Optimization of Soybean Protein Extraction with Ammonium Hydroxide (NH4OH) Using Response Surface Methodology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Proximate Composition
2.2.1. Moisture Content Determination
2.2.2. Total Protein Content
2.2.3. Total Ash Content
2.2.4. Total Fat
2.2.5. Total Carbohydrate
2.3. Experimental Design
2.4. Analytics
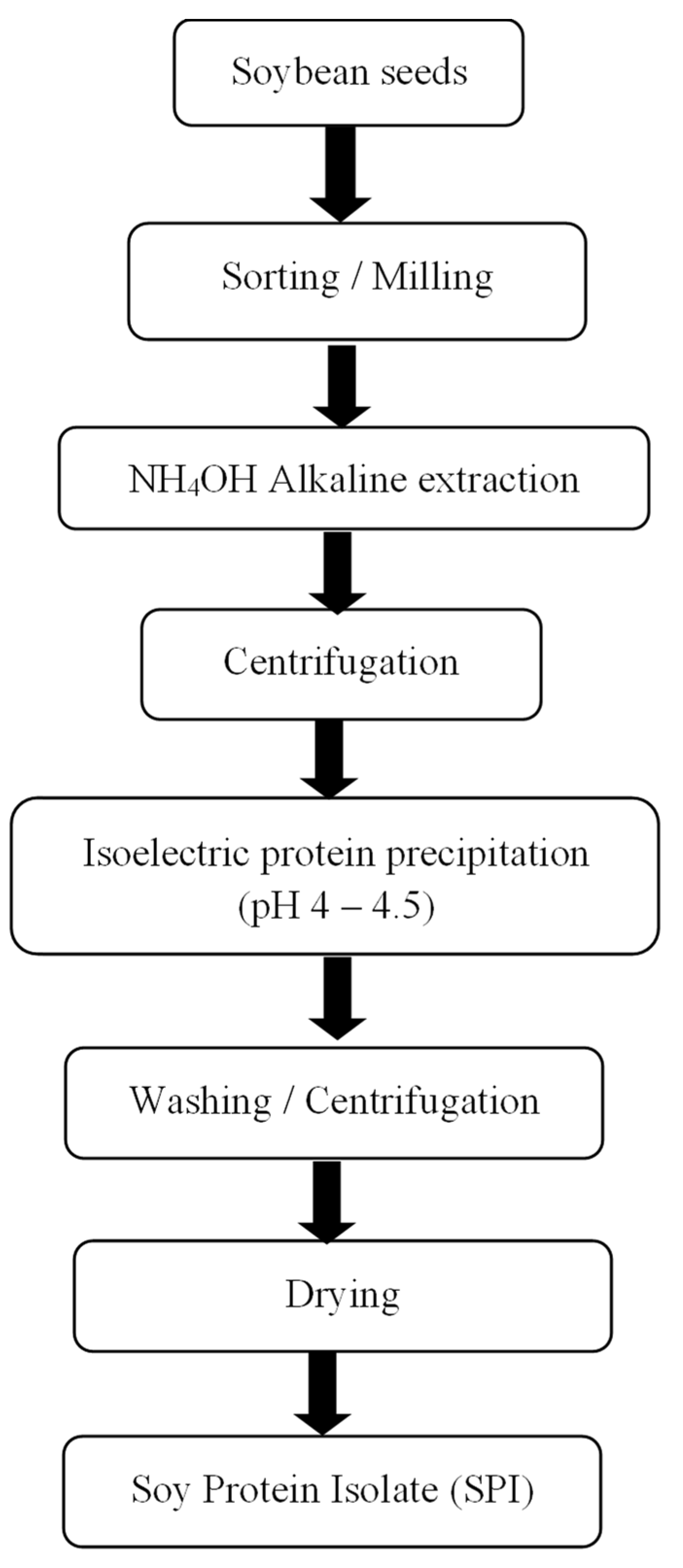
2.4.1. Protein Extraction
2.4.2. Total Amine Estimation
2.5. Statistical Analysis
3. Results and Discussion
3.1. Proximate Analysis
3.2. The Response Surface Optimization
3.2.1. Experimental of the Response Surface Methodology
3.2.2. Regression Models for Response Variables
Regression Models for Protein Extraction Yield (Y1)
Regression Model for Amine Concentration (Y2)
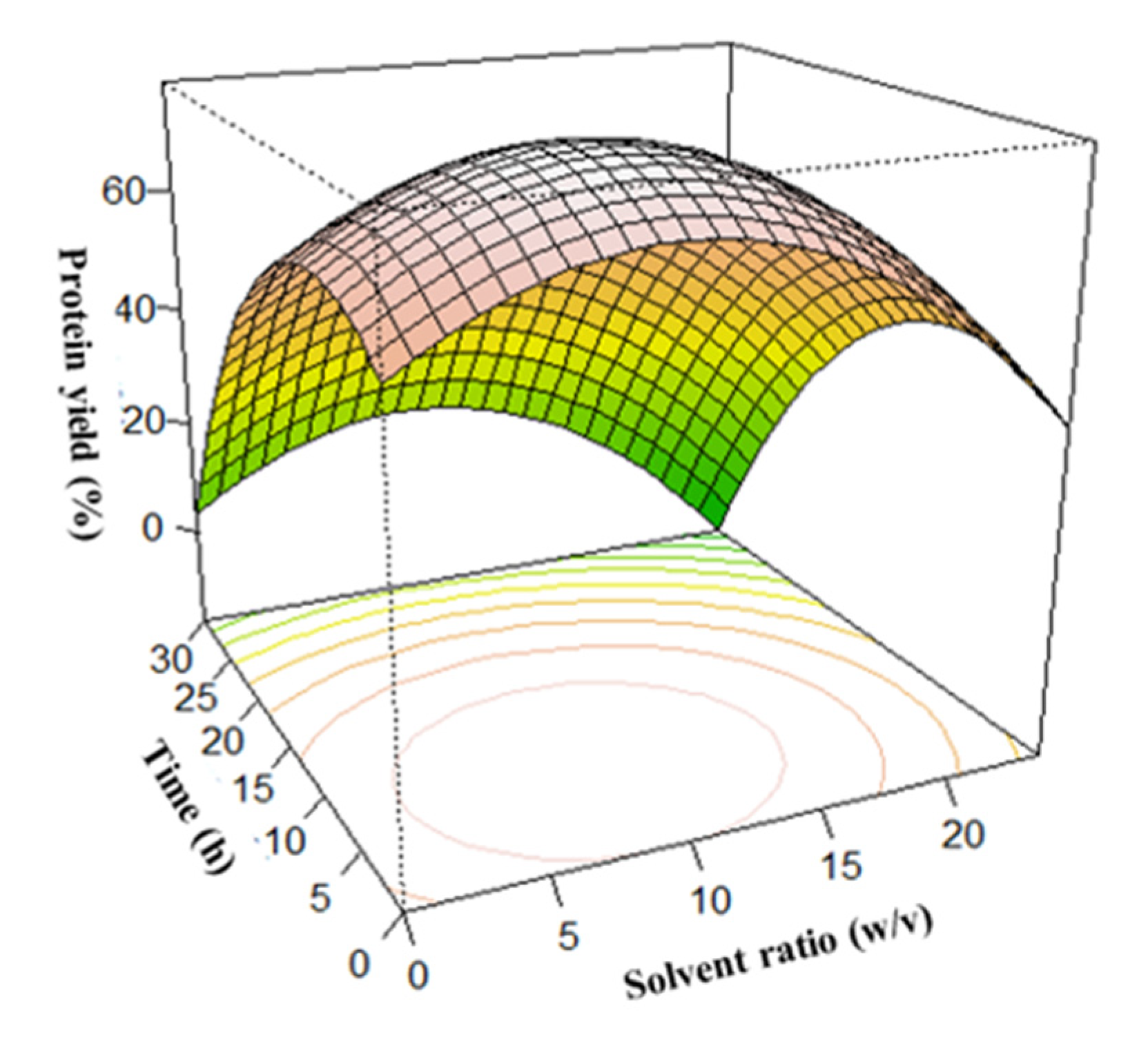
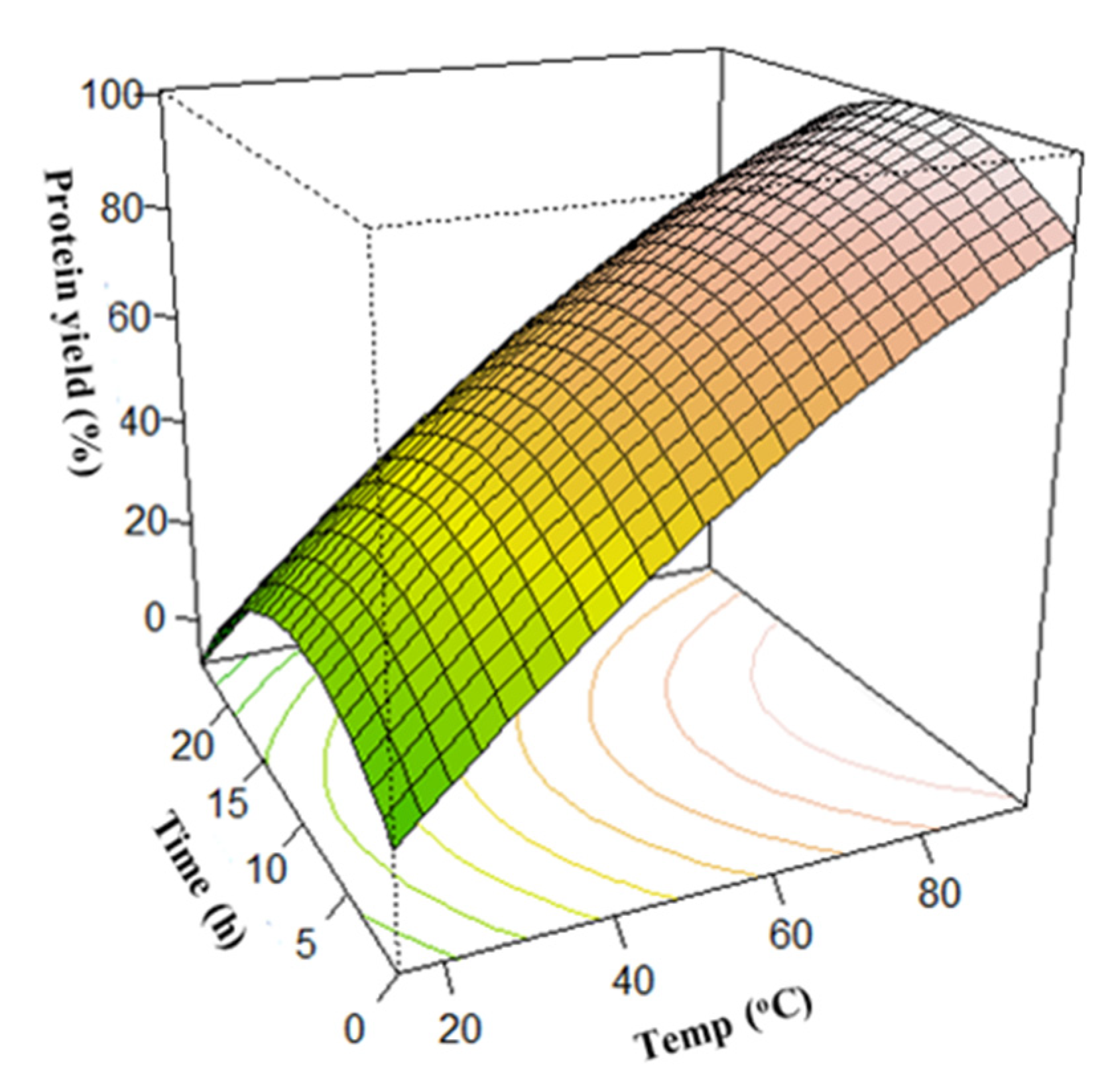
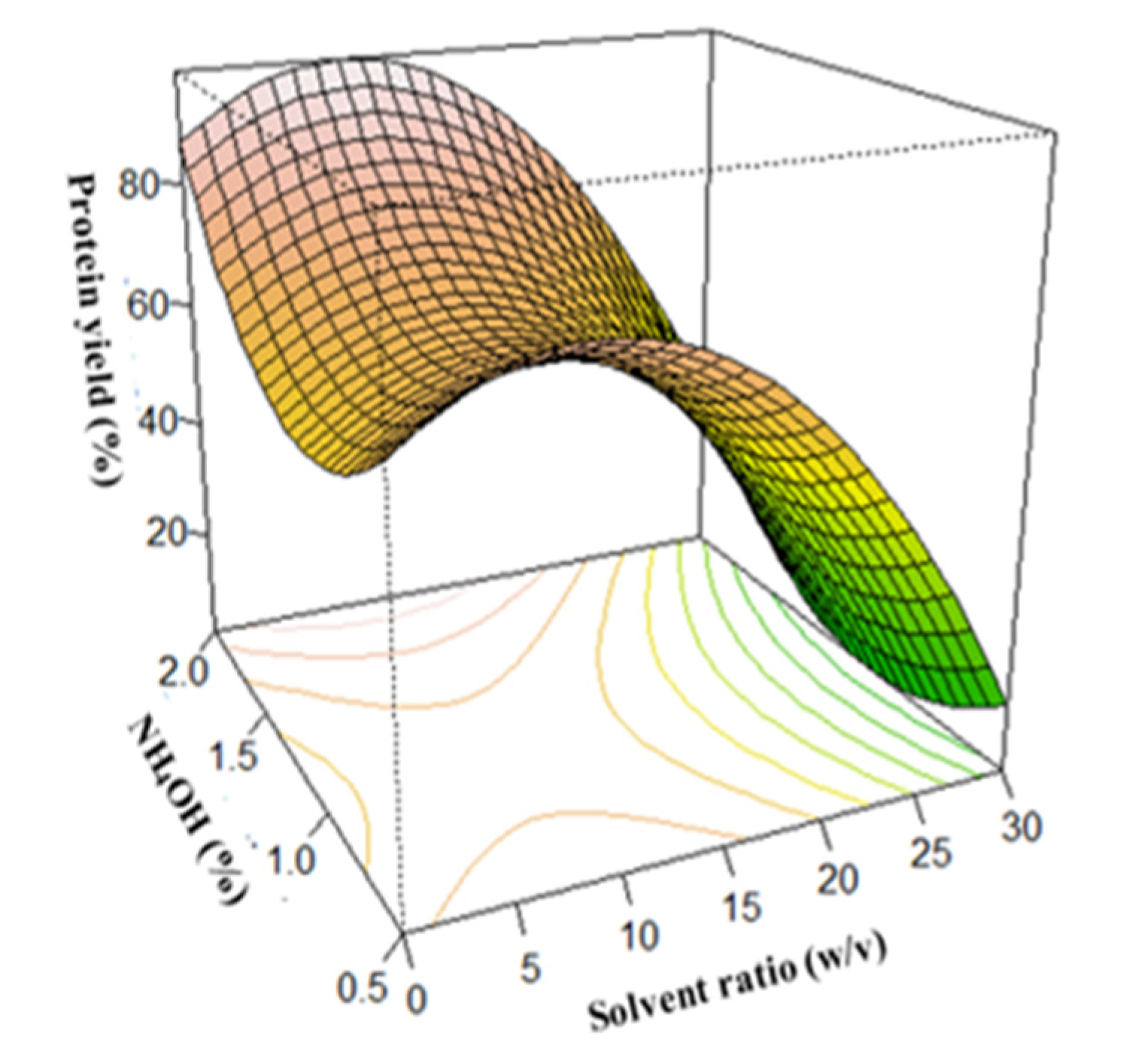
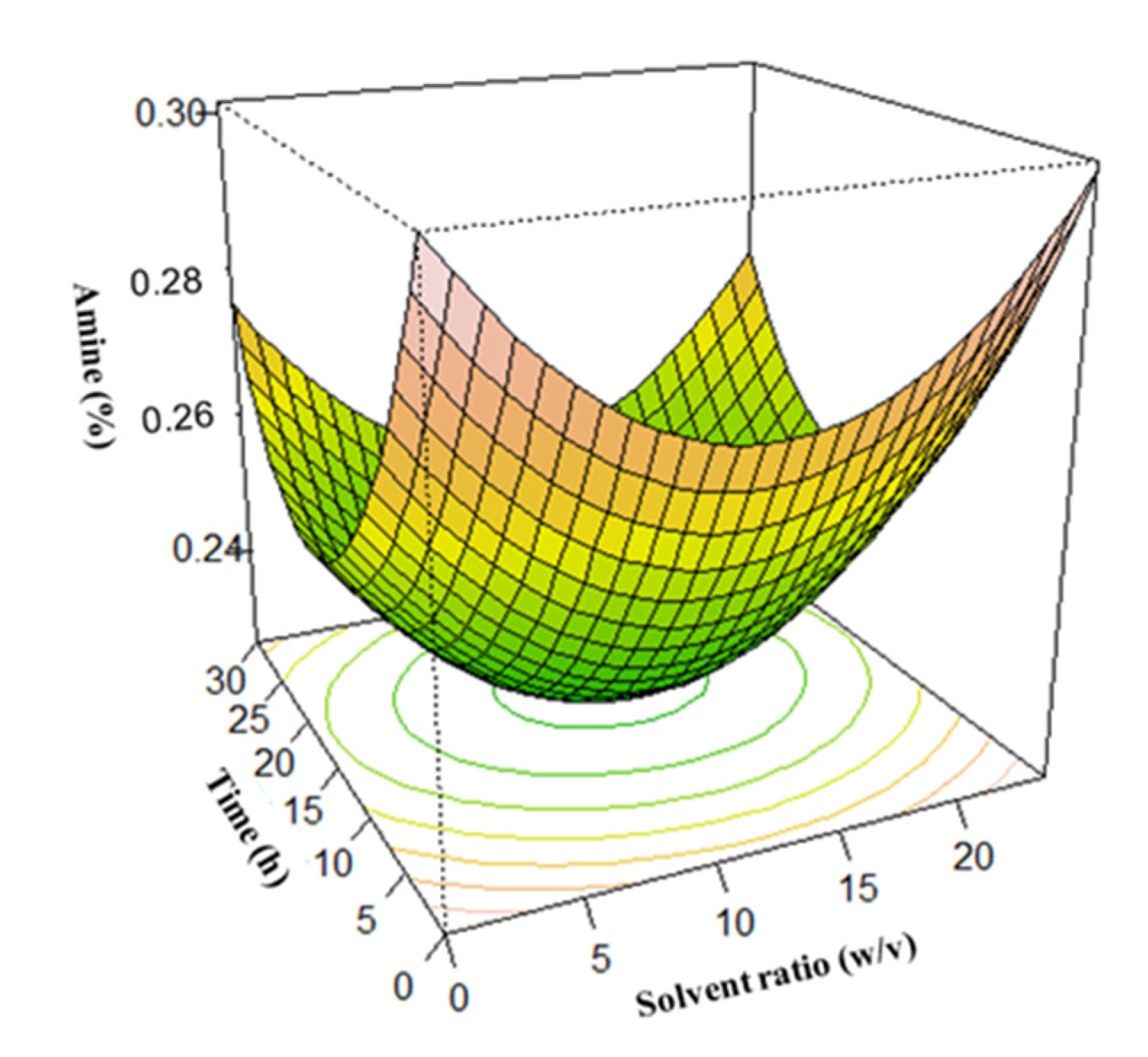
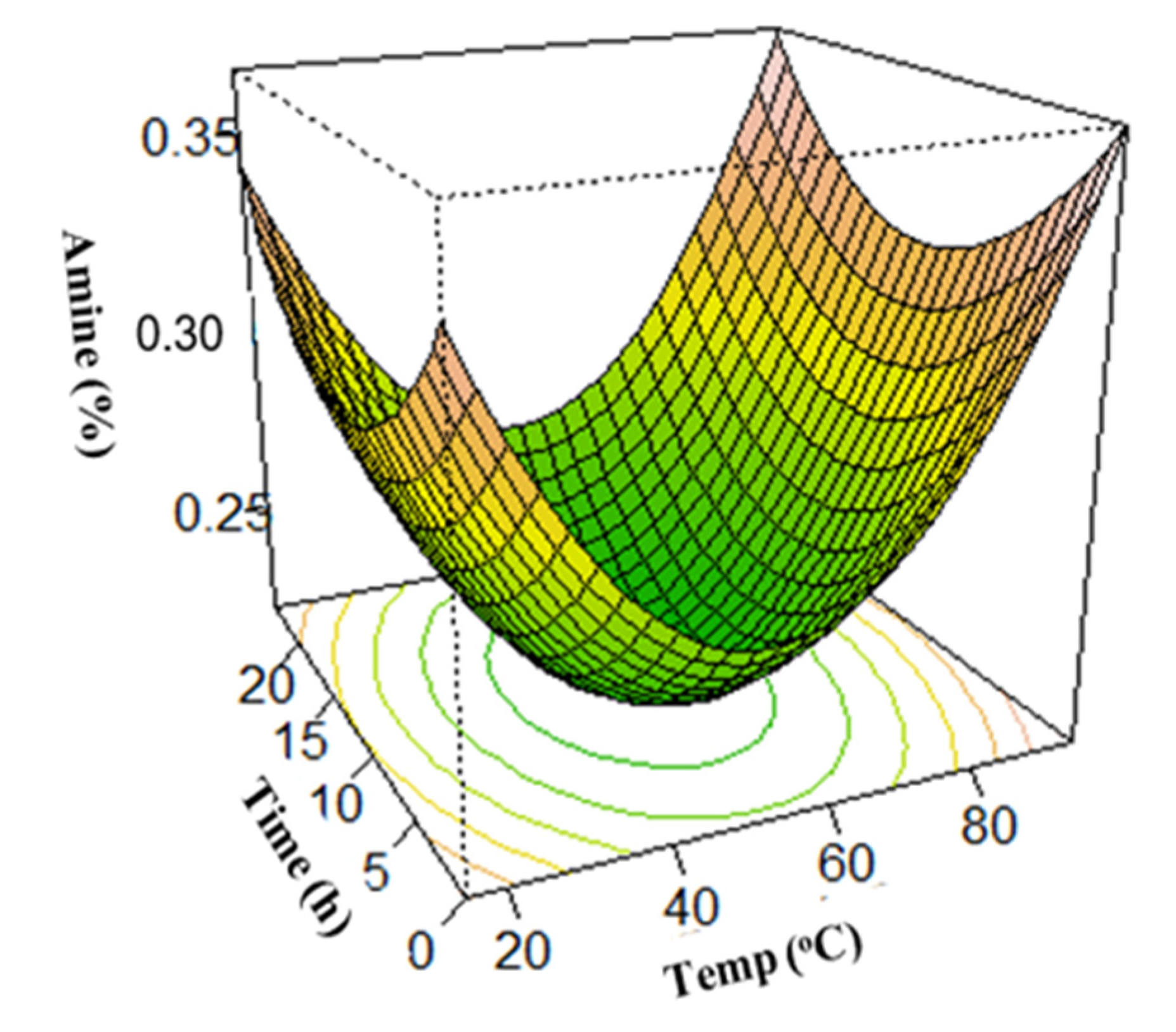
3.3. Surface Plots for Protein Extraction Yield (Y1) and Amine Concentration (Y2)
3.4. Validation Studies
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eze, C.R.; Kwofie, E.M.; Adewale, P.; Lam, E.; Ngadi, M. Advances in legume protein extraction technologies: A review. Innov. Food Sci. Emerg. Technol. 2022, 82, 103199. [Google Scholar] [CrossRef]
- Fernando, S. Production of protein-rich pulse ingredients through dry fractionation: A review. LWT 2021, 141, 110961. [Google Scholar] [CrossRef]
