Transcriptional Changes after Enniatins A, A1, B and B1 Ingestion in Rat Stomach, Liver, Kidney and Lower Intestine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
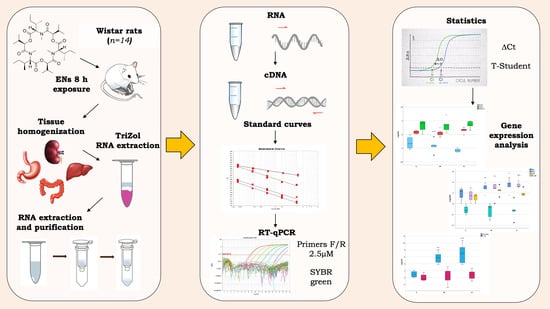
2.2. In Vivo Study Design
2.3. RNA Extraction
2.4. Gene Selection and Primer Design
2.5. Reverse Transcription and qPCR
2.6. Statyistical Analysis
3. Results
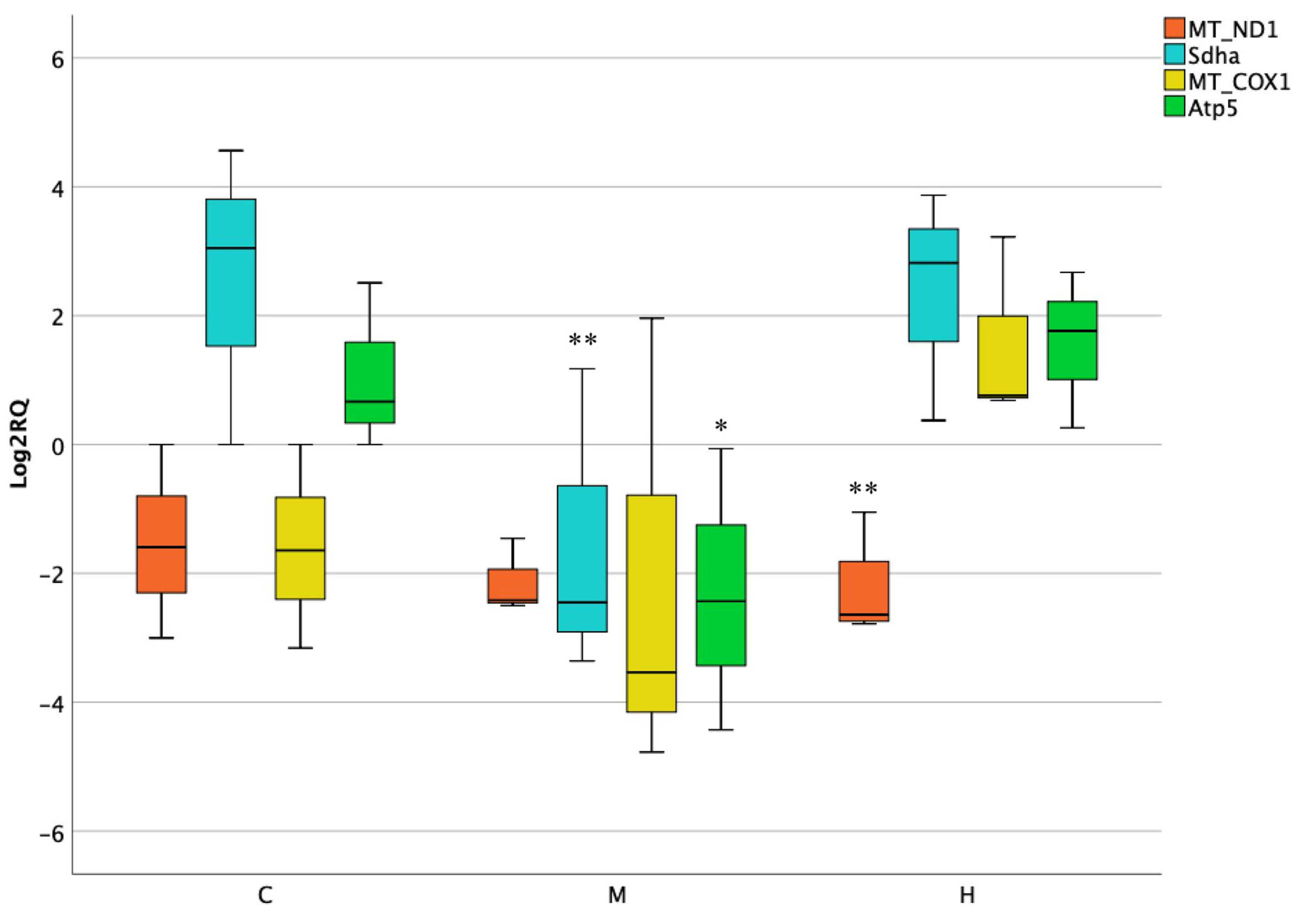
3.1. Activity of Mitochondrial and Nuclear Encoded Genes in Stomach
3.2. Activity of Mitochondrial and Nuclear Encoded Genes in Liver
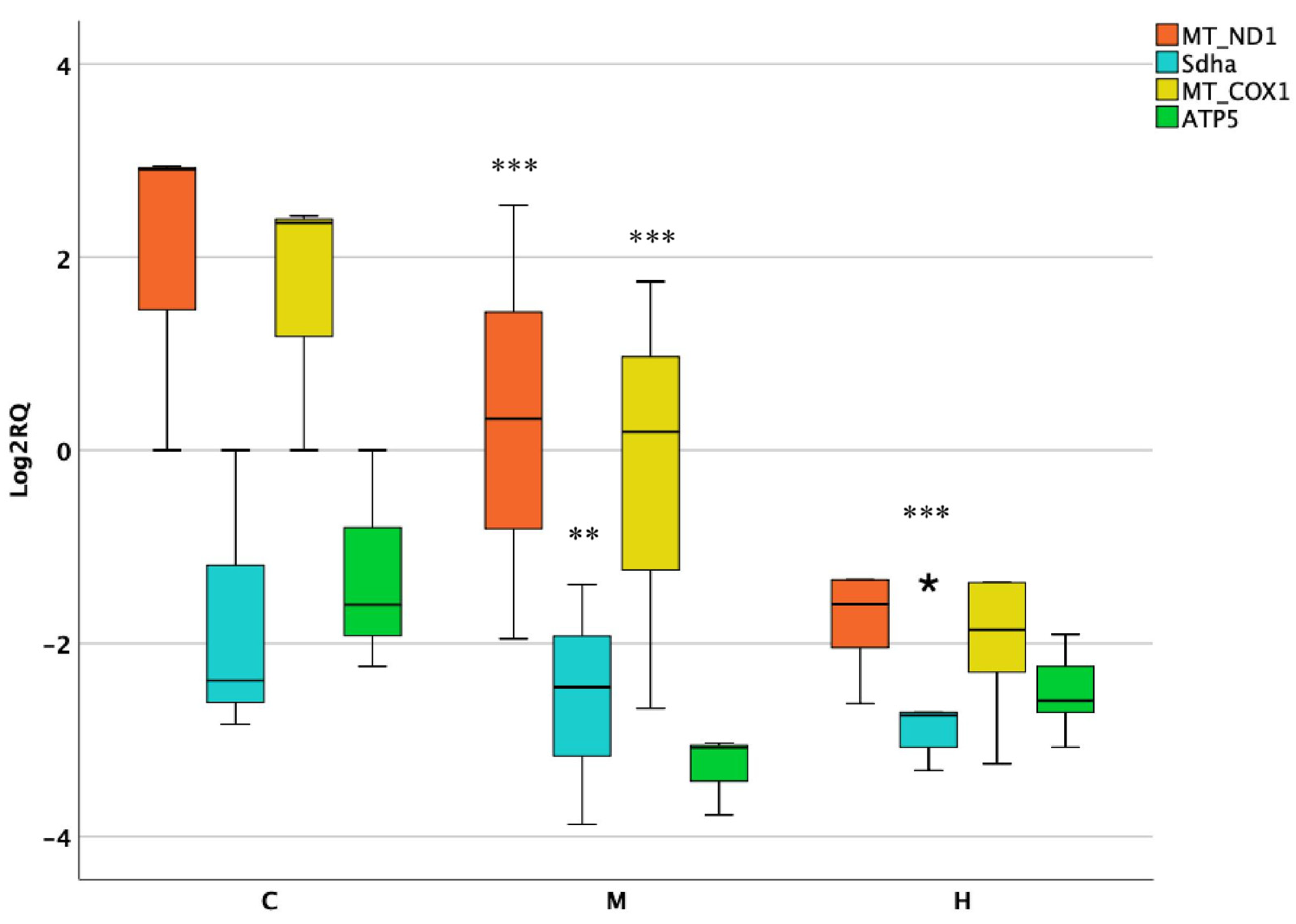
