Regulation of Eukaryote Metabolism: An Abstract Model Explaining the Warburg/Crabtree Effect
Abstract
:1. Introduction
2. Theoretical Background, Methods and Tools
2.1. Interaction Graph
2.2. Dynamic Parameters
2.3. Formal Validation
2.4. Using Biochemical Knowledge to Induce Parameter Values
2.5. Software Platforms for Validation
3. Metabolism Regulation and Warburg/Crabtree Effect
- Mitochondrial respiration: a slow degradation of glucose (time-consuming turnover) but efficient production yield (38 ATP per glucose molecule),
- Fermentation: a rapid degradation of glucose with an inefficient production yield (2 ATP per glucose molecule).
3.1. The Main Actors of Catabolism
3.2. The Main Actors of Anabolism
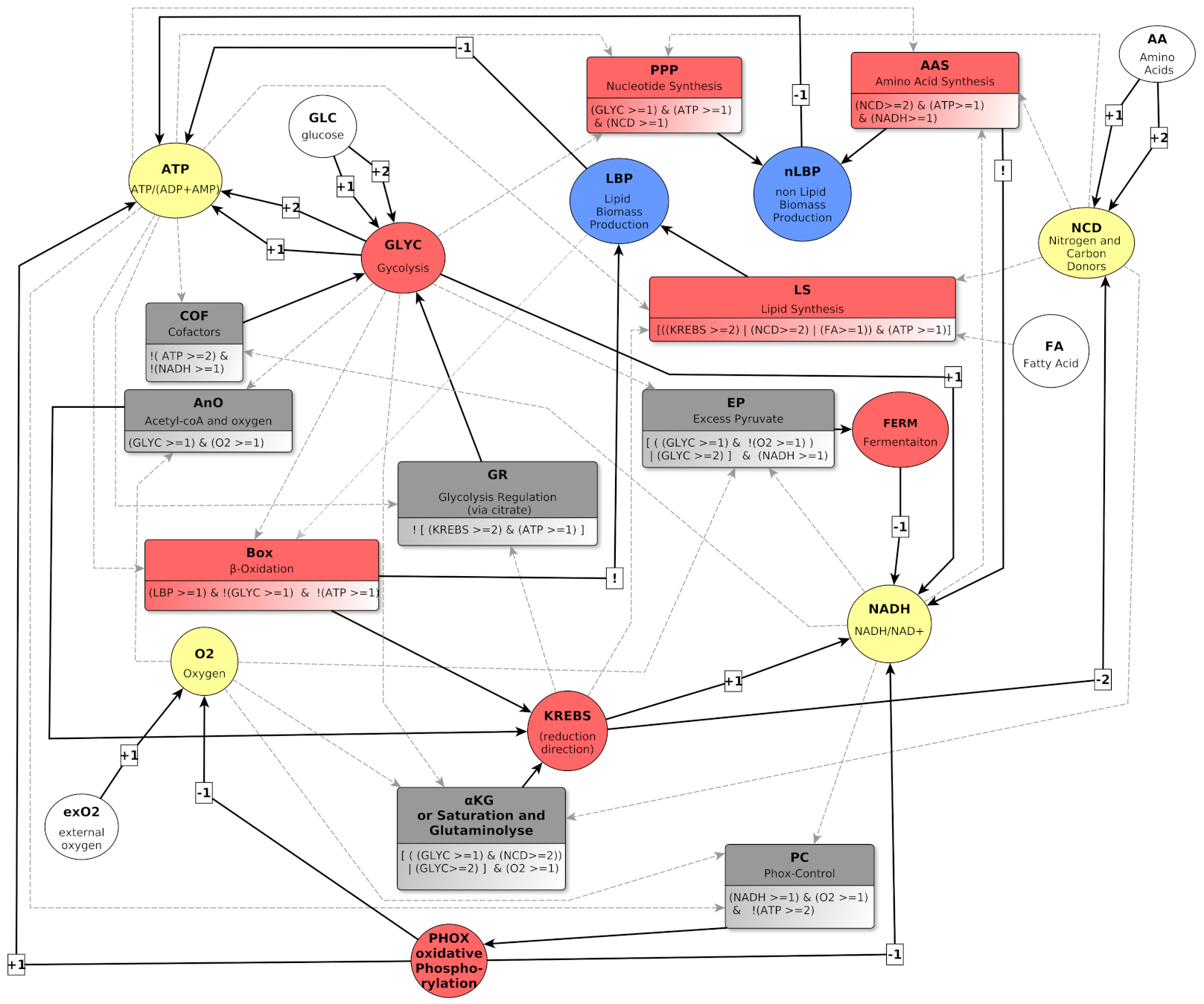
4. Graph Representation of Metabolic Regulations
4.1. Biological Regulation Graphs with Multiplexes
4.2. Variables and Their Biological Meaning
4.2.1. Environmental Variables: exO2, FA, GLC and AA
- ∘
- respectively abstract (external) oxygen and lipids (fatty acids) intakes. At level 1, FA can participate to lipid synthesis (the LS multiplex, provided that ATP is present). At level 1, exO2 activates the in-cell oxygen O2. Conversely, when their value is 0, it means that the nutrient input is not sufficient to be used normally by the cell. Please note that according to the threshold approach described above, level 0 does not mean that there is no supply but simply that it does not reach a sufficient threshold.
