Improved Model for Starch Prediction in Potato by the Fusion of Near-Infrared Spectral and Textural Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Images Collection of Potato Samples
2.3. Spectral Data Extraction from Hyperspectral Images
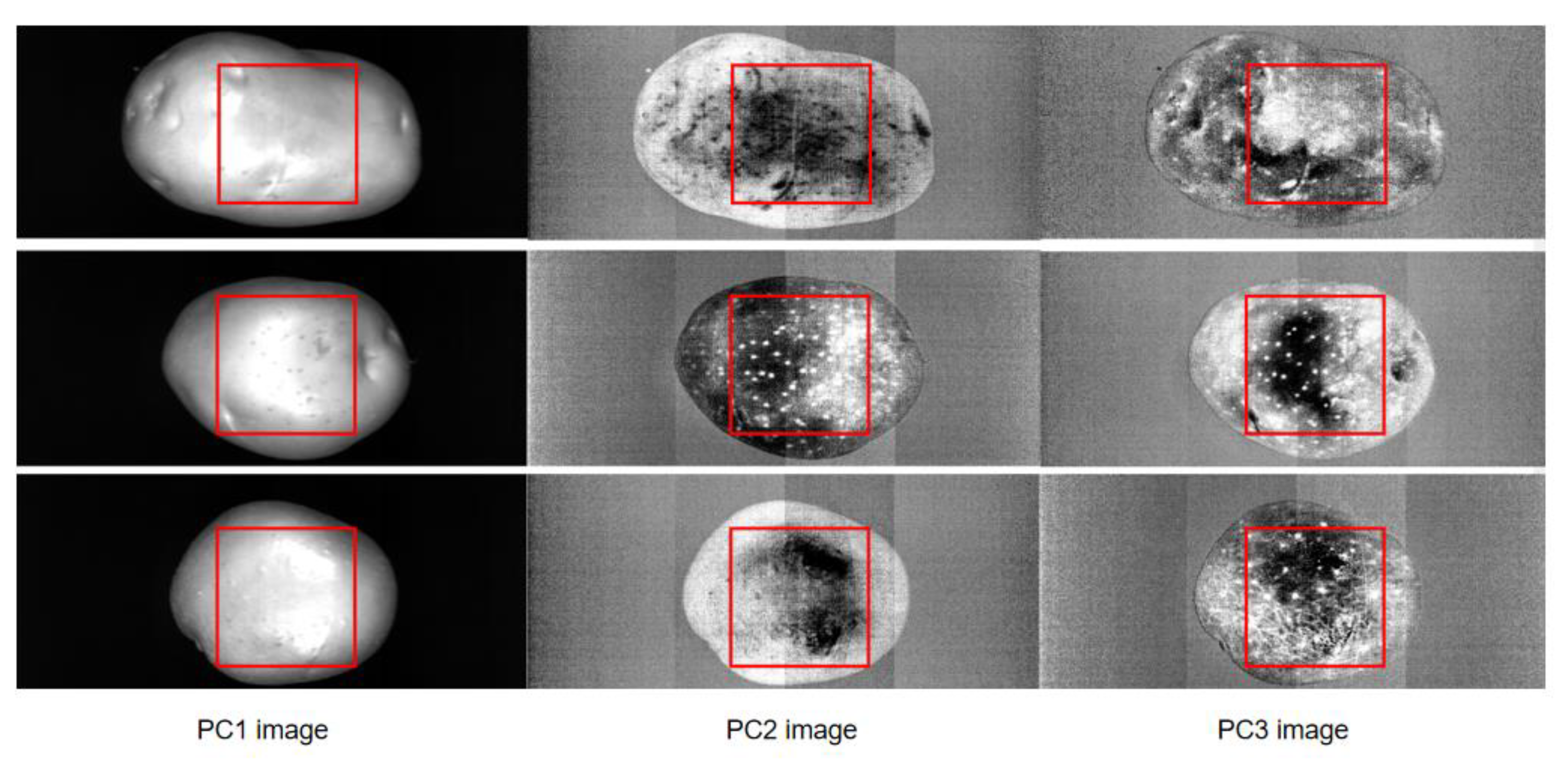
2.4. Textural Data Extraction
2.5. Chemical Analysis of the Starch Content in the Potato Samples
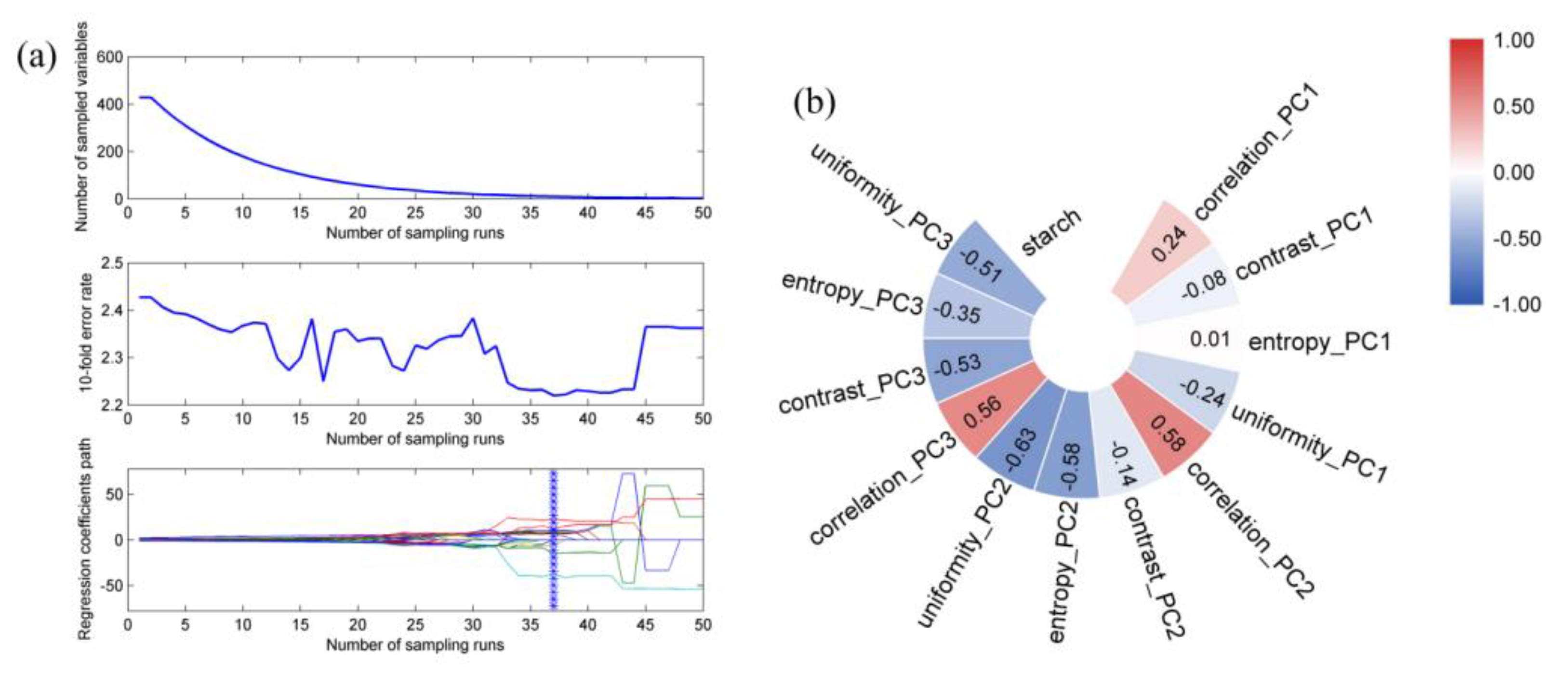
2.6. Data Fusion Strategy
2.7. Establishment of the Quantitative Model
3. Results and Discussion
3.1. Measured Starch Content in the Potato Samples
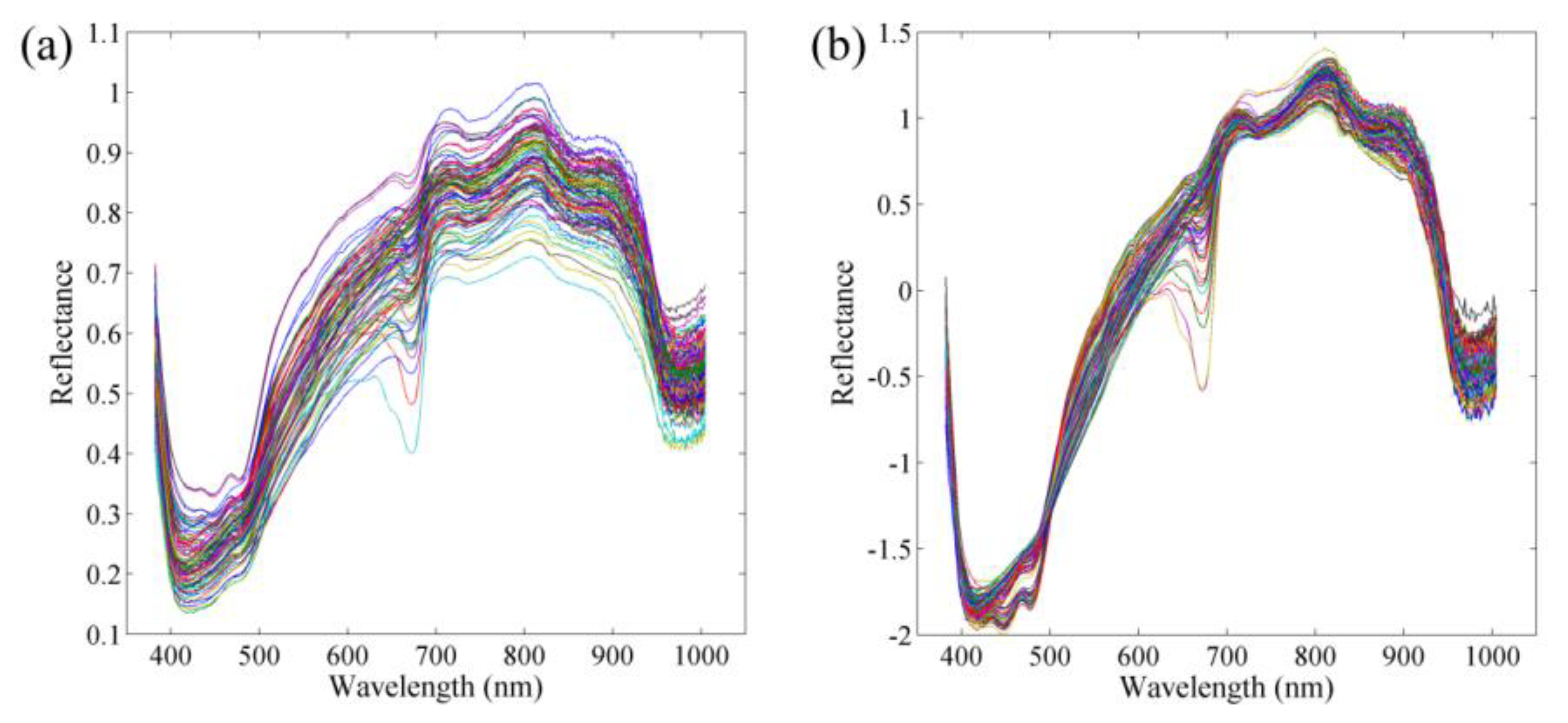
3.2. Spectral Characteristics of Potato Samples
3.3. Quantitative Model Based on a Single Type of Data
3.4. Quantitative Model Based on Fused Data
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ji, Y.M.; Sun, L.J.; Li, Y.; Li, J.; Liu, S.C.; Xie, X.; Xu, Y.Y. Non-destructive classification of defective potatoes based on hyperspectral imaging and support vector machine. Infrared Phys. Technol. 2019, 99, 71–79. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, S.W.; Fan, Y.C.; Fang, J.L. Hyperspectral detection method for starch content of potato. Adv. Sci. Technol. Lett. 2015, 111, 42–48. [Google Scholar]
- Chen, Y.; Yue, J.M.; Ding, Y. Determination of Starch Content in Potato with Acid-hydrolysis. Seed 2009, 49, 171–184. [Google Scholar]
- Amjad, W.; Crichton, S.O.; Munir, A.; Hensel, O.; Sturm, B. Hyperspectral imaging for the determination of potato slice moisture content and chromaticity during the convective hot air drying process. Biosyst. Eng. 2018, 166, 170–183. [Google Scholar] [CrossRef]