3.3. Activity of Mitochondrial and Nuclear Encoded Genes in Kidneys
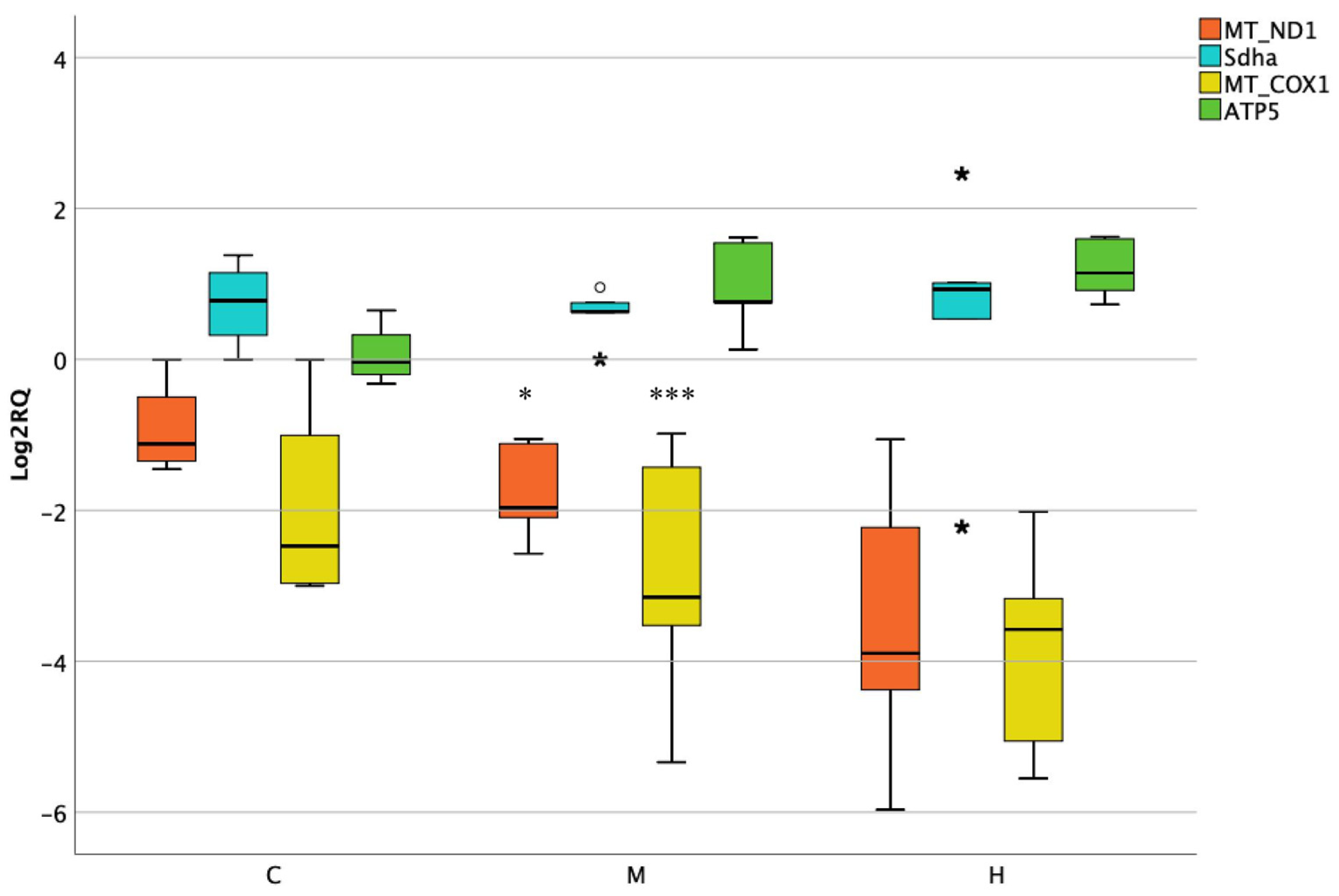
3.4. Activity of Mitochondrial and Nuclear Encoded Genes in Lower Intestine
3.5. Activity of Oxidative Stress Genes in Lower Intestine
3.6. Activity of Apoptotic Genes in Lower Intestine
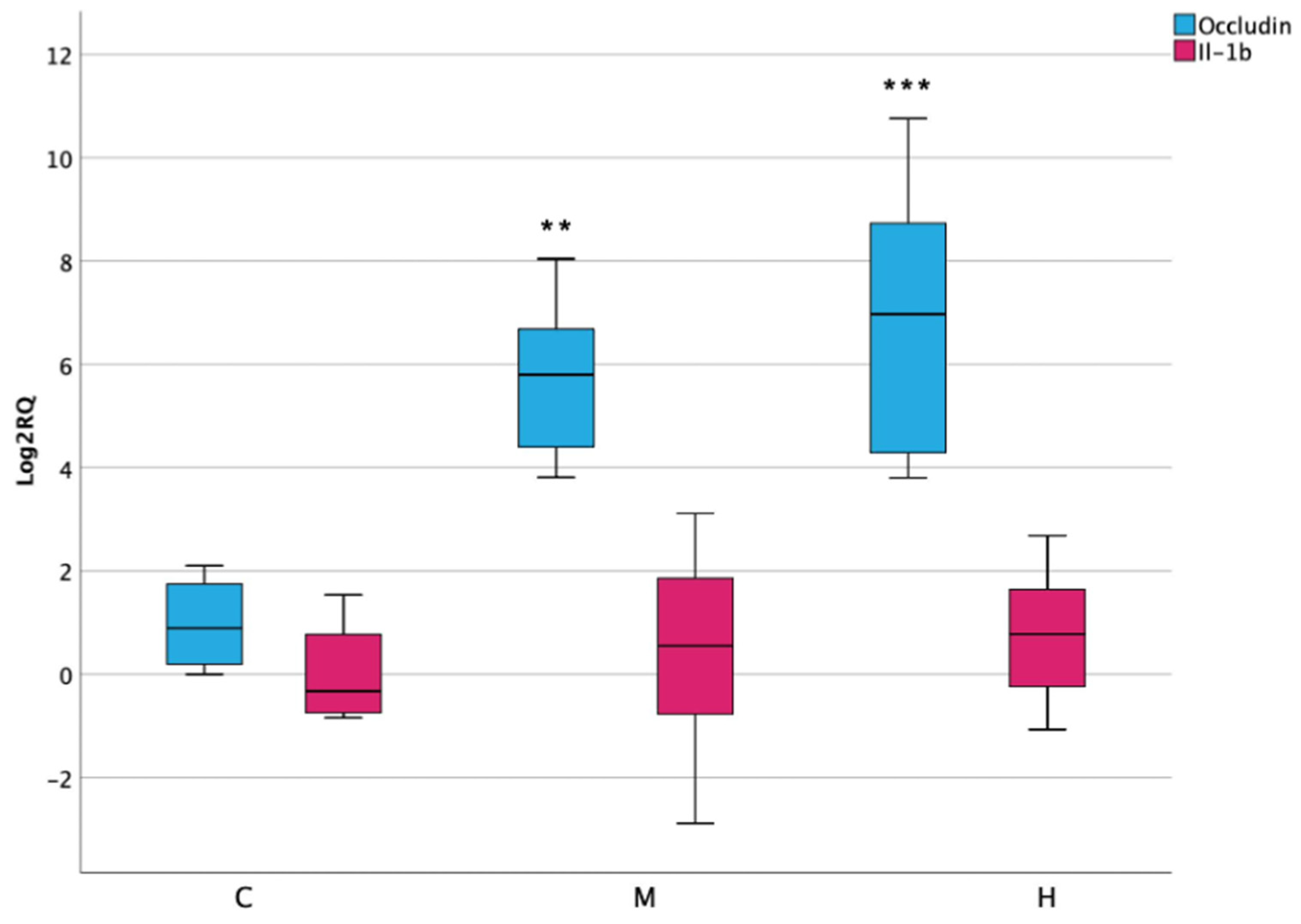
3.7. Inflammatory Response and Permeability in Lower Intestine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Gil, L.; Ruiz, P.; Font, G.; Manyes, L. An overview of the applications of hazards analysis and critical control point (HACCP) system to mycotoxins. Revista Toxicol. 2016, 33, 50–55. [Google Scholar]
