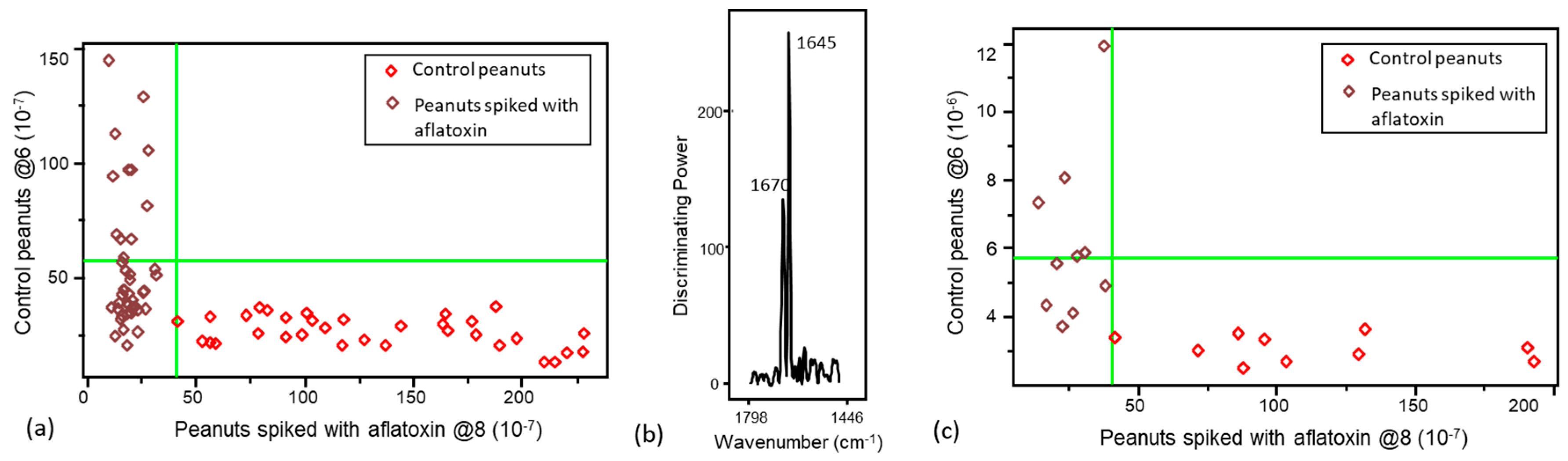
3.2. Validation of the Pattern Recognition Models
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, B.; Huai, D.; Huang, L.; Kang, Y.; Ren, X.; Chen, Y.; Zhou, X.; Luo, H.; Liu, N.; Chen, W.; et al. Identification of Genomic Regions and Diagnostic Markers for Resistance to Aflatoxin Contamination in Peanut (Arachis hypogaea L.). BMC Genet. 2019, 20, 32. [Google Scholar] [CrossRef] [PubMed]
- Perea-Moreno, M.-A.; Manzano-Agugliaro, F.; Hernandez-Escobedo, Q.; Perea-Moreno, A.-J. Peanut Shell for Energy: Properties and Its Potential to Respect the Environment. Sustainability 2018, 10, 3254. [Google Scholar] [CrossRef]
- Settaluri, V.S.; Kandala, C.V.K.; Puppala, N.; Sundaram, J. Peanuts and Their Nutritional Aspects—A Review. Food Nutr. Sci. 2012, 3, 1644–1650. [Google Scholar] [CrossRef]
- Wild, C.P.; Gong, Y.Y. Mycotoxins and Human Disease: A Largely Ignored Global Health Issue. Carcinogenesis 2010, 31, 71–82. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Mycotoxins. Available online: https://www.who.int/news-room/fact-sheets/detail/mycotoxins (accessed on 20 November 2022).
