Evaluation of Microbial Dynamics of Kombucha Consortia upon Continuous Backslopping in Coffee and Orange Juice
Abstract
:1. Introduction
2. Materials and methods
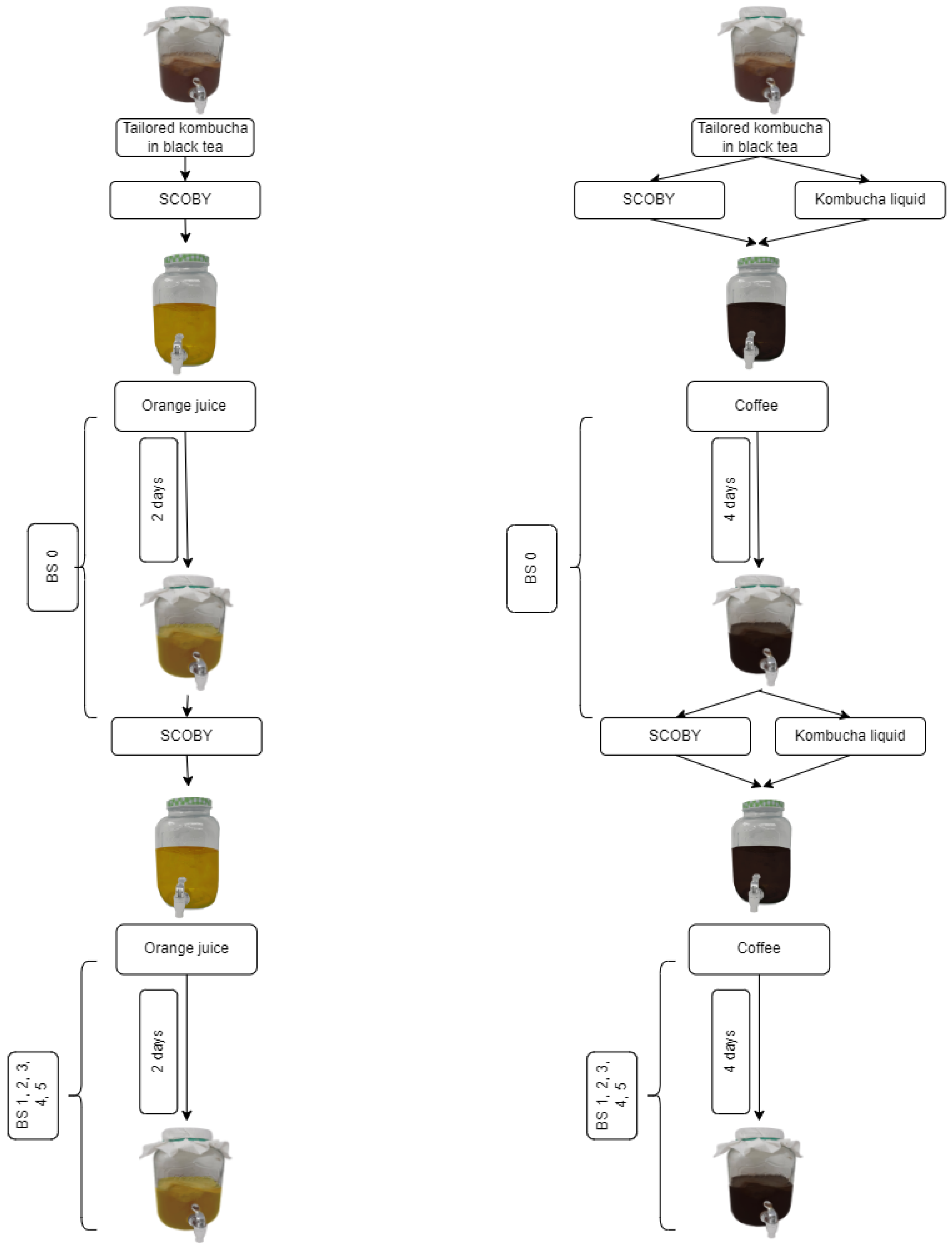
2.1. Creation of Kombucha Culture
2.2. Kombucha Cultivation in Coffee and Orange Juice
2.3. Determination of pH and Titratable Acidity
2.4. Determination of Metabolic Products in Kombucha
2.5. Microbial Quantitative Metagenomic analysis
2.5.1. Sample Preparation
2.5.2. Next-Generation Amplicon Sequencing
2.5.3. Data Quantification
2.6. Data Visualisation
3. Results
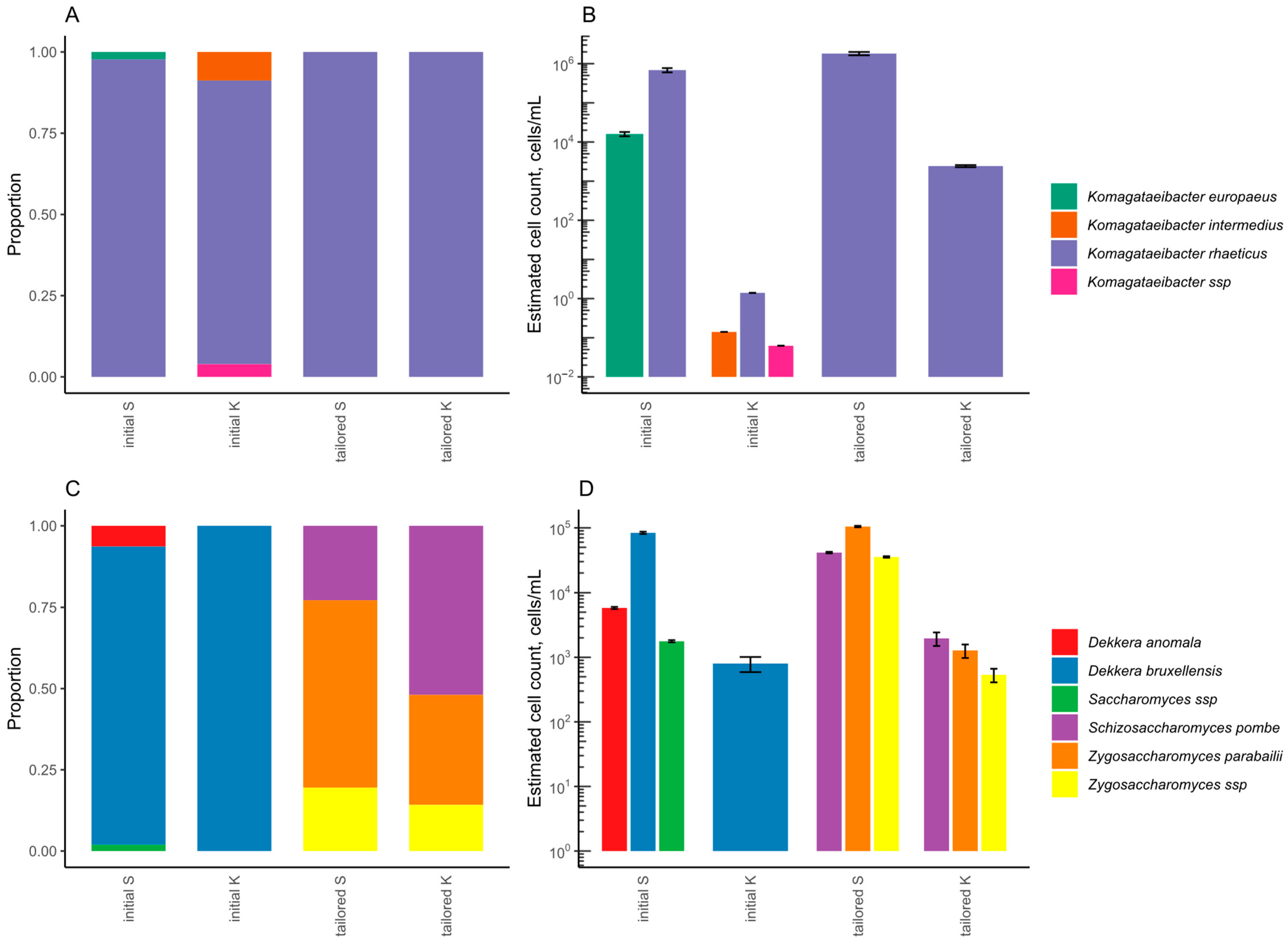
3.1. LAB-Tailored Kombucha Culture
3.2. Orange-Juice Kombucha Backslopping
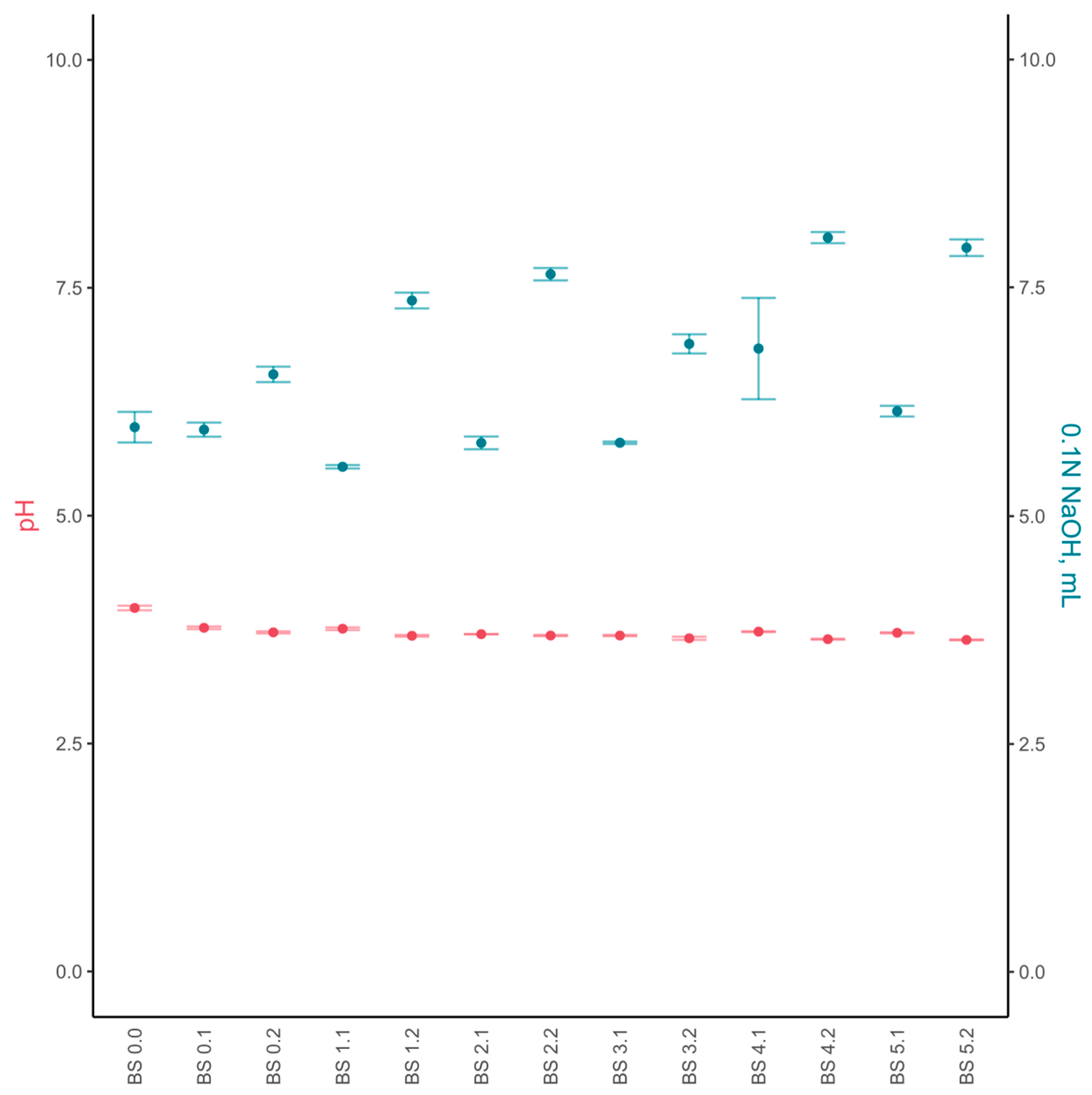
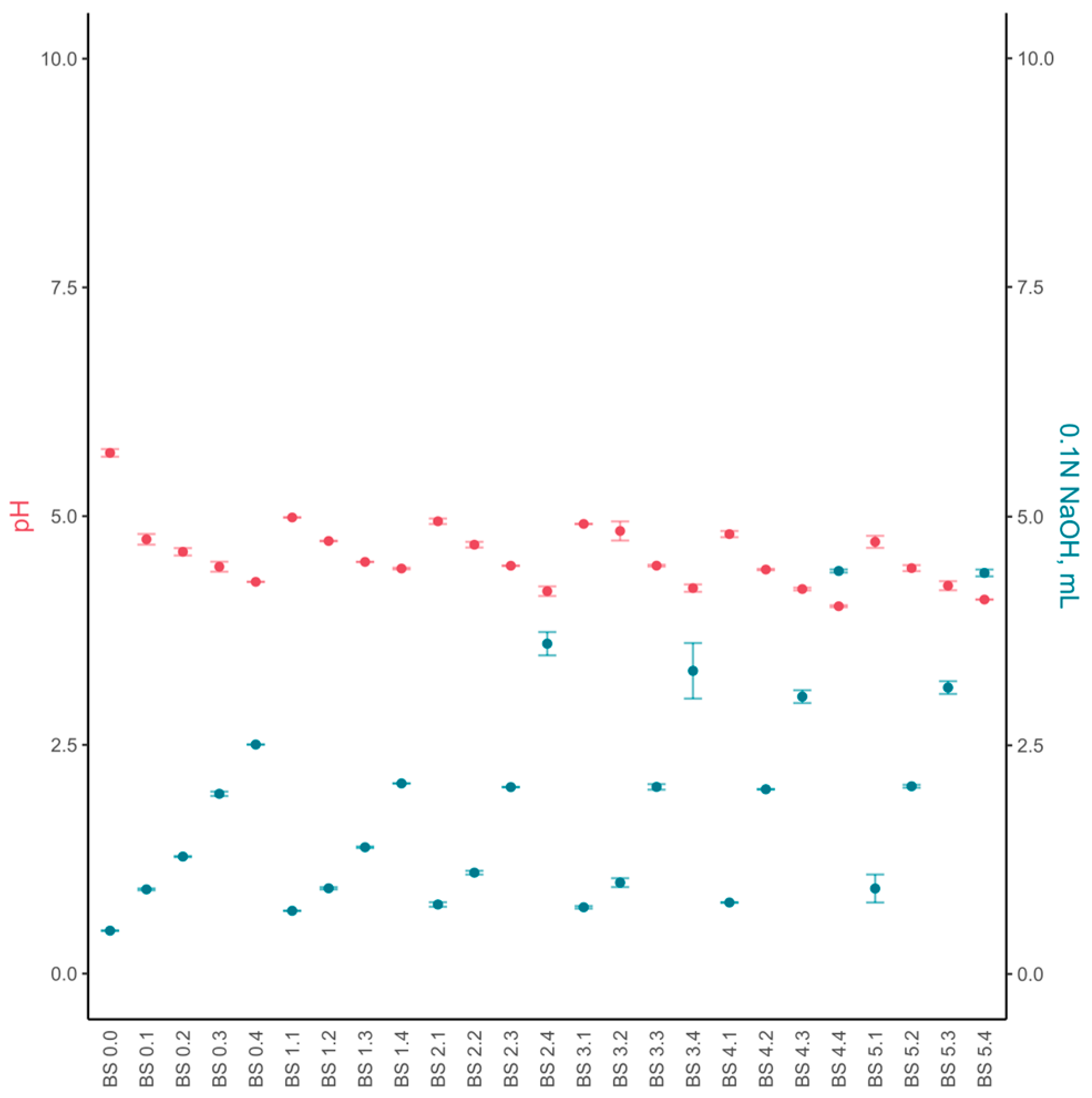
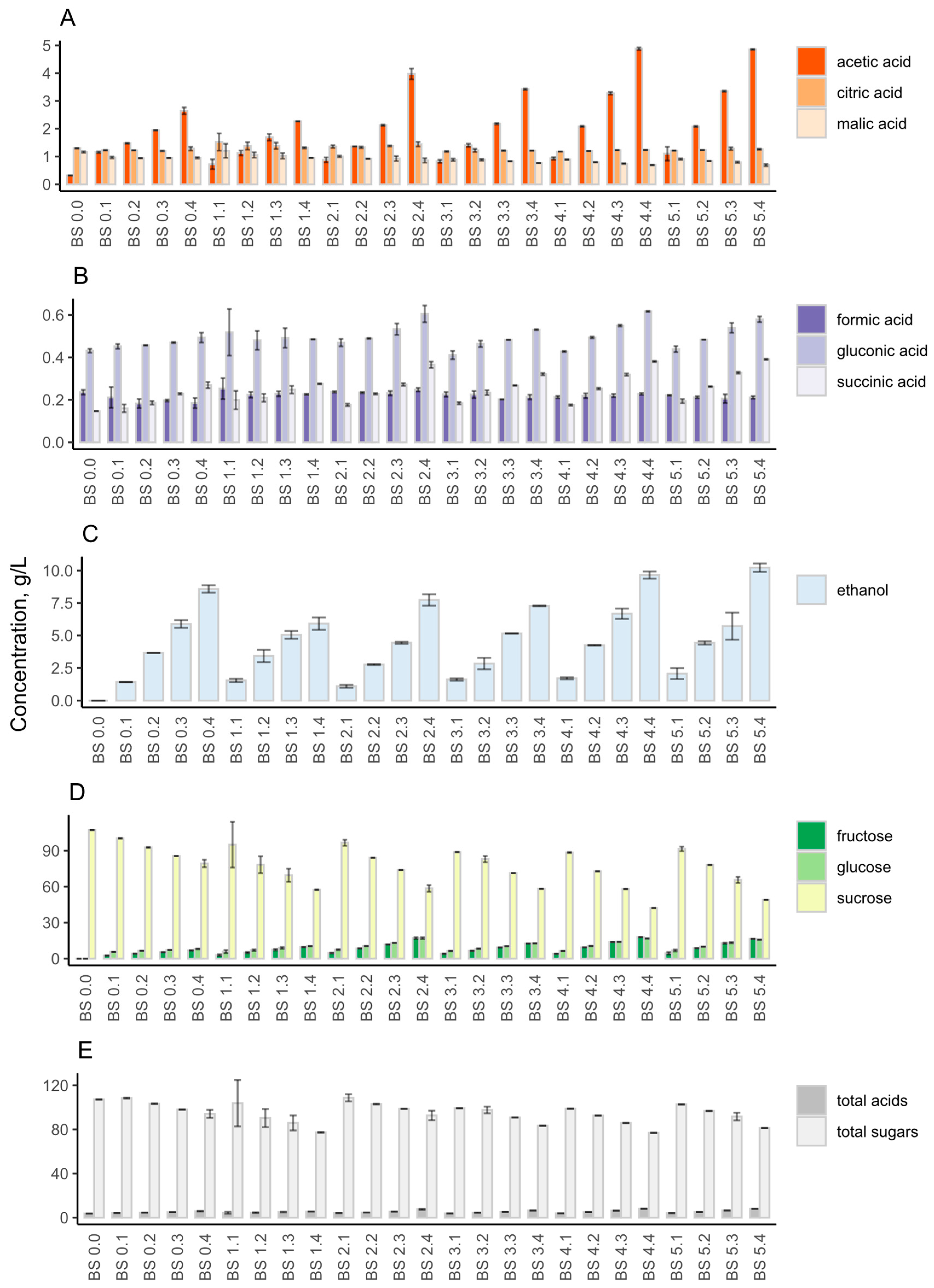
3.2.1. Chemical Composition and Sensory Evaluation
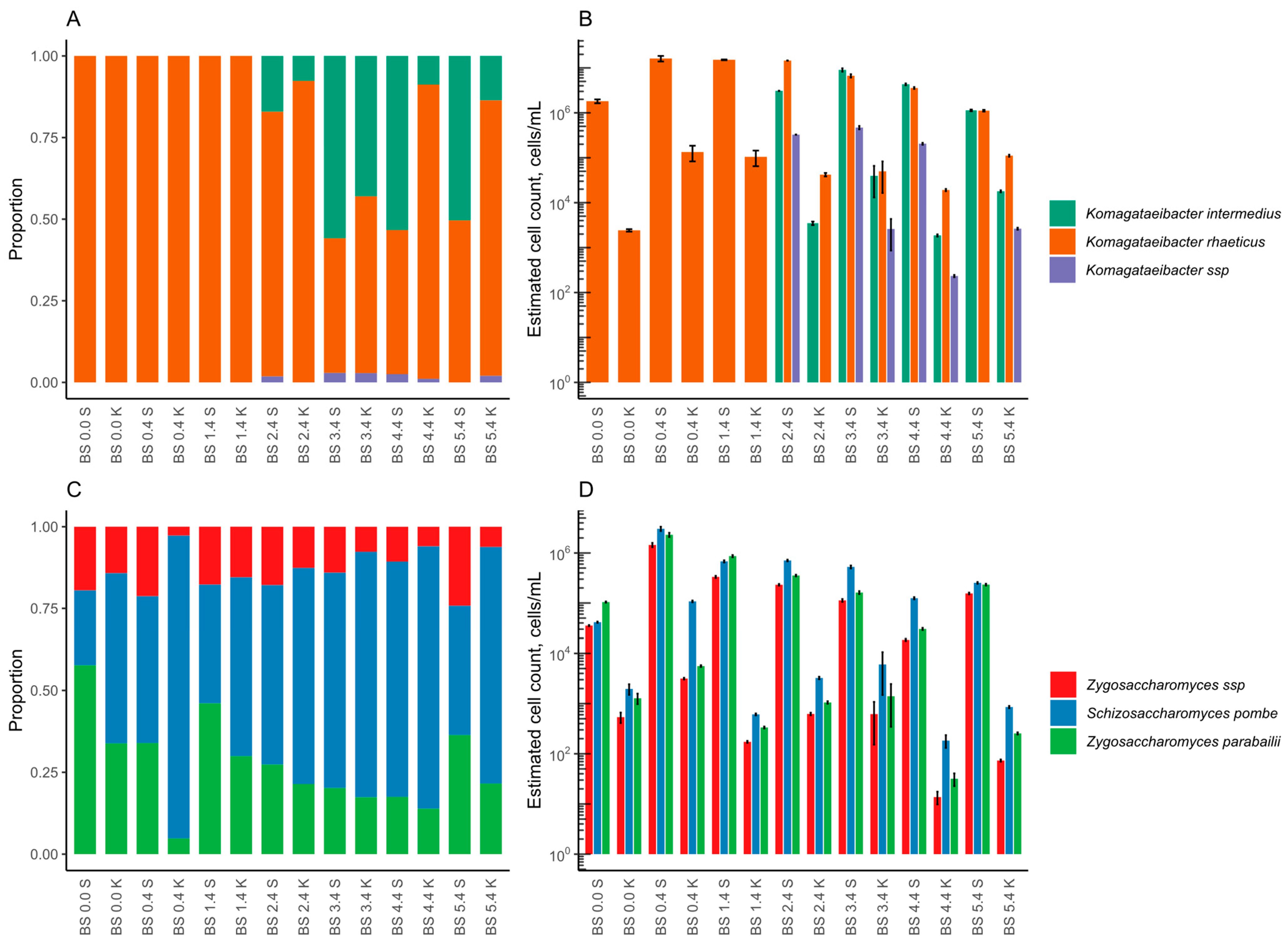
3.2.2. Microbial Composition
3.3. Coffee Kombucha Backslopping
3.3.1. Chemical Composition and Sensory Evaluation