- Escrivá, L.; Font, G.; Manyes, L. In vivo toxicity studies of fusarium mycotoxins in the last decade: A review. Food Chem. Toxicol. 2015, 78, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, O.; Juan, C.; Miere, D.; Loghin, F.; Mañes, J. Occurrence and co-occurrence of Fusarium mycotoxins in wheat grains and wheat flour from Romania. Food Control 2017, 73, 147–155. [Google Scholar] [CrossRef]
- Fraeyman, S.; Meyer, E.; Devreese, M.; Antonissen, G.; Demeyere, K.; Haesebrouck, F.; Croubels, S. Comparative in vitro cytotoxicity of the emerging Fusarium mycotoxins beauvericin and enniatins to porcine intestinal epithelial cells. Food Chem. Toxicol. 2018, 121, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Prosperini, A.; Berrada, H.; Ruiz, M.J.; Caloni, F.; Coccini, T.; Spicer, L.J.; Perego, M.C.; Lafranconi, A. A Review of the Mycotoxin Enniatin B. Front. Public Health 2017, 5, 304. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, M. The Toxicity of Fusarium Mycotoxins Enniatin and Moniliformin. 2017. Available online: https://www.ruokavirasto.fi/globalassets/yhteisot/tieteellinen-tutkimus/vaitoskirjat/2017-jonsson.pdf (accessed on 10 December 2020).