- Su, W.H.; Bakalis, S.; Sun, D.W. Fourier transform mid-infrared-attenuated total reflectance (FTMIR-ATR) microspectroscopy for determining textural property of microwave baked tuber. J. Food Eng. 2018, 218, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Su, W.H.; Sun, D.W. Chemical imaging for measuring the time series variations of tuber dry matter and starch concentration. Comput. Electron. Agric. 2017, 140, 361–373. [Google Scholar] [CrossRef]
- Ayvaz, H.; Santos, A.M.; Moyseenko, J.; Kleinhenz, M.; Rodriguez-Saona, L.E. Application of a portable infrared instrument for simultaneous analysis of sugars, asparagine and glutamine levels in raw potato tubers. Plant Foods Hum. Nutr. 2015, 70, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Khulal, U.; Zhao, J.; Hu, W.W.; Chen, Q.S. Nondestructive quantifying total volatile basic nitrogen (TVB-N) content in chicken using hyperspectral imaging (HSI) technique combined with different data dimension reduction algorithms. Food Chem. 2016, 197, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.X.; Wang, S.L.; He, X.G.; Wu, L.G.; Li, Y.; Guo, J.H. Combination of spectra and texture data of hyperspectral imaging for prediction and visualization of palmitic acid and oleic acid contents in lamb meat. Meat Sci. 2020, 169, 108194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Zhang, S.; Chen, Y.; Luo, W.; Huang, Y.F.; Tao, D.; Zhan, B.S.; Liu, X.M. Non-destructive determination of fat and moisture contents in salmon (Salmo salar) fillets using near-infrared hyperspectral imaging coupled with spectral and textural features. J. Food Compos. Anal. 2020, 92, 103567. [Google Scholar] [CrossRef]
- Wang, Y.J.; Hu, X.; Ge, J.; Hou, Z.W.; Ning, J.M. Rapid prediction of chlorophylls and carotenoids content in tea leaves under different levels of nitrogen application based on hyperspectral imaging. J. Sci. Food Agric. 