- Gnonlonfin, G.J.B.; Hell, K.; Adjovi, Y.; Fandohan, P.; Koudande, D.O.; Mensah, G.A.; Sanni, A.; Brimer, L. A Review on Aflatoxin Contamination and Its Implications in the Developing World: A Sub-Saharan African Perspective. Crit. Rev. Food Sci. Nutr. 2013, 53, 349–365. [Google Scholar] [CrossRef]
- Karlovsky, P.; Suman, M.; Berthiller, F.; De Meester, J.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recker, T.; et al. Impact of Food Processing and Detoxification Treatments on Mycotoxin Contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef]
- Martinez, L.; He, L. Detection of Mycotoxins in Food Using Surface-Enhanced Raman Spectroscopy: A Review. ACS Appl. Bio Mater. 2020, 4, 295–310. [Google Scholar] [CrossRef]
- Shen, F.; Wu, Q.; Shao, X.; Zhang, Q. Non-Destructive and Rapid Evaluation of Aflatoxins in Brown Rice by Using near-Infrared and Mid-Infrared Spectroscopic Techniques. J. Food Sci. Technol. 2018, 55, 1175–1184. [Google Scholar] [CrossRef]
- Food and Drug Administration. Sec. 683.100 Action Levels for Aflatoxins in Animal Food. Available online: https://www-fda-gov.proxy.lib.ohio-state.edu/media/121202/download (accessed on 12 December 2021).
- Fuchs, B.; Süß, R.; Teuber, K.; Eibisch, M.; Schiller, J. Lipid Analysis by Thin-Layer Chromatography—A Review of the Current State. J. Chromatogr. A 2011, 1218, 2754–2774. [Google Scholar] [CrossRef]
- Amirkhizi, B.; Arefhosseini, S.R.; Ansarin, M.; Nemati, M. Aflatoxin B1 in Eggs and Chicken Livers by Dispersive Liquid–Liquid Microextraction and HPLC. Food Addit. Contam. Part. B 2015, 8, 245–249. [Google Scholar] [CrossRef]
- Hu, X.; Yao, J.; Wang, F.; Yin, M.; Sun, Y.; Hu, M.; Shi, Q.; Zhang, G. Eu3+-labeled IgG-based Time-resolved Fluoroimmunoassay for Highly Sensitive Detection of Aflatoxin B1 in Feed. J. Sci. Food Agric. 2018, 98, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, M.; Cui, Y.; Hong, X.; Du, D. Using of Tyramine Signal Amplification to Improve the Sensitivity of ELISA for Aflatoxin B1 in Edible Oil Samples. Food Anal. Methods 2018, 11, 2553–2560. [Google Scholar] [CrossRef]
- Ong, P.; Tung, I.-C.; Chiu, C.-F.; Tsai, I.-L.; Shih, H.-C.; Chen, S.; Chuang, Y.-K. Determination of Aflatoxin B1 Level in Rice (Oryza sativa L.) through near-Infrared Spectroscopy and an Improved Simulated Annealing Variable Selection Method. Food Control 2022, 136, 108886. [Google Scholar] [CrossRef]
- Miklós, G.; Angeli, C.; Ambrus, Á.; Nagy, A.; Kardos, V.; Zentai, A.; Kerekes, K.; Farkas, Z.; Jóźwiak, Á.; Bartók, T. Detection of Aflatoxins in Different Matrices and Food-Chain Positions. Front. Microbiol. 2020, 11, 1916. [Google Scholar] [CrossRef] [PubMed]
- Krska, R.; Leslie, J.F.; Haughey, S.; Dean, M.; Bless, Y.; McNerney, O.; Spence, M.; Elliott, C. Effective Approaches for Early Identification and Proactive Mitigation of Aflatoxins in Peanuts: An EU-China Perspective. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3227–3243. [Google Scholar] [CrossRef] [PubMed]
- Kendall, C.; Isabelle, M.; Bazant-Hegemark, F.; Hutchings, J.; Orr, L.; Babrah, J.; Baker, R.; Stone, N. Vibrational Spectroscopy: A Clinical Tool for Cancer Diagnostics. Analyst 2009, 134, 1029. [Google Scholar] [CrossRef]
- Li-Chan, E.C.Y. Introduction to Vibrational Spectroscopy in Food Science. In Handbook of Vibrational Spectroscopy; Li-Chan, E.C.Y., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2010. [Google Scholar]