- Gruber-Dorninger, C.; Novak, B.; Nagl, V.; Berthiller, F. Emerging Mycotoxins: Beyond Traditionally Determined Food Contaminants. J. Agric. Food Chem. 2017, 65, 7052–7070. [Google Scholar] [CrossRef]
- Fraeyman, S.; Croubels, S.; Devreese, M.; Antonissen, G. Emerging Fusarium and Alternaria Mycotoxins: Occurrence, Toxicity and Toxicokinetics. Toxins 2017, 9, 228. [Google Scholar] [CrossRef] [Green Version]
- Maranghi, F.; Tassinari, R.; Narciso, L.; Tait, S.; Rocca, C.L.; Felice, G.D.; Butteroni, C.; Corinti, S.; Barletta, B.; Cordelli, E. In vivo toxicity and genotoxicity of beauvericin and enniatins. Combined approach to study in vivo toxicity and genotoxicity of mycotoxins beauvericin (BEA) and enniatin B (ENNB). EFSA Supporting Publ. 2018, 15, 1406E. [Google Scholar] [CrossRef]
- Cimbalo, A.; Alonso-Garrido, M.; Font, G.; Manyes, L. Toxicity of mycotoxins in vivo on vertebrate organisms: A review. Food Chem. Toxicol. 2020, 137, 111161. [Google Scholar] [CrossRef]
- Jonsson, M.; Jestoi, M.; Anthoni, M.; Welling, A.; Loivamaa, I.; Hallikainen, V.; Kankainen, M.; Lysøe, E.; Koivisto, P.; Peltonen, K. Fusarium mycotoxin enniatin B: Cytotoxic effects and changes in gene expression profile. Toxicol. In Vitro 2016, 34, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Juan-García, A.; Manyes, L.; Ruiz, M.; Font, G. Applications of flow cytometry to toxicological mycotoxin effects in cultured mammalian cells: A review. Food Chem. Toxicol. 2013, 56, 40–59. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Garrido, M.; Escrivá, L.; Manyes, L.; Font, G. Enniatin B induces expression changes in the electron transport chain pathway related genes in lymphoblastic T-cell line. Food Chem. Toxicol. 2018, 121, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Garrido, M.; Tedeschi, P.; Maietti, A.; Font, G.; Marchetti, N.; Manyes, L. Mitochondrial transcriptional study of the effect of aflatoxins, enniatins and carotenoids in vitro in a blood brain barrier model. Food Chem. Toxicol. 2020, 137, 111077. [Google Scholar] [CrossRef]
- Alonso-Garrido, M.; Manyes, L.; Pralea, I.E.; Iuga, C.A. Mitochondrial proteomics profile points oxidative phosphorylation as main target for beauvericin and enniatin B mixture. Food Chem. Toxicol. 2020, 141, 111432. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Cimbalo, A.; Frangiamone, M.; Juan, C.; Font, G.; Lozano, M.; Manyes, L. Proteomics evaluation of enniatins acute toxicity in rat liver. Food Chem. Toxicol. 2021, 151, 112130. [Google Scholar] [CrossRef] [PubMed]
- Iommarini, L.; Ghelli, A.; Tropeano, C.V.; Kurelac, I.; Leone, G.; Vidoni, S.; Lombes, A.; Zeviani, M.; Gasparre, G.; Porcelli, A.M. Unravelling the effects of the mutation m. 3571insC/MT-ND1 on respiratory complexes structural organization. Int. J. Mol. Sci. 2018, 19, 764. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Ruiz, I.; Fernández-Moreira, D.; Solis-Munoz, P.; Rodriguez-Juan, C.; Diaz-Sanjuan, T.; Muñoz-Yagüe, T.; Solis-Herruzo, J.A. Mitochondrial complex I subunits are decreased in murine nonalcoholic fatty liver disease: Implication of peroxynitrite. J. Proteome Res. 2010, 9, 2450–2459. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Bi, X.; Zhou, P.; Zhu, S.; Ding, W. NLRP3 deficiency attenuates renal fibrosis and ameliorates mitochondrial dysfunction in a mouse unilateral ureteral obstruction model of chronic kidney disease. Mediat. Inflamm. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wallace, L.; Mehrabi, S.; Bacanamwo, M.; Yao, X.; Aikhionbare, F.O. Expression of mitochondrial genes MT-ND1, MT-ND6, MT-CYB, MT-COI, MT-ATP6, and 12S/MT-RNR1 in colorectal adenopolyps. Tumor Biol. 2016, 37, 12465–12475. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Millar, A.H. Succinate dehydrogenase: The complex roles of a simple enzyme. Curr. Opin. Plant Biol. 2013, 16, 344–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miettinen, M.; Lasota, J. Succinate dehydrogenase deficient gastrointestinal stromal tumors (GISTs)–a review. Int. J. Biochem. Cell Biol. 2014, 53, 514–519. [Google Scholar] [CrossRef] [Green Version]