- ∘
- respectively abstract glucose and amino acids (derived from proteins) intakes. They are crucial nutrients for studying the Warburg/Crabtree effect [22,24], and because their action on glycolysis (GLYC) or nitrogen/carbon donors (NCD) respectively increases with their level of presence, we need to consider an “intermediate” level. Therefore, Level 0 represents a low availability of nutrient, leading to a negligible activation of metabolic processes downstream; level 1 represents a “normal” availability sufficient for basic metabolism and level 2 represents a high level of nutrients used by the cell often inducing over-activation of glycolysis.
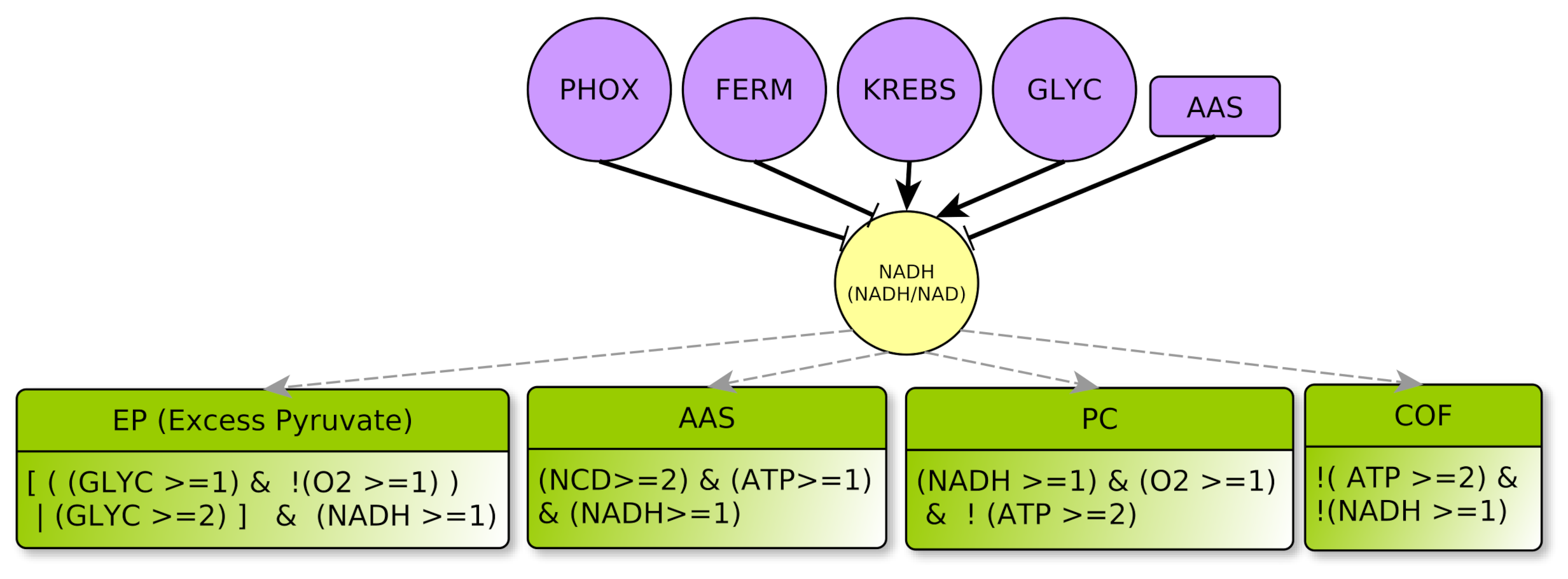
4.2.2. Metabolites: ATP, NADH, NCD and O2
- ∘
- represents the concentration ratio of . It abstracts the energetic level for the cell. During anabolic reactions, is transformed into or to release energy. ADP and AMP are regenerated during glycolysis (aerobic glycolysis) or by mitochondrial respiration. Since there are shuttles between cytoplasm and mitochondria, the , as a ratio, can be considered almost spatially homogeneous within the cell, even if individually the or + level can vary greatly from mitochondria to the cytoplasm. Notice also that a high ATP value (ATP = 2) might be equally viewed as a high level of molecules or as a low level of + molecules in the cell, and conversely for ATP = 0. Lastly, ATP = 1 represents a sort of equilibrium.
