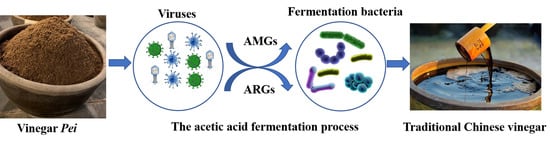
Metagenomics of Virus Diversities in Solid-State Brewing Process of Traditional Chinese Vinegar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vinegar Pei Samples Collection
2.2. Virus Purification
2.3. Viral and Bacterial DNA Extraction and Virome Sequencing
2.4. Identification of Auxiliary Carbohydrate Metabolic Genes
2.5. Antibiotic Resistance Gene Search
3. Results and Discussion
3.1. Investigation of Bacterial and Viral Metagenomes of Vinegar Pei
3.2. Taxonomic Diversities of Vinegar Pei Viral and Bacterial Communities
3.3. Dynamic Changes of Viral Communities during the Fermentation and the Interaction with Bacterial Community in Vinegar Pei
3.4. Abundant Auxiliary Carbohydrate Metabolic Genes in Vinegar Pei Viruses
3.5. Abundant Antibiotic Resistance Genes in Vinegar Pei Viruses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, T.; Duan, W.; Zhang, Z.; Fang, B.; Zhang, B.; Xu, B.; de la Cruz, C.B.V.; El-Seedi, H.; Simal-Gandara, J.; Wang, S.; et al. Polyphenol-rich extract of Zhenjiang aromatic vinegar ameliorates high glucose-induced insulin resistance by regulating JNK-IRS-1 and PI3K/Akt signaling pathways. Food Chem. 2021, 335, 127513. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Zhang, B.; Duan, W.H.; Zhang, J.; Wang, M. Nutrients and bioactive components from vinegar: A fermented and functional food. J. Funct. Foods 2020, 64, 103681. [Google Scholar] [CrossRef]
- Al-Dalali, S.; Zheng, F.P.; Xu, B.C.; Abughoush, M.; Li, L.H.; Sun, B.G. Processing technologies and flavor analysis of Chinese cereal vinegar: A comprehensive review. Food Anal. Method. 2022. [Google Scholar] [CrossRef]
- Irwin, N.A.T.; Pittis, A.A.; Richards, T.A.; Keeling, P.J. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat. Microbiol. 2022, 7, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Maske, B.L.; de Melo Pereira, G.V.; da Silva Vale, A.; Marques Souza, D.S.; de dea Lindner, J.; Soccol, C.R. Viruses in fermented foods: Are they good or bad? Two sides of the same coin. Food Microbiol. 2021, 98, 103794. [Google Scholar] [CrossRef] [PubMed]
- Agyirifo, D.S.; Wamalwa, M.; Otwe, E.P.; Galyuon, I.; Runo, S.; Takrama, J.; Ngeranwa, J. Metagenomics analysis of cocoa bean fermentation microbiome identifying species diversity and putative functional capabilities. Heliyon 2019, 5, e02170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queiroz, L.L.; Lacorte, G.A.; Isidorio, W.R.; Landgraf, M.; de Melo Franco BD, G.; Pinto, U.M.; Hoffmann, C. High Level of Interaction between phages and bacteria in an artisanal raw milk cheese microbial community. bioRxiv 2021, 454940. [Google Scholar] [CrossRef]
- Pacini, S.; Ruggiero, M. Phage composition of a fermented milk and colostrum product assessed by microbiome array; putative role of open reading frames in reference to cell signaling and neurological development. bioRxiv 2020, 714154. [Google Scholar] [CrossRef]
- Huang, X.; Jiao, N.; Zhang, R. The genomic content and context of auxiliary metabolic genes in roseophages. Environ. Microbiol. 2021, 23, 3743–3757. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anderson, R.E.; Buchan, A.; Campbell, B.J.; Hallam, S.J.; Hess, M.; Sullivan, M.B.; Walsh, D.A.; Roux, S.; et al. Ecology of inorganic sulfur auxiliary metabolism in widespread bacteriophages. Nat. Commun. 2021, 12, 3503. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, R.; Zhang, T.; Sun, C. Deep-sea bacteriophages facilitate their host in utilizing polysaccharide. bioRxiv 2022, 504481. [Google Scholar] [CrossRef]