- Hashimoto, K.; Badarla, V.R.; Kawai, A.; Ideguchi, T. Complementary Vibrational Spectroscopy. Nat. Commun. 2019, 10, 4411. [Google Scholar] [CrossRef]
- Yao, S.; Aykas, D.P.; Rodriguez-Saona, L. Rapid Authentication of Potato Chip Oil by Vibrational Spectroscopy Combined with Pattern Recognition Analysis. Foods 2020, 10, 42. [Google Scholar] [CrossRef]
- Song, H.; LI, F.; Guang, P.; Yang, X.; Pan, H.; Huang, F. Detection of Aflatoxin B1 in Peanut Oil Using Attenuated Total Reflection Fourier Transform Infrared Spectroscopy Combined with Partial Least Squares Discriminant Analysis and Support Vector Machine Models. J. Food Prot. 2021, 84, 1315–1320. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; He, C.; Xie, M.; Luo, H.; Wang, Y.; Zhang, J. Rapid Screen of Aflatoxin-Contaminated Peanut Oil Using Fourier Transform Infrared Spectroscopy Combined with Multivariate Decision Tree. Int. J. Food Sci. Technol. 2018, 53, 2386–2393. [Google Scholar] [CrossRef]
- Mirghani, M.E.S.; Che Man, Y.B.; Jinap, S.; Baharin, B.S.; Bakar, J. A New Method for Determining Aflatoxins in Groundnut and Groundnut Cake Using Fourier Transform Infrared Spectroscopy with Attenuated Total Reflectance. J. Am. Oil Chem. Soc. 2001, 78, 985–992. [Google Scholar] [CrossRef]
- Lin, B.; Kannan, P.; Qiu, B.; Lin, Z.; Guo, L. On-Spot Surface Enhanced Raman Scattering Detection of Aflatoxin B1 in Peanut Extracts Using Gold Nanobipyramids Evenly Trapped into the AAO Nanoholes. Food Chem. 2020, 307, 125528. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-H.; Wen, B.-Y.; Lin, J.-S.; Yang, Z.-W.; Luo, S.-Y.; Li, J.-F. Rapid and Quantitative Detection of Aflatoxin B 1 in Grain by Portable Raman Spectrometer. Appl. Spectrosc. 2020, 74, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-Enhanced Raman Spectroscopy: Benefits, Trade-Offs and Future Developments. Chem. Sci. 2020, 11, 4563–4577. [Google Scholar] [CrossRef]
- Reyes, N.; Myers, C. Analysis of Pesticides and Mycotoxins in Cannabis Brownies. Available online: https://www.restek.com/en/technical-literature-library/articles/analysis-of-pesticides-and-mycotoxins-in-cannabis-brownies/ (accessed on 20 November 2022).
- SHIMAZU Excellence in Science. A High Sensitivity LC/MS/MS Method with QuEChERS Sample Pre-Treatment for Analysis of Aflatoxins in Peanut Butter Samples. 2015. Available online: https://www.shimadzu.com/an/literature/lcms/apo115045.html/ (accessed on 20 November 2022).
- Solfrizzo, M.; Gambacorta, L.; Lattanzio, V.M.T.; Powers, S.; Visconti, A. Simultaneous LC–MS/MS Determination of Aflatoxin M1, Ochratoxin A, Deoxynivalenol, de-Epoxydeoxynivalenol, α and β-Zearalenols and Fumonisin B1 in Urine as a Multi-Biomarker Method to Assess Exposure to Mycotoxins. Anal. Bioanal. Chem. 2011, 401, 2831–2841. [Google Scholar] [CrossRef]
- Boffo, E.F.; Tavares, L.A.; Ferreira, M.M.C.; Ferreira, A.G. Classification of Brazilian Vinegars According to Their 1H NMR Spectra by Pattern Recognition Analysis. LWT Food Sci. Technol. 2009, 42, 1455–1460. [Google Scholar] [CrossRef]
- Feng, L.; Zhu, S.; Chen, S.; Bao, Y.; He, Y. Combining Fourier Transform Mid-Infrared Spectroscopy with Chemometric Methods to Detect Adulterations in Milk Powder. Sensors 2019, 19, 2934. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised Pattern Recognition in Food Analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef]