- Boikos, S.A.; Pappo, A.S.; Killian, J.K.; LaQuaglia, M.P.; Weldon, C.B.; George, S.; Trent, J.C.; Von Mehren, M.; Wright, J.A.; Schiffman, J.D. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: A report from the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol. 2016, 2, 922–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamai, T.; Higashi, S.; Murakami, S.; Arai, K.; Namatame, T.; Kijima, T.; Abe, H.; Jamiyan, T.; Ishida, K.; Shirataki, H.; et al. Single nucleotide variants of succinate dehydrogenase A gene in renal cell carcinoma. Cancer Sci. 2021. [CrossRef]
- Abdulhag, U.N.; Soiferman, D.; Schueler-Furman, O.; Miller, C.; Shaag, A.; Elpeleg, O.; Edvardson, S.; Saada, A. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur. J. Hum. Genet. 2015, 23, 159–164. [Google Scholar] [CrossRef]
- Jimenez, P.; Piazuelo, E.; Cebrian, C.; Ortego, J.; Strunk, M.; Garcia-Gonzalez, M.A.; Santander, S.; Alcedo, J.; Lanas, A. Prostaglandin EP2 receptor expression is increased in Barrett’s oesophagus and oesophageal adenocarcinoma. Aliment. Pharmacol. Ther. 2010, 31, 440–451. [Google Scholar] [CrossRef]
- Cathcart, M.; O’Byrne, K.J.; Reynolds, J.V.; O’Sullivan, J.; Pidgeon, G.P. COX-derived prostanoid pathways in gastrointestinal cancer development and progression: Novel targets for prevention and intervention. Biochim. Biophys. Acta BBA Rev. Cancer 2012, 1825, 49–63. [Google Scholar] [CrossRef]
- Martín-Sanz, P.; Mayoral, R.; Casado, M.; Boscá, L. COX-2 in liver, from regeneration to hepatocarcinogenesis: What we have learned from animal models? World J. Gastroenterol. WJG 2010, 16, 1430. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Liong, E.C.; Huang, H.; On Tse, W.; Lau, K.S.; Pan, J.; Nanji, A.A.; Fung, M.L.; Xing, F.; Tipoe, G.L. Cyclooxygenase-1 serves a vital hepato-protective function in chemically induced acute liver injury. Toxicol. Sci. 2015, 143, 430–440. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.C. Physiologic and pathophysiologic roles of cyclooxygenase-2 in the kidney. Trans. Am. Clin. Climatol. Assoc. 2013, 124, 139. [Google Scholar]
- Sarkar, S.; Mukherjee, S.; Chattopadhyay, A.; Bhattacharya, S. Differential modulation of cellular antioxidant status in zebrafish liver and kidney exposed to low dose arsenic trioxide. Ecotoxicol. Environ. Saf. 2017, 135, 173–182. [Google Scholar] [CrossRef]
- Jonckheere, A.I.; Smeitink, J.A.; Rodenburg, R.J. Mitochondrial ATP synthase: Architecture, function and pathology. J. Inherit. Metab. Dis. 2012, 35, 211–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiyar, R.S.; Bohnert, M.; Duvezin-Caubet, S.; Voisset, C.; Gagneur, J.; Fritsch, E.S.; Couplan, E.; Von Der Malsburg, K.; Funaya, C.; Soubigou, F. Mitochondrial protein sorting as a therapeutic target for ATP synthase disorders. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Schmidt, C.J.; Lamont, S.J. Distinct genes and pathways associated with transcriptome differences in early cardiac development between fast-and slow-growing broilers. PLoS ONE 2018, 13, e0207715. [Google Scholar] [CrossRef]