- Henchion, M.; Hayes, M.; Mullen, A.M.; Fenelon, M.; Tiwari, B. Future protein supply and demand: Strategies and factors influencing a sustainable equilibrium. Foods 2017, 6, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sijpestijn, G.F.; Wezel, A.; Chriki, S. Can agroecology help in meeting our 2050 protein requirements? Livest. Sci. 2022, 256, 104822. [Google Scholar] [CrossRef]
- Detzel, A.; Krüger, M.; Busch, M.; Blanco-Gutiérrez, I.; Varela, C.; Manners, R.; Bez, J.; Zannini, E. Life cycle assessment of animal-based foods and plant-based protein-rich alternatives: An environmental perspective. J. Sci. Food Agric. 2022, 102, 5098–5110. [Google Scholar] [CrossRef] [PubMed]
- Järviö, N.; Maljanen, N.-L.; Kobayashi, Y.; Ryynänen, T.; Tuomisto, H.L. An attributional life cycle assessment of microbial protein production: A case study on using hydrogen-oxidizing bacteria. Sci. Total Environ. 2021, 776, 145764. [Google Scholar] [CrossRef] [PubMed]
- Saw, H.S.; Sankaran, R.; Khoo, K.S.; Chew, K.W.; Phong, W.N.; Tang, M.S.; Lim, S.S.; Mohd Zaid, H.F.; Naushad, M.; Show, P.L. Application of a liquid biphasic flotation (LBF) system for protein extraction from Persiscaria tenulla leaf. Processes 2020, 8, 247. [Google Scholar] [CrossRef] [Green Version]