- Heyerhoff, B.; Engelen, B.; Bunse, C. Auxiliary metabolic gene functions in pelagic and benthic viruses of the Baltic Sea. Front. Microbiol. 2022, 13, 863620. [Google Scholar] [CrossRef] [PubMed]
- Rong, C.; Zhou, K.; Li, S.; Xiao, K.; Xu, Y.; Zhang, R.; Yang, Y.; Zhang, Y. Isolation and characterization of a novel cyanophage encoding multiple auxiliary metabolic genes. Viruses 2022, 14, 887. [Google Scholar] [CrossRef] [PubMed]
- Gazitua, M.C.; Vik, D.R.; Roux, S.; Gregory, A.C.; Bolduc, B.; Widner, B.; Mulholland, M.R.; Hallam, S.J.; Ulloa, O.; Sullivan, M.B. Potential virus-mediated nitrogen cycling in oxygen-depleted oceanic waters. ISME J. 2021, 15, 981–998. [Google Scholar] [CrossRef]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef]
- Huang, D.; Yu, P.F.; Ye, M.; Schwarz, C.; Jiang, X.; Alvarez, P.J.J. Enhanced mutualistic symbiosis between soil phages and bacteria with elevated chromium-induced environmental stress. Microbiome 2021, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.X.; Jahn, M.T.; Sun, M.M.; Friman, V.P.; Balcazar, J.L.; Wang, J.F.; Shi, Y.; Gong, X.; Hu, F.; Zhu, Y.G. Organochlorine contamination enriches virus-encoded metabolism and pesticide degradation associated auxiliary genes in soil microbiomes. ISME J. 2022, 16, 1397–1408. [Google Scholar] [CrossRef]
- Kou, R.; Li, M.; Xing, J.; He, Y.; Wang, H.; Fan, X. Exploring of seasonal dynamics of microbial community in multispecies fermentation of Shanxi mature vinegar. J. Biosci. Bioeng. 2022, 133, 375–381. [Google Scholar] [CrossRef]
- Dugat-Bony, E.; Lossouarn, J.; De Paepe, M.; Sarthou, A.S.; Fedala, Y.; Petit, M.A.; Chaillou, S. Viral metagenomic analysis of the cheese surface: A comparative study of rapid procedures for extracting viral particles. Food Microbiol. 2020, 85, 103278. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.M.; Leung, C.M.; Ting, H.F.; Sadakane, K.; Yamashita, H.; Lam, T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lomsadze, A.; Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic. Acids. Res. 2010, 38, e132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.; Guo, X.; Zhang, R.; Qu, W.; Gao, B.; Zeng, R. Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome 2019, 7, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic. Acids. Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Omata, K.; Hibi, N.; Nakano, S.; Komoto, S.; Sato, K.; Nunokawa, K.; Amano, S.; Ueda, K.; Takano, H. Distribution and genome structures of temperate phages in acetic acid bacteria. Sci. Rep. 2021, 11, 21567. [Google Scholar] [CrossRef] [PubMed]
- Rihtman, B.; Bowman-Grahl, S.; Millard, A.; Corrigan, R.M.; Clokie, M.R.J.; Scanlan, D.J. Cyanophage MazG is a pyrophosphohydrolase but unable to hydrolyse magic spot nucleotides. Environ. Microbiol. Rep. 2019, 11, 448–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.M.; Zhang, Y.; Hong, H.S. Classification of acetic acid bacteria and their acid resistant mechanism. Amb. Express. 2021, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Ichita, J.; Kato, Y. Production of vinegar from apple pomace and its physiological function. J. Jpn. Soc. Food Sci. 2011, 58, 37–42. [Google Scholar] [CrossRef] [Green Version]