3.3.2. Microbial Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reva, O.N.; Zaets, I.E.; Ovcharenko, L.P.; Kukharenko, O.E.; Shpylova, S.P.; Podolich, O.V.; de Vera, J.P.; Kozyrovska, N.O. Metabarcoding of the kombucha microbial community grown in different microenvironments. AMB Express 2015, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Andreson, M.; Kazantseva, J.; Kuldjärv, R.; Malv, E.; Vaikma, H.; Kaleda, A.; Kütt, M.-L.; Vilu, R. Characterisation of chemical, microbial and sensory profiles of commercial kombuchas. Int. J. Food Microbiol. 2022, 373, 109715. [Google Scholar] [CrossRef] [PubMed]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.P.; Taillandier, P. Understanding Kombucha Tea Fermentation: A Review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93, fix048. [Google Scholar] [CrossRef]
- May, A.; Narayanan, S.; Alcock, J.; Varsani, A.; Maley, C.; Aktipis, A. Kombucha: A novel model system for cooperation and conflict in a complex multi-species microbial ecosystem. PeerJ 2019, 2019, e7565. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbaša, R.V.; Lončar, E.S.; Vitas, J.S.; Sathishkumar, M. A review on kombucha tea-microbiology, composition, fermentation, beneficial effects, toxicity, and tea fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef]
- Nionelli, L.; Rizzello, C.G. Sourdough-based biotechnologies for the production of gluten-free foods. Foods 2016, 5, 65. [Google Scholar] [CrossRef]
- Emiljanowicz, K.E.; Malinowska-Pańczyk, E. Kombucha from alternative raw materials—The review. Crit. Rev. Food Sci. Nutr. 2020, 60, 3185–3194. [Google Scholar] [CrossRef]
- Sun, T.Y.; Li, J.S.; Chen, C. Effects of blending wheatgrass juice on enhancing phenolic compounds and antioxidant activities of traditional kombucha beverage. J. Food Drug Anal. 2015, 23, 709–718. [Google Scholar] [CrossRef]
- Zofia, N.-Ł.; Aleksandra, Z.; Tomasz, B.; Martyna, Z.; Tomasz, W. Effect of fermentation time on antioxidant and anti-ageing properties of green coffee kombucha ferments. Molecules 2020, 25, 5394. [Google Scholar] [CrossRef]