- Feichtinger, R.G.; Schäfer, G.; Seifarth, C.; Mayr, J.A.; Kofler, B.; Klocker, H. Reduced levels of ATP synthase subunit ATP5F1A correlate with earlier-onset prostate cancer. Oxidative Med. Cell. Longev. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Li, J.Y. ATP5A1 and ATP5B are highly expressed in glioblastoma tumor cells and endothelial cells of microvascular proliferation. J. Neurooncol. 2016, 126, 405–413. [Google Scholar] [CrossRef]
- Bertero, A.; Fossati, P.; Tedesco, D.E.A.; Caloni, F. Beauvericin and Enniatins: In Vitro Intestinal Effects. Toxins 2020, 12, 686. [Google Scholar] [CrossRef]
- Eleutherio, E.C.A.; Magalhães, R.S.S.; de Araújo Brasil, A.; Neto, J.R.M.; de Holanda Paranhos, L. SOD1, more than just an antioxidant. Arch. Biochem. Biophys. 2021, 697, 108701. [Google Scholar] [CrossRef]
- Gonzalez-Menendez, P.; Hevia, D.; Alonso-Arias, R.; Alvarez-Artime, A.; Rodriguez-Garcia, A.; Kinet, S.; Gonzalez-Pola, I.; Taylor, N.; Mayo, J.C.; Sainz, R.M. GLUT1 protects prostate cancer cells from glucose deprivation-induced oxidative stress. Redox Biol. 2018, 17, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Chen, B.; Shen, Z.; Liu, Y.; Li, H.; Xie, X.; Xu, X.; Li, H.; Huang, Z.; Chen, J. High GPX1 expression promotes esophageal squamous cell carcinoma invasion, migration, proliferation and cisplatin-resistance but can be reduced by vitamin D. Int. J. Clin. Exp. Med. 2014, 7, 2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, R.; Qiu, H.; Xu, J.; Mo, J.; Liu, Y.; Gui, Y.; Huang, G.; Zhang, S.; Yao, H.; Huang, X. Expression and prognostic potential of GPX1 in human cancers based on data mining. Ann. Transl. Med. 2020, 8, 124. [Google Scholar] [CrossRef]
- Ribeiro, J.; Malta, M.; Galaghar, A.; Silva, F.; Afonso, L.P.; Medeiros, R.; Sousa, H. P53 deregulation in Epstein-Barr virus-associated gastric cancer. Cancer Lett. 2017, 404, 37–43. [Google Scholar] [CrossRef]
- Ryter, S.W. Heme Oxgenase-1, a Cardinal Modulator of Regulated Cell Death and Inflammation. Cells 2021, 10, 515. [Google Scholar] [CrossRef]
- Kim, K.W.; Kim, N.; Choi, Y.; Kim, W.S.; Yoon, H.; Shin, C.M.; Park, Y.S.; Lee, D.H.; Park, Y.S.; Ahn, S. Different effects of p53 protein overexpression on the survival of gastric cancer patients according to Lauren histologic classification: A retrospective study. Gastric Cancer 2021, 24, 844–857. [Google Scholar] [CrossRef]
- Dashzeveg, N.; Yoshida, K. Cell death decision by p53 via control of the mitochondrial membrane. Cancer Lett. 2015, 367, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Balkwill, F.; Chonchol, M.; Cominelli, F.; Donath, M.Y.; Giamarellos-Bourboulis, E.J.; Golenbock, D.; Gresnigt, M.S.; Heneka, M.T.; Hoffman, H.M. A guiding map for inflammation. Nat. Immunol. 2017, 18, 826–831. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zeng, H.; Lei, L.; Tong, X.; Yang, L.; Yang, Y.; Li, S.; Zhou, Y.; Luo, L.; Huang, J. Tight junctions and their regulation by non-coding RNAs. Int. J. Biol. Sci. 2021, 17, 712. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Bian, C.; Luo, Z.; Guille, C.; Ogunrinde, E.; Wu, J.; Zhao, M.; Fitting, S.; Kamen, D.L.; Oates, J.C. Progesterone decreases gut permeability through upregulating occludin expression in primary human gut tissues and Caco-2 cells. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Sample | RNA (ng/μL) | 260/280 | 260/230 | |
|---|---|---|---|---|
| Stomach | C1 | 738.4 | 1.93 | 2.07 |
| C2 | 370.8 | 1.90 | 2.26 | |
| C3 | 246.7 | 1.85 | 1.95 | |
| C4 | 1051.2 | 2.01 | 2.15 | |
| M1 | 546.4 | 1.89 | 1.76 | |
| M2 | 647.1 | 1.96 | 2.07 | |
| M3 | 923.2 | 1.95 | 2.10 | |
| M4 | 1003.8 | 1.98 | 2.21 | |