- Godfray, H.C.J.; Aveyard, P.; Garnett, T.; Hall, J.W.; Key, T.J.; Lorimer, J.; Pierrehumbert, R.T.; Scarborough, P.; Springmann, M.; Jebb, S.A. Meat consumption, health, and the environment. Science 2018, 361, eaam5324. [Google Scholar] [CrossRef] [Green Version]
- Aiking, H.; de Boer, J. The next protein transition. Trends Food Sci. Technol. 2020, 105, 515–522. [Google Scholar] [CrossRef]
- de Boer, J.; Aiking, H. Prospects for pro-environmental protein consumption in Europe: Cultural, culinary, economic and psychological factors. Appetite 2018, 121, 29–40. [Google Scholar] [CrossRef]
- Graça, J.; Godinho, C.A.; Truninger, M. Reducing meat consumption and following plant-based diets: Current evidence and future directions to inform integrated transitions. Trends Food Sci. Technol. 2019, 91, 380–390. [Google Scholar] [CrossRef] [Green Version]
- Adeniyi, A.; Bello, I.; Mukaila, T.; Hammed, A. A Review of Microbial Molecular Profiling during Biomass Valorization. Biotechnol. Bioprocess Eng. 2022, 27, 515–532. [Google Scholar] [CrossRef]
- Bekhit, A.E.-D.A.; Riley, W.W.; Hussain, M.A. Alternative Proteins: Safety and Food Security Considerations; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Preece, K.; Hooshyar, N.; Zuidam, N. Whole soybean protein extraction processes: A review. Innov. Food Sci. Emerg. Technol. 2017, 43, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Medic, J.; Atkinson, C.; Hurburgh, C.R. Current knowledge in soybean composition. J. Am. Oil Chem. Soc. 2014, 91, 363–384. [Google Scholar] [CrossRef]