- ∘
- similarly to ATP, NADH represents the mean concentration ratio of , as well as , which belong to the same electron carrier molecular group. The modeling choice of abstracting together these three different types of ratios deserves some comments. For sure, the couple is commonly used in catabolic processes whereas is mostly used in anabolic processes. Additionally, in Figure 3, glycolysis activates the ratio (“” plain arrow) and it does not address the same ratio as oxidative phosphorylation, which acts on . Nevertheless, according to our very abstract level of description: (i) At the temporal resolution granularity, we consider their mean values to have the same signature as shown for example in the context of the cell cycle [25]. (ii) The thresholds between 0 and 1 are purely symbolic in the Thomas’ framework so that it is not required for the and thresholds to be quantitatively equal. The same reasoning applies to the quotient. Therefore, we had no valuable reasons neither to distinguish between these three ratios nor to distinguish more than two different levels for the NADH targets (principle of parsimony [26]). All in all, level 0 simply represents a too low ratio to act on its targets (low or, equally, high ), contrarily to level 1.
- ∘
- represents the Nitrogen and Carbon Donors, useful to the cell and derived from amino acids. These elements are used to supply metabolic pathways such as KREBS. At level 0 NCD action is too low to undergo anabolic processes; at level 1 it can participate in the activation of the Pentose Phosphate Pathway (PPP), and at level 2 it allows the production of -KG through glutaminolysis even if is absent (-KG) and it also participates in amino acids and lipid synthesis (AAS and LS).
- ∘