| M5 | 1312.1 | 1.96 | 2.07 | |
| H1 | 607.8 | 1.92 | 2.08 | |
| H2 | 1177.1 | 1.99 | 2.21 | |
| H3 | 1388.9 | 2.02 | 2.01 | |
| H4 | 652.0 | 1.92 | 2.08 | |
| H5 | 1259.3 | 1.99 | 2.21 | |
| Liver | C1 | 664.1 | 2.10 | 2.24 |
| C2 | 695.8 | 2.10 | 2.20 | |
| C3 | 493.9 | 2.07 | 2.10 | |
| C4 | 638.0 | 2.11 | 2.28 | |
| M1 | 914.3 | 2.16 | 2.24 | |
| M2 | 2199.2 | 2.12 | 2.05 | |
| M3 | 1613.9 | 2.17 | 2.22 | |
| M4 | 1592.0 | 2.15 | 2.14 | |
| M5 | 1340.0 | 2.18 | 2.14 | |
| H1 | 1875.7 | 2.13 | 2.17 | |
| H2 | 1017.6 | 2.15 | 2.32 | |
| H3 | 1199.6 | 2.16 | 2.35 | |
| H4 | 1502.6 | 2.16 | 2.31 | |
| H5 | 1334.5 | 2.15 | 2.31 | |
| Kidneys | C1 | 1197.6 | 2.07 | 2.27 |
| C2 | 2359.5 | 2.01 | 2.16 | |
| C3 | 1550.3 | 2.12 | 2.14 | |
| C4 | 1615.1 | 2.1 | 2.25 | |
| M1 | 1995.0 | 2.10 | 2.18 | |
| M2 | 1585.0 | 2.07 | 2.26 | |
| M3 | 1606.0 | 2.07 | 2.22 | |
| M4 | 1211.6 | 2.11 | 2.16 | |
| M5 | 600.8 | 2.06 | 2.26 | |
| H1 | 329.6 | 2.06 | 2.25 | |
| H2 | 247.4 | 2.11 | 2.30 | |
| H3 | 715.7 | 2.10 | 2.15 | |
| H4 | 615.0 | 2.10 | 2.15 | |
| H5 | 2407.3 | 2.12 | 2.25 | |
| Colon | C1 | 1728.0 | 2.16 | 2.20 |
| C2 | 1581.0 | 2.18 | 2.20 | |
| C3 | 759.0 | 2.17 | 2.22 | |
| C4 | 2306 | 2.10 | 2.14 | |
| M1 | 1838.0 | 2.15 | 2.20 | |
| M2 | 1891.0 | 2.14 | 2.17 | |
| M3 | 1407.0 | 2.16 | 2.23 | |
| M4 | 1360.0 | 2.18 | 2.23 | |
| M5 | 1619.0 | 2.17 | 2.19 | |
| H1 | 1961.0 | 2.17 | 2.19 | |
| H2 | 1736.0 | 2.16 | 2.19 | |
| H3 | 838.0 | 2.17 | 2.21 | |
| H4 | 1084.0 | 2.17 | 2.21 | |
| H5 | 859.0 | 2.16 | 2.20 |
| Gene | Sequence | Efficiency | Linearity |
|---|---|---|---|
| MT-ND1 | F: CGAGCTCCCTTCGACTTAAC R: GAATAGGGCGAATGGTCCTG | 101.806 | 0.991 |
| Sdha | F: GACGATCTCTGCGGTATGAC R: TCGGTGTATGGACCCATCTT | 107.658 | 0.975 |
| MT-COX1 | F: GCTGGAGCATCCGTAGATTT R: ATTGGGTTATAGCAGGGGGT | 106.425 | 0.985 |
| ATP5 | F: GTGATGTGTCCGCCTACATT R: ACAAGCCCACATTAATGGCA | 107.423 | 0.978 |
| SOD1 | F: ACACAAGGCTGTACCACTGC R: CCACATTGCCCAGGTCTCC | 124.573 | 0.991 |
| GPx1 | F: GTCCACCGTGTATGCCTTCTCC R: TCTCCTGATGTCCGAACTGATTGC | 105.269 | 0.990 |
| Hmox1 | F: CACGCATATACCCGCTACCT R: AAGGCGGTCTTAGCCTCTTC | 146.284 | 0.981 |
| Bcl2 | F: ACTGAGTACCTGAACCGGCATC R: GGAGAAATCAAACAGAGGTCGC | 148.237 | 0.990 |
| Bax | F: AAGAAGCTGAGCGAGTGTCT R: CAAAGATGGTCACTGTCTGC | 117.003 | 0.981 |
| p53 | F: GTTCCGAGAGCTGAATGAGG R: TTTTATGGCGGGACGTAGAC | 111.153 | 0.990 |
| Nf-κB | F: CTTCTCGGAGTCCCTCACTG R: CCAATAGCAGCTGGAAAAGC | 102.480 | 0.996 |
| Il-1β | F: CTTGTCGAGAATGGGCAGTCT R: TGTGCCACGGTTTTCTTATGG | 105.269 | 0.990 |
| Occludin | F: AGTACATGGCTGCTGCTGATG R: CCCACCATCCTCTTGATGTGT | 143.830 | 0.990 |
| β-actin | F: AACCTTCTTGCAGCTCCTCCG R: CCATACCCACCATCACACCCT | 94.242 | 0.996 |
| 18S rRNA | F: GAGCGTGTGATCACCATCAT R: TCCTTCACGTCCTTCTGTCT | 105.487 | 0.979 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cimbalo, A.; Alonso-Garrido, M.; Font, G.; Frangiamone, M.; Manyes, L. Transcriptional Changes after Enniatins A, A1, B and B1 Ingestion in Rat Stomach, Liver, Kidney and Lower Intestine. Foods 2021, 10, 1630. https://doi.org/10.3390/foods10071630
Cimbalo A, Alonso-Garrido M, Font G, Frangiamone M, Manyes L. Transcriptional Changes after Enniatins A, A1, B and B1 Ingestion in Rat Stomach, Liver, Kidney and Lower Intestine. Foods. 2021; 10(7):1630. https://doi.org/10.3390/foods10071630
Chicago/Turabian StyleCimbalo, Alessandra, Manuel Alonso-Garrido, Guillermina Font, Massimo Frangiamone, and Lara Manyes. 2021. "Transcriptional Changes after Enniatins A, A1, B and B1 Ingestion in Rat Stomach, Liver, Kidney and Lower Intestine" Foods 10, no. 7: 1630. https://doi.org/10.3390/foods10071630