- Dale, B.E.; Allen, M.S.; Laser, M.; Lynd, L.R. Protein feeds coproduction in biomass conversion to fuels and chemicals. Biofuels Bioprod. Biorefin. 2009, 3, 219–230. [Google Scholar] [CrossRef]
- Grieshop, C.M.; Kadzere, C.T.; Clapper, G.M.; Flickinger, E.A.; Bauer, L.L.; Frazier, R.L.; Fahey, G.C. Chemical and nutritional characteristics of United States soybeans and soybean meals. J. Agric. Food Chem. 2003, 51, 7684–7691. [Google Scholar] [CrossRef]
- Liu, Z.; Li, L.; Eizo, T. Nutritional quality and physiological functions of soy protein: A review. Soybean Sci. 2000, 19, 263–268. [Google Scholar]
- Qin, P.; Wang, T.; Luo, Y. A review on plant-based proteins from soybean: Health benefits and soy product development. J. Agric. Food Res. 2022, 7, 100265. [Google Scholar] [CrossRef]
- Rizzo, G.; Baroni, L. Soy, soy foods and their role in vegetarian diets. Nutrients 2018, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Freitas, M.L.F.; Albano, K.M.; Telis, V.R.N. Characterization of biopolymers and soy protein isolate-high-methoxyl pectin complex. Polímeros 2017, 27, 62–67. [Google Scholar] [CrossRef]
- Bjarnadóttir, M.; Aðalbjörnsson, B.V.; Nilsson, A.; Slizyte, R.; Roleda, M.Y.; Hreggviðsson, G.Ó.; Friðjónsson, Ó.H.; Jónsdóttir, R. Palmaria palmata as an alternative protein source: Enzymatic protein extraction, amino acid composition, and nitrogen-to-protein conversion factor. J. Appl. Phycol. 2018, 30, 2061–2070. [Google Scholar] [CrossRef]