- Debroas, D.; Siguret, C. Viruses as key reservoirs of antibiotic resistance genes in the environment. ISME J. 2019, 13, 2856–2867. [Google Scholar] [CrossRef] [PubMed]
- Sagrillo, C.; Changey, F.; Bellanger, X. Bacteriophages vehiculate a high amount of antibiotic resistance determinants of bacterial origin in the Orne River ecosystem. Environ. Microbiol. 2022, 24, 4317–4328. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Rana, A.P.; Wenzler, E.; Ozer, E.A.; Krapp, F.; Bulitta, J.B.; Hauser, A.R.; Bulman, Z.P. Aminoglycoside-resistance gene signatures are predictive of aminoglycoside MICs for carbapenem-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2022, 77, 356–363. [Google Scholar] [CrossRef]
- Wachino, J.I.; Doi, Y.; Arakawa, Y. Aminoglycoside resistance: Updates with a focus on acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 2020, 34, 887–902. [Google Scholar] [CrossRef]
- Moon, K.; Jeon, J.H.; Kang, I.N.; Park, K.S.; Lee, K.Y.; Cha, C.J.; Lee, S.H.; Cho, J.C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef]
- Hou, W.T.; Xu, D.; Wang, L.; Chen, Y.; Chen, Z.P.; Zhou, C.Z.; Chen, Y. Plastic structures for diverse substrates: A revisit of human ABC transporters. Proteins 2022, 90, 1749–1765. [Google Scholar] [CrossRef]
- Kikuchi, T.; Hayashi, A.; Ikeda, N.; Morita, O.; Tasaki, J. Multidrug resistance-associated protein 2 (MRP2) is an efflux transporter of EGCG and its metabolites in the human small intestine. J. Nutr. Biochem. 2022, 107, 109071. [Google Scholar] [CrossRef]
- Ray, M.; Manjunath, A.; Halami, P.M. Prevalence of polymyxin resistance through the food chain, the global crisis. J. Antibiot. 2022, 75, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.P.; Pang, W.H.; Dou, C.L.; Yin, D.Q. Sulfamethoxazole and COD increase abundance of sulfonamide resistance genes and change bacterial community structures within sequencing batch reactors. Chemosphere 2017, 175, 21–27. [Google Scholar] [CrossRef]
- Rindi, L. Efflux pump inhibitors against nontuberculous mycobacteria. Int. J. Mol. Sci. 2020, 21, 4191. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, A.; Avison, M.B. Aph(3′)-IIc, an aminoglycoside resistance determinant from Stenotrophomonas maltophilia. Antimicrob. Agents. Chemother. 2007, 51, 359–360. [Google Scholar] [CrossRef] [PubMed]




| Samples | Bacterial Metagenomes | Viral Metagenomes | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of Clean Reads | Q20% | Number of Contigs | Predicted ORF Number | Number of Clean Reads | Q20% | Number of Contigs | Predicted ORF Number | |
| 0d-1 | 61,503,358 | 98.23 | 84,185 | 57,456 | 15,100,950 | 96.54 | 5169 | 3224 |
| 0d-2 | 65,015,640 | 98.19 | 87,846 | 60,122 | 7,127,744 | 96.64 | 1002 | 626 |
| 0d-3 | 62,704,114 | 98.09 | 124,169 | 85,929 | 31,426,924 | 97.05 | 24,646 | 16,876 |
| 8d-1 | 106,916,406 | 97.53 | 141,863 | 230,229 | 4,713,670 | 93.42 | 15,783 | 27,164 |
| 8d-2 | 84,764,580 | 91.23 | 128,904 | 208,635 | 4,543,128 | 91.23 | 16,241 | 27,219 |
| 8d-3 | 103,547,844 | 97.26 | 142,724 | 233,312 | 12,537,328 | 93.49 | 15,203 | 36,663 |
| 12d-1 | 84,039,874 | 96.89 | 102,676 | 175,735 | 9,204,336 | 93.89 | 15,562 | 26,444 |
| 12d-2 | 17,228,660 | 96.63 | 18,150 | 43,128 | 17,228,660 | 93.73 | 18,183 | 43,145 |