- represents the intracellular oxygen concentration. Once again the Thomas’ framework not being quantitative, distinguishing several thresholds for the O2 concentration can only be motivated by several targets of O2 which cannot share the same regulation thresholds. According to Figure 3, we had no valuable reason to distinguish different levels of activation for the O2 targets, so a present/absent state is sufficient in our model.
4.2.3. Metabolic pathways: GLYC, FERM, KREBS and PHOX
- ∘
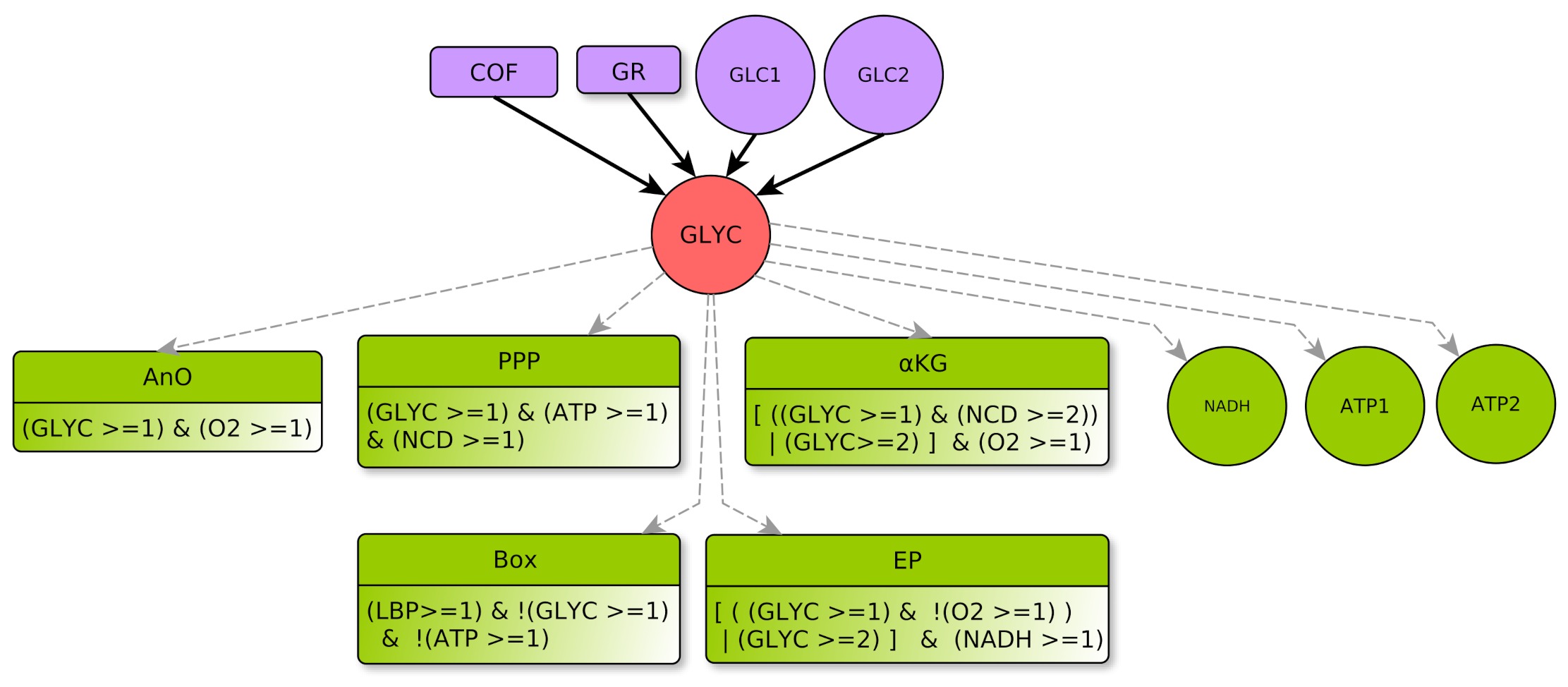
- represents glycolysis, which degrades glucose and produces pyruvate and (GLYC activates ATP) using ten chain reactions [27]. Three of these reactions are limiting and carried out using different enzymes, such as Phospho-Fructo-Kinase (PFK) which has a major role in glycolysis regulation. Level 0 represents glycolysis that does not produce enough intermediates, e.g., pyruvate, useful to other metabolic pathways (such as the Krebs cycle), nor any noticeable ATP. Level 1 represents a glycolytic flux sufficient to activate the related metabolic pathways (PPP). In terms of flux, PPP can be considered to be a competitor of glycolysis. However, in terms of regulation, because the variable GLYC abstracts all intermediate metabolites involved in glycolysis, GLYC is an activator of PPP (through one of its early metabolites), Krebs, etc., as well as fermentation in the absence of oxygen). Finally, level 2 represents a high level of activity where glycolysis can be considered to be over-functioning, compared to the needs of a healthy cell under optimal nutrient conditions. In such a case, glycolysis promotes the accumulation of pyruvate which in turn promotes the production of -KG and glycolysis also fosters the fermentation process if NADH is present, even in the presence of oxygen.
- ∘
- represents fermentation. This metabolic pathway becomes important when the oxygen supply is no longer sufficient. As production is the only target of FERM in the model, FERM is obviously a boolean variable.