- Jung, S. Aqueous extraction of oil and protein from soybean and lupin: A comparative study. J. Food Process. Preserv. 2009, 33, 547–559. [Google Scholar] [CrossRef]
- Latif, S.; Anwar, F. Aqueous enzymatic sesame oil and protein extraction. Food Chem. 2011, 125, 679–684. [Google Scholar] [CrossRef]
- Gerliani, N.; Hammami, R.; Aïder, M. Extraction of protein and carbohydrates from soybean meal using acidic and alkaline solutions produced by electro-activation. Food Sci. Nutr. 2020, 8, 1125–1138. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-Y.; Lee, H.D.; Lee, C.-H. Characterization of hydrolysates produced by mild-acid treatment and enzymatic hydrolysis of defatted soybean flour. Food Res. Int. 2001, 34, 217–222. [Google Scholar] [CrossRef]
- Collier, P.D.; Cromie, D.D.O.; Davies, A.P. Mechanism of Formation of Chloropropanols Present in Protein Hydrolysates. J. Am. Oil Chem. Soc. 1991, 68, 785–790. [Google Scholar] [CrossRef]
- Andres, S.; Appel, K.E.; Lampen, A. Toxicology, Occurrence and Risk Characterisation of the Chloropropanols in Food: 2-Monochloro-1, 3-Propanediol, 1, 3-Dichloro-2-Propanol and 2, 3-Dichloro-1-Propanol. Food Chem. Toxicol. 2013, 58, 467–478. [Google Scholar] [CrossRef]
- FAO Joint; WHO Expert Committee on Food Additives. Summary of the Fifty-Seventh Meeting of the Joint FAO; WHO Expert Committee on Food Additives (JECFA): Rome, Italy, 2001; Volume 514.