- Akpolat, H.; Barineau, M.; Jackson, K.A.; Akpolat, M.Z.; Francis, D.M.; Chen, Y.J.; Rodriguez-Saona, L.E. High-Throughput Phenotyping Approach for Screening Major Carotenoids of Tomato by Handheld Raman Spectroscopy Using Chemometric Methods. Sensors 2020, 20, 3723. [Google Scholar] [CrossRef]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of Multidimensional Data Processing Approaches for Raman and Infrared Spectroscopy. EPJ Tech. Instrum. 2015, 2, 8. [Google Scholar] [CrossRef]
- Nunes, K.M.; Andrade, M.V.O.; Almeida, M.R.; Sena, M.M. A Soft Discriminant Model Based on Mid-Infrared Spectra of Bovine Meat Purges to Detect Economic Motivated Adulteration by the Addition of Non-Meat Ingredients. Food Anal. Methods 2020, 13, 1699–1709. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Q.; Zhao, H. Sub-Regional Identification of Peanuts from Shandong Province of China Based on Fourier Transform Infrared (FT-IR) Spectroscopy. Food Control 2021, 124, 107879. [Google Scholar] [CrossRef]
- Aykas, D.P.; Rodriguez-Saona, L.E. Assessing Potato Chip Oil Quality Using a Portable Infrared Spectrometer Combined with Pattern Recognition Analysis. Anal. Methods 2016, 8, 731–741. [Google Scholar] [CrossRef]
- Ruiz, M.; Beriain, M.J.; Beruete, M.; Insausti, K.; Lorenzo, J.M.; Sarriés, M.V. Application of MIR Spectroscopy to the Evaluation of Chemical Composition and Quality Parameters of Foal Meat: A Preliminary Study. Foods 2020, 9, 583. [Google Scholar] [CrossRef] [PubMed]
- Manrich, A.; Martins, M.A.; Mattoso, L.H.C. Manufacture and Performance of Peanut Skin Cellulose Nanocrystals. Sci. Agric. 2022, 79, 6. [Google Scholar] [CrossRef]
- Xia, K.; Daws, M.I.; Hay, F.R.; Chen, W.-Y.; Zhou, Z.-K.; Pritchard, H.W. A Comparative Study of Desiccation Responses of Seeds of Asian Evergreen Oaks, Quercus Subgenus Cyclobalanopsis and Quercus Subgenus Quercus. S. Afr. J. Bot. 2012, 78, 47–54. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.H.; Liu, H.H.; Zeng, C. Rapid Identification of Black Peanut Seeds by Confocal Micro-Raman and near-Infrared FT-Raman Spectroscopy. Anal. Methods 2014, 6, 2537–2544. [Google Scholar] [CrossRef]
- Devitt, G.; Howard, K.; Mudher, A.; Mahajan, S. Raman Spectroscopy: An Emerging Tool in Neurodegenerative Disease Research and Diagnosis. ACS Chem. Neurosci. 2018, 9, 404–420. [Google Scholar] [CrossRef]
- Farber, C.; Sanchez, L.; Rizevsky, S.; Ermolenkov, A.; McCutchen, B.; Cason, J.; Simpson, C.; Burow, M.; Kurouski, D. Raman Spectroscopy Enables Non-Invasive Identification of Peanut Genotypes and Value-Added Traits. Sci. Rep. 2020, 10, 7730. [Google Scholar] [CrossRef]
- Peica, N. Erlangung Vibrational Spectroscopy and Density Functional Theory Calculations on Biological Molecules. Ph.D. Thesis, Bayerischen Julius-Maximilians-Universität Würzburg, Würzburg, Germany, 2006. [Google Scholar]
- Yao, S.; Ball, C.; Miyagusuku-Cruzado, G.; Giusti, M.M.; Aykas, D.P.; Rodriguez-Saona, L.E. A Novel Handheld FT-NIR Spectroscopic Approach for Real-Time Screening of Major Cannabinoids Content in Hemp. Talanta 2022, 247, 123559. [Google Scholar] [CrossRef]
- Rigaku Analytical Devices ProgenyTM and ProgenyTM LT Analyzer User Manual; 2017. Available online: https://data2.manualslib.com/pdf7/157/15696/1569590-rigaku/progeny.pdf?3fdfa8d3859a47a31e8f993cc830b5cc (accessed on 20 November 2022).