| 12d-3 | 96,679,336 | 96.29 | 126,429 | 212,888 | 6,556,868 | 93.41 | 14,005 | 22,960 |
| 18d-1 | 61,944,434 | 97.27 | 53,651 | 90,339 | 12,858,800 | 93.46 | 10,056 | 17,343 |
| 18d-2 | 94,410,756 | 97.48 | 72,331 | 121,053 | 5,626,462 | 93.46 | 7917 | 13,722 |
| 18d-3 | 76,201,500 | 97.42 | 53,490 | 88,684 | 5,692,556 | 91.30 | 6936 | 10,804 |
| Viral Auxiliary CAZymes | Viral Auxiliary CAZymes |
|---|---|
| AAs family | Peptidoglycan hydrolase with Endo-beta-N-acetylglucosaminidase specificity (GH73) |
| Copper-dependent lytic polysaccharide monooxygenases (AA10) | Amylomaltase or 4-alpha-glucanotransferase (GH77) |
| Pyrroloquinoline quinone-dependent oxidoreductase (AA12) | Alpha-L-rhamnosidase (GH78) |
| Class II lignin-modifying peroxidases (AA2) | Chitosanase (GH80) |
| Glucose-methanol-choline (GMC) oxidoreductases (AA3) | Glycoprotein endo-alpha-1,2-mannosidase (GH99) |
| Vanillyl-alcohol oxidases (AA4) | GTs family |
| 1,4-benzoquinone reductases (AA6) | Lipid-A-disaccharide synthase (GT19) |
| CEs family | Cellulose synthase (GT2) |
| UDP-3-0-acyl N-acetylglucosamine deacetylase (CE11) | Alpha, alpha-trehalose-phosphate synthase (GT20) |
| Acetylesterase (CE16) | Ceramide beta-glucosyltransferase (GT21) |
| Acetyl xylan esterase (CE2, CE3, CE6) | Polypeptide alpha-N-acetylgalactosaminyltransferase (GT27) |
| GHs familyCellulase (GH5) | Alpha-3-deoxy-D-manno-octulosonic-acid (KDO) Transferase (GT30) |
| Peptidoglycan lytic transglycosylases (GH23, GH102, GH103, GH104) | Glycogen or starch phosphorylase (GT35) |
| Unsaturated rhamnogalacturonyl hydrolase (GH105) | Peptide beta-N-acetylglucosaminyltransferase (GT41) |
| N-acetylmuramidase (GH108) | Murein polymerase (GT51) |
| Alpha-N-acetylgalactosaminidase (GH109) | Lipid II Fuc4NAc transferase (GT56) |
| Beta-mannanase (GH113) | Alpha-3-deoxy-D-manno-octulosonic-acid (KDO) Transferase (GT73) |
| Endo-alpha-1,4-polygalactosaminidase (GH114) | Cyclic beta-1,2-glucan synthase (GT84) |
| β-L-arabinofuranosidase (GH127) | PLs family |
| Chitinase (GH19) | Pectate lyase (PL10) |
| Lysozyme (GH24, GH25) | Heparin-sulfate lyase (PL12) |
| Alpha-L-fucosidase (GH29) | Oligo-alginate lyase (PL15) |
| Alpha, alpha-trehalase (GH37) | Alginate lyase (PL17, PL5) |
| Beta-agarase (GH50) | Oligogalacturonate lyase/oligogalacturonide lyase (PL22) |
| Target Antibiotics | 0d-1 | 0d-2 | 0d-3 | 8d-1 | 8d-2 | 8d-3 | 12d-1 | 12d-2 | 12d-3 | 18d-1 | 18d-2 | 18d-3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sulfonamide | 0 | 0 | 0 | 36 | 23 | 5 | 8 | 3 | 10 | 2 | 11 | 3 |
| multidrug transport | 0 | 0 | 0 | 49 | 54 | 41 | 18 | 36 | 23 | 9 | 21 | 19 |
| aminoglycoside | 0 | 0 | 1 | 53 | 39 | 40 | 12 | 37 | 20 | 5 | 11 | 13 |
| fluoroquinolone | 0 | 0 | 0 | 9 | 11 | 10 | 3 | 9 | 3 | 1 | 4 | 4 |
| elfamycin | 0 | 0 | 0 | 14 | 11 | 20 | 7 | 14 | 5 | 3 | 6 | 4 |
| polymyxin | 0 | 0 | 0 | 29 | 30 | 26 | 10 | 20 | 14 | 5 | 10 | 11 |
| mupirocin | 0 | 0 | 0 | 13 | 12 | 9 | 4 | 8 | 4 | 1 | 2 | 3 |
| novobiocin | 0 | 0 | 0 | 69 | 37 | 21 | 13 | 14 | 15 | 8 | 12 | 20 |
| quinolone | 0 | 0 | 0 | 20 | 8 | 19 | 5 | 17 | 5 | 2 | 2 | 3 |
| Total | 0 | 0 | 1 | 292 | 225 | 191 | 80 | 158 | 99 | 36 | 79 | 80 |
| % in virome reads a | 0 | 0 | 0 | 0.0062% | 0.005% | 0.0015% | 0.0009% | 0.0009% | 0.0015% | 0.0003% | 0.0014% | 0.0014% |
| Protein ID a | ARO Category b | ARG Name | e Value | % Identity |