- Qing, Q.; Guo, Q.; Zhou, L.; Gao, X.; Lu, X.; Zhang, Y. Comparison of alkaline and acid pretreatments for enzymatic hydrolysis of soybean hull and soybean straw to produce fermentable sugars. Ind. Crops Prod. 2017, 109, 391–397. [Google Scholar] [CrossRef]
- Chen, Y.; Stevens, M.A.; Zhu, Y.; Holmes, J.; Xu, H. Understanding of alkaline pretreatment parameters for corn stover enzymatic saccharification. Biotechnol. Biofuels 2013, 6, 8. [Google Scholar] [CrossRef]
- Maurya, D.P.; Singla, A.; Negi, S. An overview of key pretreatment processes for biological conversion of lignocellulosic biomass to bioethanol. 3 Biotech 2015, 5, 597–609. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Sanders, J.P.; Bruins, M.E. Critical parameters in cost-effective alkaline extraction for high protein yield from leaves. Biomass Bioenergy 2014, 67, 466–472. [Google Scholar] [CrossRef]
- Huang, X.; Li, J.; Chang, C.; Gu, L.; Su, Y.; Yang, Y. Effects of NaOH/NaCl pickling on heat-induced gelation behaviour of egg white. Food Chem. 2019, 297, 124939. [Google Scholar] [CrossRef] [PubMed]
- Beukes, N.; Pletschke, B.I. Effect of alkaline pre-treatment on enzyme synergy for efficient hemicellulose hydrolysis in sugarcane bagasse. Bioresour. Technol. 2011, 102, 5207–5213. [Google Scholar] [CrossRef] [PubMed]
- Hamman, C.; Schmidt Jr, D.E.; Wong, M.; Hayes, M. The use of ammonium hydroxide as an additive in supercritical fluid chromatography for achiral and chiral separations and purifications of small, basic medicinal molecules. J. Chromatogr. A 2011, 1218, 7886–7894. [Google Scholar] [CrossRef]
- Beler-Baykal, B.; Bayram, S.; Akkaymak, E.; Cinar, S. Removal of ammonium from human urine through ion exchange with clinoptilolite and its recovery for further reuse. Water Sci. Technol. 2004, 50, 149–156. [Google Scholar] [CrossRef]
- Mafart, P.; Béliard, E. Food Industry Engineering. Volume II. Separation Techniques; Lavoisier: Paris, France, 1992. [Google Scholar]
- Wang, Y.; Wang, Z.; Handa, C.L.; Xu, J. Effects of ultrasound pre-treatment on the structure of β-conglycinin and glycinin and the antioxidant activity of their hydrolysates. Food Chem. 2017, 218, 165–172. [Google Scholar] [CrossRef]
- Wani, A.A.; Sogi, D.S.; Grover, L.; Saxena, D.C. Effect of temperature, alkali concentration, mixing time and meal/solvent ratio on the extraction of watermelon seed proteins—A response surface approach. Biosyst. Eng. 2006, 94, 67–73. [Google Scholar] [CrossRef]
- Rustom, I.Y.; Ópez-leiva, L.M.; Nair, B.M. Optimization of extraction of peanut proteins with water by response surface methodology. J. Food Sci. 1991, 56, 1660–1663. [Google Scholar] [CrossRef]
- Kang, P.Y.; Ishak, N.H.; Sarbon, N.M. Optimization of enzymatic hydrolysis of shortfin scad (Decapterus macrosoma) myofibrillar protein with antioxidant effect using alcalase. Int. Food Res. J. 2018, 25, 1808–1817. [Google Scholar]
- Mudgil, P.; Omar, L.S.; Kamal, H.; Kilari, B.P.; Maqsood, S. Multi-functional bioactive properties of intact and enzymatically hydrolysed quinoa and amaranth proteins. LWT 2019, 110, 207–213. [Google Scholar] [CrossRef]
- Kamal, H.; Le, C.F.; Salter, A.M.; Ali, A. Extraction of protein from food waste: An overview of current status and opportunities. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2455–2475. [Google Scholar] [CrossRef] [PubMed]
- Urribarrí, L.; Ferrer, A.; Colina, A. Leaf protein from ammonia-treated dwarf elephant grass (Pennisetum purpureum) schum cv. mott. Appl. Biochem. Biotechnol. 2005, 122, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Atasie, V.N.; Akinhanmi, T.F.; Ojiodu, C.C. Proximate analysis and physico-chemical properties of groundnut (Arachis hypogaea L.). Pak. J. Nutr. 2009, 8, 194–197. [Google Scholar] [CrossRef] [Green Version]
- García, R.; Pizarro, C.; Lavín, A.G.; Bueno, J.L. Biomass proximate analysis using thermogravimetry. Bioresour. Technol. 2013, 139, 1–4. [Google Scholar] [CrossRef]
- Odebunmi, E.O.; Oluwaniyi, O.O.; Bashiru, M.O. Comparative proximate analysis of some food condiments. J. Appl. Sci. Res. 2010, 6, 272–274. [Google Scholar]
- Telmo, C.; Lousada, J.; Moreira, N. Proximate analysis, backwards stepwise regression between gross calorific value, ultimate and chemical analysis of wood. Bioresour. Technol. 2010, 101, 3808–3815. [Google Scholar] [CrossRef]
- Ismail, B.P. Ash content determination. In Food Analysis Laboratory Manual; Springer: Berlin/Heidelberg, Germany, 2017; pp. 117–119. [Google Scholar]
- Luthria, D.; Vinjamoori, D.; Noel, K.; Ezzell, J. Accelerated solvent extraction. In Oil Extraction and Analysis; AOCS Publishing: Urbana, IL, USA, 2019; pp. 25–38. [Google Scholar]
- Merrill, A.L.; Watt, B.K. Energy Value of Foods: Basis and Derivation; U.S. Government Printing Office: Washington, DC, USA, 1955.