- Rácz, A.; Gere, A.; Bajusz, D.; Héberger, K. Is Soft Independent Modeling of Class Analogies a Reasonable Choice for Supervised Pattern Recognition? RSC Adv. 2018, 8, 10–21. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA Cross Validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Shotts, M.L.; Plans Pujolras, M.; Rossell, C.; Rodriguez-Saona, L. Authentication of Indigenous Flours (Quinoa, Amaranth and Kañiwa) from the Andean Region Using a Portable ATR-Infrared Device in Combination with Pattern Recognition Analysis. J. Cereal Sci. 2018, 82, 65–72. [Google Scholar] [CrossRef]
- Nandiyanto, A.B.D.; Oktiani, R.; Ragadhita, R. How to Read and Interpret FTIR Spectroscope of Organic Material. Indones. J. Sci. Technol. 2019, 4, 97. [Google Scholar] [CrossRef]
- Parker, F.S. Amides and Amines. In Applications of Infrared Spectroscopy in Biochemistry, Biology, and Medicine; Springer: Boston, MA, USA, 1971; pp. 165–172. [Google Scholar]
- Lee, K.-M.; Davis, J.; Herrman, T.J.; Murray, S.C.; Deng, Y. An Empirical Evaluation of Three Vibrational Spectroscopic Methods for Detection of Aflatoxins in Maize. Food Chem. 2015, 173, 629–639. [Google Scholar] [CrossRef]
- Lee, K.-M.; Herrman, T.J.; Yun, U. Application of Raman Spectroscopy for Qualitative and Quantitative Analysis of Aflatoxins in Ground Maize Samples. J. Cereal Sci. 2014, 59, 70–78. [Google Scholar] [CrossRef]
- Kos, G.; Sieger, M.; McMullin, D.; Zahradnik, C.; Sulyok, M.; Öner, T.; Mizaikoff, B.; Krska, R. A Novel Chemometric Classification for FTIR Spectra of Mycotoxin-Contaminated Maize and Peanuts at Regulatory Limits. Food Addit. Contam. Part A 2016, 33, 1596–1607. [Google Scholar] [CrossRef]
| Spiked Concentration (ppb) | Sample Number | LC-MS/MS Results (ppb) | ||||
|---|---|---|---|---|---|---|
| Min | Max | Mean | STD | SE | ||
| 50 | 4 | 36.49 | 61.99 | 46.28 | 11.19 | 5.60 |
| 100 | 4 | 123.02 | 141.79 | 130.57 | 8.15 | 4.08 |
| 200 | 4 | 194.11 | 248.84 | 227.18 | 23.52 | 11.76 |
| 400 | 4 | 341.35 | 394.05 | 372.34 | 24.33 | 12.17 |
| Model Types | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| FT-IR | 100 | 100 | 100 |
| Raman | 80.8 | 62.5 | 88.9 |
| Sample | Aflatoxins | Instrument | Chemometrics | Results | Reference |
|---|---|---|---|---|---|
| Ground peanut paste | Aflatoxin B1 (<10,624 ppb) | Benchtop FT-IR with ATR accessory | Bagged decision tree | Accuracy = 77% | [55] |
| Brown rice | Aflatoxins (B1, B2, G1, G2) (<2406 ppb) | Benchtop FT-IR with ATR accessory | LDA a | Accuracy = 90.6% | [9] |
| Ground maize | Aflatoxins (B1, B2, G1, G2) (<1206 ppb) | Benchtop FT-IR with a diffuse reflectance holder | KNN b LDA PLS-DA c | KNN: CCR d = 84% LDA: CCR d = 84% PLS-DA: CCR = 72% | [53] |
| Ground maize | Aflatoxins (B1, B2, G1, G2) (<1206 ppb) | Benchtop Raman (785 nm) | LDA PCDA e PLS-DA | LDA: False negative rate = 0% PCDA: False negative rate = 13.3% PLS-DA: False negative rate = 6.7% | [54] |
| Ground maize | Aflatoxins (B1, B2, G1, G2) (<1206 ppb) | Benchtop Raman (785 nm) | KNN LDA PLS-DA | KNN: CCR = 91.4% LDA: CCR = 91.4% PLS-DA: CCR = 53.4% | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, S.; Miyagusuku-Cruzado, G.; West, M.; Nwosu, V.; Dowd, E.; Fountain, J.; Giusti, M.M.; Rodriguez-Saona, L.E. Nondestructive and Rapid Screening of Aflatoxin-Contaminated Single Peanut Kernels Using Field-Portable Spectroscopy Instruments (FT-IR and Raman). Foods 2024, 13, 157. https://doi.org/10.3390/foods13010157
Yao S, Miyagusuku-Cruzado G, West M, Nwosu V, Dowd E, Fountain J, Giusti MM, Rodriguez-Saona LE. Nondestructive and Rapid Screening of Aflatoxin-Contaminated Single Peanut Kernels Using Field-Portable Spectroscopy Instruments (FT-IR and Raman). Foods. 2024; 13(1):157. https://doi.org/10.3390/foods13010157
Chicago/Turabian StyleYao, Siyu, Gonzalo Miyagusuku-Cruzado, Megan West, Victor Nwosu, Eric Dowd, Jake Fountain, M. Monica Giusti, and Luis E. Rodriguez-Saona. 2024. "Nondestructive and Rapid Screening of Aflatoxin-Contaminated Single Peanut Kernels Using Field-Portable Spectroscopy Instruments (FT-IR and Raman)" Foods 13, no. 1: 157. https://doi.org/10.3390/foods13010157