- ∘
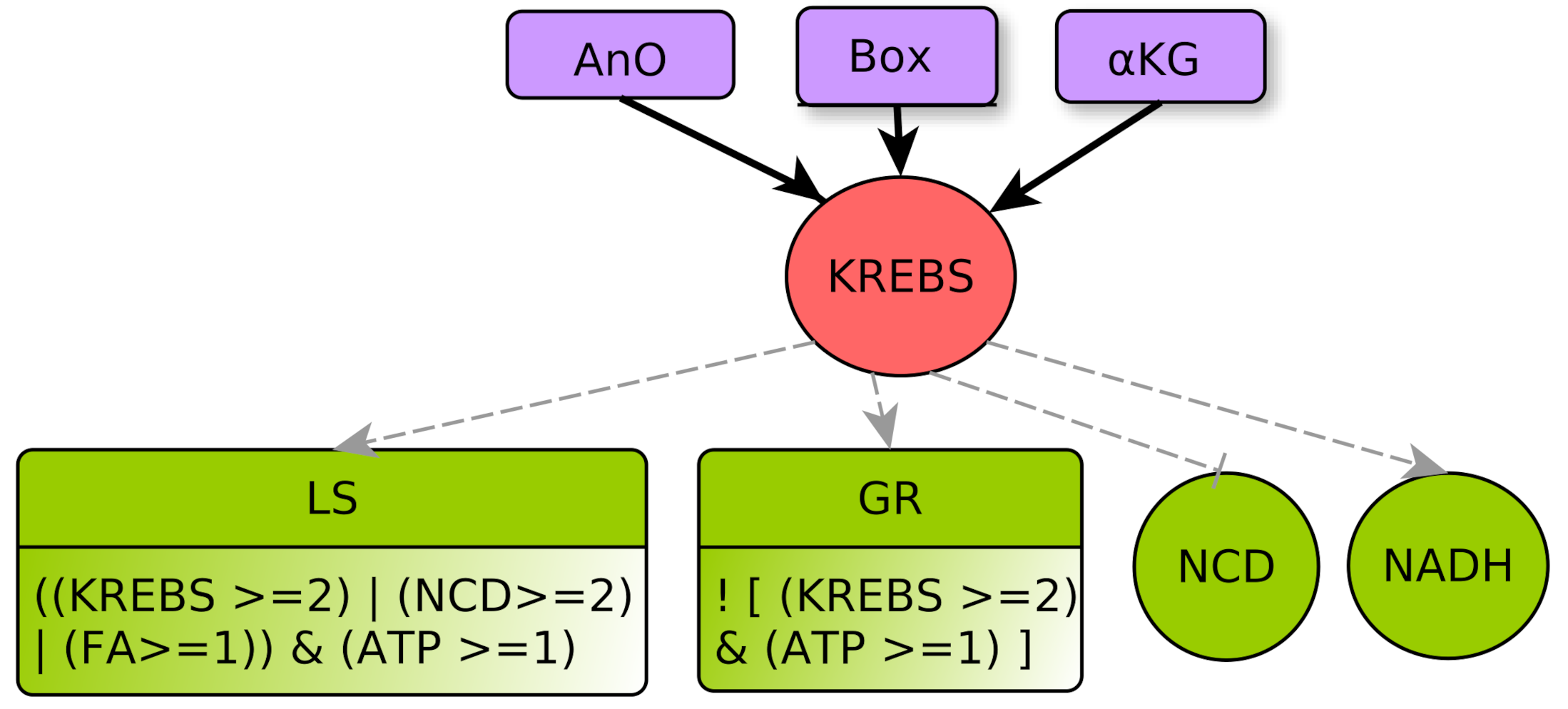
- represents the TCA cycle (tricarboxylic acid cycle) or Krebs cycle. It does not represent the reverse reactions (reductive branch of the Krebs cycle) that are implicitly taken into account within multiplexes. This central metabolic cycle allows the oxidation of groups of molecules resulting from different catabolic processes (glycolysis, -oxidation, degradation of amino acids [28]). A sufficient flow on this pathway will allow the cell to make the oxidative phosphorylation and reduce to (KREBS activates NADH). Its flux is dependent on the level of cellular oxygen but also on the quantity of precursors available [29]. Level 0 represents a low flux that does not allow one to noticeably obtain . Level 1 represents a Krebs cycle flow capable of reducing to from basic catabolic processes (glycolysis and -oxidation). This is the normal flow for healthy cells with an adapted supply of nutrients. At level 2 Krebs cycle is over-functioning and alarms the cell to lower catabolic processes, such as glycolysis (via GR), and promotes anabolic processes such as lipid synthesis (via LS).