- Tömösközi, S.; Lásztity, R.; Haraszi, R.; Baticz, O. Isolation and study of the functional properties of pea proteins. Food/Nahrung 2001, 45, 399–401. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford method for protein quantitation. In The Protein Protocols Handbook; Humana Press: Totowa, NJ, USA, 2009; pp. 17–24. [Google Scholar]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Qutob, D.; Ma, F.; Peterson, C.A.; Bernards, M.A.; Gijzen, M. Structural and permeability properties of the soybean seed coat. Botany 2008, 86, 219–227. [Google Scholar] [CrossRef]
- Wu, J.; Johnson, L.A.; Jung, S. Demulsification of oil-rich emulsion from enzyme-assisted aqueous extraction of extruded soybean flakes. Bioresour. Technol. 2009, 100, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Bello, I.; Rasaq, N.; Adeniyi, A.; Hammed, A. Enzyme Aided Processing of Oil. Int. J. Halal Res. 2021, 3, 60–72. [Google Scholar] [CrossRef]
- Bayero, A.S.; Datti, Y.; Abdulhadi, M.; Yahya, A.T.; Salihu, I.; Lado, U.A.; Nura, T.; Imrana, B. Proximate composition and the mineral contents of soya beans (Glycine max) available in Kano State, Nigeria. ChemSearch J. 2019, 10, 62–65. [Google Scholar]
- Chiozza, M.V.; Burachik, M.; Miranda, P.V. Compositional analysis of soybean event IND-ØØ41Ø-5. GM Crops Food 2020, 11, 154–163. [Google Scholar] [CrossRef]
- Eden, W.T.; Rumambarsari, C.O. Proximate analysis of soybean and red beans cookies according to the Indonesian National Standard. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2020; Volume 1567, p. 022033. [Google Scholar]
- Etiosa, O.R.; Chika, N.B.; Benedicta, A. Mineral and proximate composition of soya bean. Asian J. Phys. Chem. Sci. 2017, 4, 1–6. [Google Scholar] [CrossRef]
- Smith, E.P.; Rose, K.A. Model goodness-of-fit analysis using regression and related techniques. Ecol. Model. 1995, 77, 49–64. [Google Scholar] [CrossRef]
- Ekpenyong, M.; Antai, S.; Asitok, A.; Ekpo, B. Response surface modeling and optimization of major medium variables for glycolipopeptide production. Biocatal. Agric. Biotechnol. 2017, 10, 113–121. [Google Scholar] [CrossRef]
- Halim, N.R.A.; Sarbon, N.M. A response surface approach on hydrolysis condition of eel (Monopterus sp.) protein hydrolysate with antioxidant activity. Int. Food Res. J. 2017, 24, 1081–1093. [Google Scholar]
- Skipnes, D.; Van der Plancken, I.; Van Loey, A.; Hendrickx, M.E. Kinetics of heat denaturation of proteins from farmed Atlantic cod (Gadus morhua). J. Food Eng. 2008, 85, 51–58. [Google Scholar] [CrossRef]
- Xie, Y.-Y.; Shu, Y.-L.; Li, D.-L.; Li, B.; Sun, Z.; Zou, Y.; Qiao, C.-S.; Jia, S.-R.; Li, Z.-X.; Zhong, C. Modeling Kinetics of the Water Extraction of Protein from Caragana korshinskii Kom. BioResources 2020, 15, 5032–5048. [Google Scholar] [CrossRef]
- Ovando, E.; Rodríguez-Sifuentes, L.; Martínez, L.M.; Chuck-Hernández, C. Optimization of Soybean Protein Extraction Using By-Products from NaCl Electrolysis as an Application of the Industrial Symbiosis Concept. Appl. Sci. 2022, 12, 3113. [Google Scholar] [CrossRef]
- Jarpa-Parra, M.; Bamdad, F.; Wang, Y.; Tian, Z.; Temelli, F.; Han, J.; Chen, L. Optimization of lentil protein extraction and the influence of process pH on protein structure and functionality. LWT-Food Sci. Technol. 2014, 57, 461–469. [Google Scholar] [CrossRef]
- Quidilig, J.R.B.; Guerrero, G.A.M.; Sanchez, D.E.S.; Gatdula, K.M.; Ventura, R.L.G.; Escobar, E.C.; Ventura, J.-R.S. Optimization of Protein Extraction from Chlorella vulgaris sp. using Response Surface Methodology. Philipp. e-J. Appl. Res. Dev. 2018, 8, 15–23. [Google Scholar]

| Parameters | Quantity (%) |
|---|---|
| Moisture | 10.78 ± 0.07 |
| Protein (Ν × 6.25) | 31.50 ± 2.61 |
| Fat | 19.34 ± 0.11 |
| Carbohydrate (by difference) | 33.84 ± 1.67 |
| Ash | 4.53 ± 0.07 |
| Variables | Levels | |||||
|---|---|---|---|---|---|---|
| Codes | −2 | −1 | 0 | 1 | 2 | |
| Extraction time (h) | X1 | 0 | 6 | 12 | 18 | 24 |
| Temperature (°C) | X2 | 25 | 40 | 55 | 70 | 85 |