2019, 99, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Y.; Jin, S.; Li, M.; Zhang, Z. Evaluation of black tea by using smartphone imaging coupled with micro-near-infrared spectrometer. Spectrochim. Acta Part A 2021, 246, 118991. [Google Scholar] [CrossRef]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmuga, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, F.; Aguilera, J.M. Application of image analysis for classification of ripening bananas. J. Food Sci. 2004, 69, 471–477. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, C.T.; Liu, F.; Kong, W.W.; He, Y. Hyperspectral imaging analysis for ripeness evaluation of strawberry with support vector machine. J. Food Eng. 2016, 179, 11–18. [Google Scholar] [CrossRef]
- Ma, J.C. How to measure starch content by amylase hydrolysis method. Occupation 2011, 23, 141. [Google Scholar]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of electronic nose, electronic tongue and computer vision for animal source food authentication and quality assessment—A review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Li, L.; Ning, J.; Zhang, Z. Green analytical assay for the quality assessment of tea by using pocket-sized nir spectrometer—Sciencedirect. Food Chem. 2021, 345, 128816. [Google Scholar] [CrossRef] [PubMed]
- Wold, S.; Sjostrom, M.; Eriksson, L. PLS-regression:a basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Wang, Y.J.; Li, T.H.; Li, L.Q.; Ning, J.M.; Zhang, Z.Z. Evaluating taste-related attributes of black tea by micro-nirs. J. Food Eng. 2021, 290, 110181. [Google Scholar] [CrossRef]
- Sugiyama, J. Visualization of sugar content in the flesh of a melon by near-infrared imaging. J. Agric. Food Chem. 1999, 47, 2715–2718. [Google Scholar] [CrossRef]
- Rady, A.; Guyer, D.; Lu, R. Evaluation of Sugar Content of Potatoes using Hyperspectral Imaging. Food Bioprocess Technol. 2015, 8, 995–1010. [Google Scholar] [CrossRef]