|---|---|---|---|---|
| 12d-2-k141-25303 gene 30071 | efflux pump | lrfA | 2.19 × 10−33 | 27.59 |
| 12d-2-k141-25303 gene 30077 | efflux pump | lrfA | 7.04 × 10−33 | 30.08 |
| 12d-2-k141-8532 gene 10306 | efflux pump | lrfA | 2.29 × 10−67 | 37.23 |
| 12d-2-k141-23839 gene 28370 | efflux pump | lrfA | 2.41 × 10−39 | 31.50 |
| 8d-3-k141-5725 gene 7838 | efflux pump | lrfA | 1.02 × 10−66 | 37.23 |
| 18d-1-k141-10283 gene 7834 | efflux pump | macA | 1.62 × 10−52 | 47.12 |
| 12d-2-k141-15026 gene 18130 | efflux pump | macA | 2.48 × 10−64 | 37.40 |
| 12d-2-k141-13619 gene 16396 | efflux pump | macA | 1.72 × 10−64 | 36.41 |
| 12d-1-k141-16328 gene 12997 | efflux pump | macA | 3.76 × 10−85 | 39.53 |
| 12d-2-k141-26375 gene 31413 | efflux pump | macA | 7.51 × 10−84 | 41.19 |
| 12d-2-k141-28030 gene 33312 | efflux pump | macA | 1.28 × 10−81 | 41.58 |
| 18d-3-k141-6633 gene 4643 | efflux pump | macB | 8.90 × 10−133 | 54.19 |
| 8d-3-k141-28586 gene 37717 | efflux pump | macB | 6.36 × 10−161 | 40.54 |
| 12d-2-k141-17472 gene 21035 | antibiotic inactivation | APH (3’)-IIc | 2.77 × 10−161 | 84.64 |
| 12d-2-k141-18203 gene 22034 | antibiotic inactivation | APH (3’)-IIc | 8.53 × 10−153 | 80.52 |
| 12d-1-k141-16136 gene 12816 | antibiotic resistant gene variant or mutant | EF-Tu | 1.27 × 10−109 | 70.56 |
| 8d-3-k141-1508 gene 1984 | antibiotic resistant gene variant or mutant | parC | 6.32 × 10−119 | 49.74 |
| 8d-3-k141-4044 gene 5547 | antibiotic resistant gene variant or mutant | gyrB | 5.97 × 10−171 | 47.53 |
| 12d-3-k141-25307 gene 18959 | aminocoumarin resistance gene | alaS | 5.88 × 10−167 | 66.57 |
| 8d-2-k141-36863 gene 29116 | aminocoumarin resistance gene | alaS | 5.04 × 10−118 | 87.57 |
| 18d-1-k141-10925 gene 8326 | aminocoumarin resistance gene | alaS | 1.61 × 10−63 | 55.17 |
| 8d-1-k141-631 gene 476 | aminocoumarin resistance gene | alaS | 8.61 × 10−78 | 62.96 |
| 8d-1-k141-34699 gene 28239 | aminocoumarin resistance gene | alaS | 8.77 × 10−110 | 83.62 |
| 8d-3-k141-6438 gene 8856 | aminocoumarin resistance gene | alaS | 8.53 × 10−97 | 61.64 |
| 8d-2-k141-3704 gene 3047 | aminocoumarin resistance gene | alaS | 1.78 × 10−167 | 59.60 |
| 18d-3-k141-15560 gene 10446 | aminocoumarin resistance gene | alaS | 5.77 × 10−57 | 59.12 |
| 8d-1-k141-5267 gene 4197 | mupirocin resistance gene | ileS | 1.13 × 10−59 | 24.94 |
| 12d-1-k141-32581 gene 25448 | mupirocin resistance gene | ileS | 9.06 × 10−58 | 25.50 |
| 8d-3-k141-12723 gene 17311 | mupirocin resistance gene | ileS | 2.07 × 10−64 | 25.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Ma, Y.; Guan, Y.; Zhu, Y.; Wang, K.; Wang, Y.; Liu, P.; Chen, J.; Yu, Y. Metagenomics of Virus Diversities in Solid-State Brewing Process of Traditional Chinese Vinegar. Foods 2022, 11, 3296. https://doi.org/10.3390/foods11203296
Yu Z, Ma Y, Guan Y, Zhu Y, Wang K, Wang Y, Liu P, Chen J, Yu Y. Metagenomics of Virus Diversities in Solid-State Brewing Process of Traditional Chinese Vinegar. Foods. 2022; 11(20):3296. https://doi.org/10.3390/foods11203296
Chicago/Turabian StyleYu, Zhen, Yan Ma, Yingfen Guan, Yuanyuan Zhu, Ke Wang, Yuqin Wang, Peng Liu, Juan Chen, and Yongjian Yu. 2022. "Metagenomics of Virus Diversities in Solid-State Brewing Process of Traditional Chinese Vinegar" Foods 11, no. 20: 3296. https://doi.org/10.3390/foods11203296