| NH4OH Concentration (%) | X3 | 0.5 | 0.5 | 1 | 1.5 | 2 |
| Solvent ratio (w/v) | X4 | 1:5 | 1:5 | 1:10 | 1:15 | 1:15 |
| Coded Variables | Uncoded Variables | Responses | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Runs | X1 | X2 | X3 | X4 | Time, (X1) (h) | Temperature (X2), (°C) | NH4OH (X3), (%) | Solvent Ratio (X4) | Y1 | Y2 |
| (%) | (mM) | |||||||||
| 1 | −1 | −1 | −1 | −1 | 6 | 40 | 0.5 | 5 | 44.51 | 0.25 |
| 2 | 1 | −1 | −1 | −1 | 18 | 40 | 0.5 | 5 | 32.24 | 0.28 |
| 3 | −1 | 1 | −1 | −1 | 6 | 70 | 0.5 | 5 | 84.10 | 0.23 |
| 4 | 1 | 1 | −1 | −1 | 18 | 70 | 0.5 | 5 | 99.88 | 0.23 |
| 5 | −1 | −1 | 1 | −1 | 6 | 40 | 1.5 | 5 | 53.05 | 0.35 |
| 6 | 1 | −1 | 1 | −1 | 18 | 40 | 1.5 | 5 | 33.87 | 0.34 |
| 7 | −1 | 1 | 1 | −1 | 6 | 70 | 1.5 | 5 | 91.99 | 0.32 |
| 8 | 1 | 1 | 1 | −1 | 18 | 70 | 1.5 | 5 | 79.09 | 0.28 |
| 9 | −1 | −1 | −1 | 1 | 6 | 40 | 0.5 | 15 | 56.14 | 0.15 |
| 10 | 1 | −1 | −1 | 1 | 18 | 40 | 0.5 | 15 | 18.60 | 0.19 |
| 11 | −1 | 1 | −1 | 1 | 6 | 70 | 0.5 | 15 | 92.97 | 0.22 |
| 12 | 1 | 1 | −1 | 1 | 18 | 70 | 0.5 | 15 | 78.56 | 0.21 |
| 13 | −1 | −1 | 1 | 1 | 6 | 40 | 1.5 | 15 | 41.99 | 0.27 |
| 14 | 1 | −1 | 1 | 1 | 18 | 40 | 1.5 | 15 | 19.22 | 0.31 |
| 15 | −1 | 1 | 1 | 1 | 6 | 70 | 1.5 | 15 | 94.22 | 0.30 |
| 16 | 1 | 1 | 1 | 1 | 18 | 70 | 1.5 | 15 | 53.65 | 0.28 |
| 17 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 70.46 | 0.23 |
| 18 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 75.50 | 0.17 |
| 19 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 64.89 | 0.19 |
| 20 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 83.04 | 0.22 |
| 21 | −2 | 0 | 0 | 0 | 0 | 55 | 1 | 10 | 42.51 | 0.27 |
| 22 | 2 | 0 | 0 | 0 | 24 | 55 | 1 | 10 | 52.67 | 0.23 |
| 23 | 0 | −2 | 0 | 0 | 12 | 25 | 1 | 10 | 41.82 | 0.24 |
| 24 | 0 | 2 | 0 | 0 | 12 | 85 | 1 | 10 | 83.16 | 0.35 |
| 25 | 0 | 0 | 2 | 0 | 12 | 55 | 2 | 10 | 99.23 | 0.23 |
| 26 | 0 | 0 | 0 | 2 | 12 | 55 | 1 | 20 | 3.90 | 0.23 |
| 27 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 73.67 | 0.24 |
| 28 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 73.31 | 0.22 |
| 29 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 80.78 | 0.18 |
| 30 | 0 | 0 | 0 | 0 | 12 | 55 | 1 | 10 | 66.67 | 0.27 |
| Source | DF | Sum of Squares | Mean Squares | F-Value | p-Value |
|---|---|---|---|---|---|
| Regression Linear | 14 4 | 16,736.4 11,063.7 | 1195.46 2765.93 | 7.672 18.2034 | 0.0001677 0.0000014 |
| Two-way interaction | 6 | 853.4 | 142.23 | 0.9127 | 0.512141 |
| Quadratic | 4 | 4819.3 | 1204.83 | 7.7317 | 0.0004647 |
| Residuals | 15 | 2337.5 | 155.83 | - | - |
| Lack of fit | 14 | 2912.7 | 208.05 | 5.2357 | 0.4965 |
| Pure error | 7 | 278.2 | 39.74 | - | - |
| Total | 35 | 19,927.3 |
| Source | DF | Sum of Squares | Mean Squares | F-Value | p-Value |
|---|---|---|---|---|---|
| Regression Linear | 14 4 | 0.0616806 0.0295495 | 0.0044 0.007387 | 3.855 6.4634 | 0.006919 0.003132 |
| Two-way interaction | 6 | 0.0075330 | 0.001255 | 1.0985 | 0.407582 |
| Quadratic | 4 | 0.0245981 | 0.006149 | 5.3804 | 0.006848 |
| Residuals | 15 | 0.0171443 | 0.001143 | - | - |
| Lack of fit | 14 | 0.0089026 | 0.001113 | 1.0000 | 0.531090 |
| Pure error | 7 | 0.0082416 | 0.001177 | - | - |
| Total | 35 | 0.0788224 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bello, I.; Adeniyi, A.; Mukaila, T.; Hammed, A. Optimization of Soybean Protein Extraction with Ammonium Hydroxide (NH4OH) Using Response Surface Methodology. Foods 2023, 12, 1515. https://doi.org/10.3390/foods12071515
Bello I, Adeniyi A, Mukaila T, Hammed A. Optimization of Soybean Protein Extraction with Ammonium Hydroxide (NH4OH) Using Response Surface Methodology. Foods. 2023; 12(7):1515. https://doi.org/10.3390/foods12071515
Chicago/Turabian StyleBello, Ibrahim, Adewale Adeniyi, Taofeek Mukaila, and Ademola Hammed. 2023. "Optimization of Soybean Protein Extraction with Ammonium Hydroxide (NH4OH) Using Response Surface Methodology" Foods 12, no. 7: 1515. https://doi.org/10.3390/foods12071515