- Farhadi, R.; Afkari-Sayyah, A.H.; Jamshidi, B.; Gorji, A.M. Prediction of internal compositions change in potato during storage using visible/near-infrared(vis/nir) spectroscopy. Int. J. Food Eng. 2020, 16, 395–424. [Google Scholar] [CrossRef]
- Williams, P.J.; Kucheryavskiy, S. Classification of maize kernels using NIR hyperspectral imaging. Food Chem. 2016, 209, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, S.; Fan, Y. Hyperspectral detection method for starch content of potato. Next Gener. Comput. Inf. Technol. 2015, 111, 42–48. [Google Scholar]
| Dataset | Number | Min | Max | Mean | SD |
|---|---|---|---|---|---|
| Kexin No.1 | 68 | 1.77 | 18.31 | 10.23 | 2.95 |
| Holland No.15 | 28 | 17.01 | 22.81 | 18.96 | 1.29 |
| Calibration set | 64 | 1.77 | 22.81 | 12.78 | 4.80 |
| Prediction set | 32 | 5.04 | 21.01 | 12.77 | 4.71 |
| Data | NVs | LVs | Calibration Set | Prediction Set | |||
|---|---|---|---|---|---|---|---|
| Rc | RMSEC | Rp | RMSEP | RPD | |||
| Raw spectra | 428 | 5 | 0.9052 | 2.03 | 0.8102 | 2.81 | 1.67 |
| SNV preprocessed | 428 | 5 | 0.8719 | 2.34 | 0.8584 | 2.48 | 1.89 |
| Texture | 12 | 3 | 0.5451 | 4.02 | 0.7271 | 3.29 | 1.43 |
| Low-level fusion | 440 | 5 | 0.8606 | 2.44 | 0.8467 | 2.53 | 1.86 |
| Mid-level fusion | 17 | 5 | 0.8911 | 2.17 | 0.8832 | 2.29 | 2.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Wang, C. Improved Model for Starch Prediction in Potato by the Fusion of Near-Infrared Spectral and Textural Data. Foods 2022, 11, 3133. https://doi.org/10.3390/foods11193133
Wang F, Wang C. Improved Model for Starch Prediction in Potato by the Fusion of Near-Infrared Spectral and Textural Data. Foods. 2022; 11(19):3133. https://doi.org/10.3390/foods11193133
Chicago/Turabian StyleWang, Fuxiang, and Chunguang Wang. 2022. "Improved Model for Starch Prediction in Potato by the Fusion of Near-Infrared Spectral and Textural Data" Foods 11, no. 19: 3133. https://doi.org/10.3390/foods11193133