- Tu, C.; Tang, S.; Azi, F.; Hu, W.; Dong, M. Use of kombucha consortium to transform soy whey into a novel functional beverage. J. Funct. Foods 2019, 52, 81–89. [Google Scholar] [CrossRef]
- Chakravorty, S.; Bhattacharya, S.; Chatzinotas, A.; Chakraborty, W.; Bhattacharya, D.; Gachhui, R. Kombucha tea fermentation: Microbial and biochemical dynamics. Int. J. Food Microbiol. 2016, 220, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Savary, O.; Mounier, J.; Thierry, A.; Poirier, E.; Jourdren, J.; Maillard, M.B.; Penland, M.; Decamps, C.; Coton, E.; Coton, M. Tailor-made microbial consortium for Kombucha fermentation: Microbiota-induced biochemical changes and biofilm formation. Food Res. Int. 2021, 147, 110549. [Google Scholar] [CrossRef] [PubMed]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbiol. 2014, 38, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Bessmeltseva, M.; Viiard, E.; Simm, J.; Paalme, T.; Sarand, I. Evolution of Bacterial Consortia in Spontaneously Started Rye Sourdoughs during Two Months of Daily Propagation. PLoS ONE 2014, 9, e95449. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Taylor, D.L.; Walters, W.A.; Lennon, N.J.; Bochicchio, J.; Krohn, A.; Caporaso, J.G.; Pennanen, T. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl. Environ. Microbiol. 2016, 82, 7217–7226. [Google Scholar] [CrossRef]
- Kazantseva, J.; Malv, E.; Kaleda, A.; Kallastu, A.; Meikas, A. Optimisation of sample storage and DNA extraction for human gut microbiota studies. BMC Microbiol. 2021, 21, 158. [Google Scholar] [CrossRef]
- Espinosa-Gongora, C.; Larsen, N.; Schønning, K.; Fredholm, M.; Guardabassi, L. Differential Analysis of the Nasal Microbiome of Pig Carriers or Non-Carriers of Staphylococcus aureus. PLoS ONE 2016, 11, e0160331. [Google Scholar] [CrossRef]
- McDonald, J.E.; Larsen, N.; Pennington, A.; Connolly, J.; Wallis, C.; Rooks, D.J.; Hall, N.; McCarthy, A.J.; Allison, H.E. Characterising the Canine Oral Microbiome by Direct Sequencing of Reverse-Transcribed rRNA Molecules. PLoS ONE 2016, 11, e0157046. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Fiedorová, K.; Radvanský, M.; Němcová, E.; Grombiříková, H.; Bosák, J.; Černochová, M.; Lexa, M.; Šmajs, D.; Freiberger, T. The Impact of DNA Extraction Methods on Stool Bacterial and Fungal Microbiota Community Recovery. Front. Microbiol. 2019, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Yavari, N.; Mazaheri-Assadi, M.; Mazhari, Z.H.; Moghadam, M.B.; Larijani, K. Glucuronic Acid Rich Kombucha-fermented Pomegranate Juice. J. Food Res. 2017, 7, 61. [Google Scholar] [CrossRef]
- Ayed, L.; Ben Abid, S.; Hamdi, M. Development of a beverage from red grape juice fermented with the Kombucha consortium. Ann. Microbiol. 2017, 67, 111–121. [Google Scholar] [CrossRef]
- Nguyen, N.K.; Dong, N.T.N.; Nguyen, H.T.; Le, P.H. Lactic acid bacteria: Promising supplements for enhancing the biological activities of kombucha. Springerplus 2015, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.K.; Dong, N.T.N.; Le, P.H.; Nguyen, H.T. Evaluation of the glucuronic acid production and other biological activities of fermented sweeten-black tea by kombucha layer and the co-culture with different Lactobacillus sp. strains. Int. J. Mod. Eng. Res. 2014, 4, 12–17. [Google Scholar]
- Chen, C.; Liu, B.Y. Changes in major components of tea fungus metabolites during prolonged fermentation. J. Appl. Microbiol. 2000, 89, 834–839. [Google Scholar] [CrossRef]
- Tan, L.; Sun, Z.Y.; Okamoto, S.; Takaki, M.; Tang, Y.Q.; Morimura, S.; Kida, K. Production of ethanol from raw juice and thick juice of sugar beet by continuous ethanol fermentation with flocculating yeast strain KF-7. Biomass Bioenergy 2015, 81, 265–272. [Google Scholar] [CrossRef]
- Liu, S.; Qureshi, N. How microbes tolerate ethanol and butanol. Nat. Biotechnol. 2009, 26, 117–121. [Google Scholar] [CrossRef]
- Alcoholic Beverages | Knowledge for Policy. Available online: https://knowledge4policy.ec.europa.eu/health-promotion-knowledge-gateway/alcoholic-beverages_en (accessed on 12 July 2023).
- Kim, J.; Adhikari, K. Current trends in kombucha: Marketing perspectives and the need for improved sensory research. Beverages 2020, 6, 15. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreson, M.; Kazantseva, J.; Malv, E.; Kuldjärv, R.; Priidik, R.; Kütt, M.-L. Evaluation of Microbial Dynamics of Kombucha Consortia upon Continuous Backslopping in Coffee and Orange Juice. Foods 2023, 12, 3545. https://doi.org/10.3390/foods12193545
Andreson M, Kazantseva J, Malv E, Kuldjärv R, Priidik R, Kütt M-L. Evaluation of Microbial Dynamics of Kombucha Consortia upon Continuous Backslopping in Coffee and Orange Juice. Foods. 2023; 12(19):3545. https://doi.org/10.3390/foods12193545
Chicago/Turabian StyleAndreson, Maret, Jekaterina Kazantseva, Esther Malv, Rain Kuldjärv, Reimo Priidik, and Mary-Liis Kütt. 2023. "Evaluation of Microbial Dynamics of Kombucha Consortia upon Continuous Backslopping in Coffee and Orange Juice" Foods 12, no. 19: 3545. https://doi.org/10.3390/foods12193545