- ∘
- represents oxidative phosphorylation, a mitochondrial metabolic pathway that allows the creation of (PHOX activates ATP) by consuming oxygen and (PHOX inhibits NADH and O2). Several chemical reactions allow the reduction of an oxygen molecule into a water molecule. These steps release energy, which is used to form . This pathway depends entirely on oxygen [30]. PHOX activates all its targets at the same level, so, it is a boolean variable.
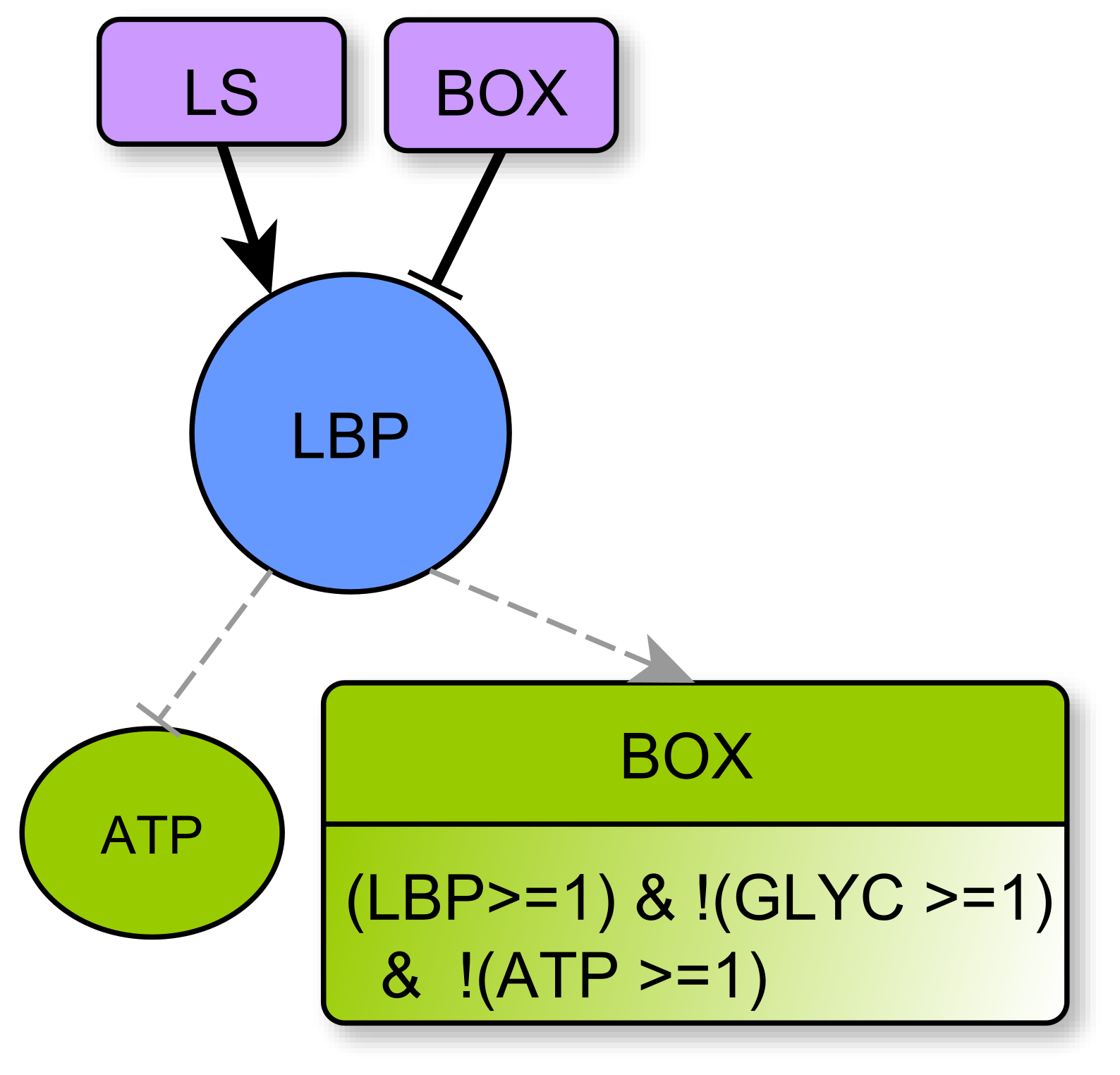
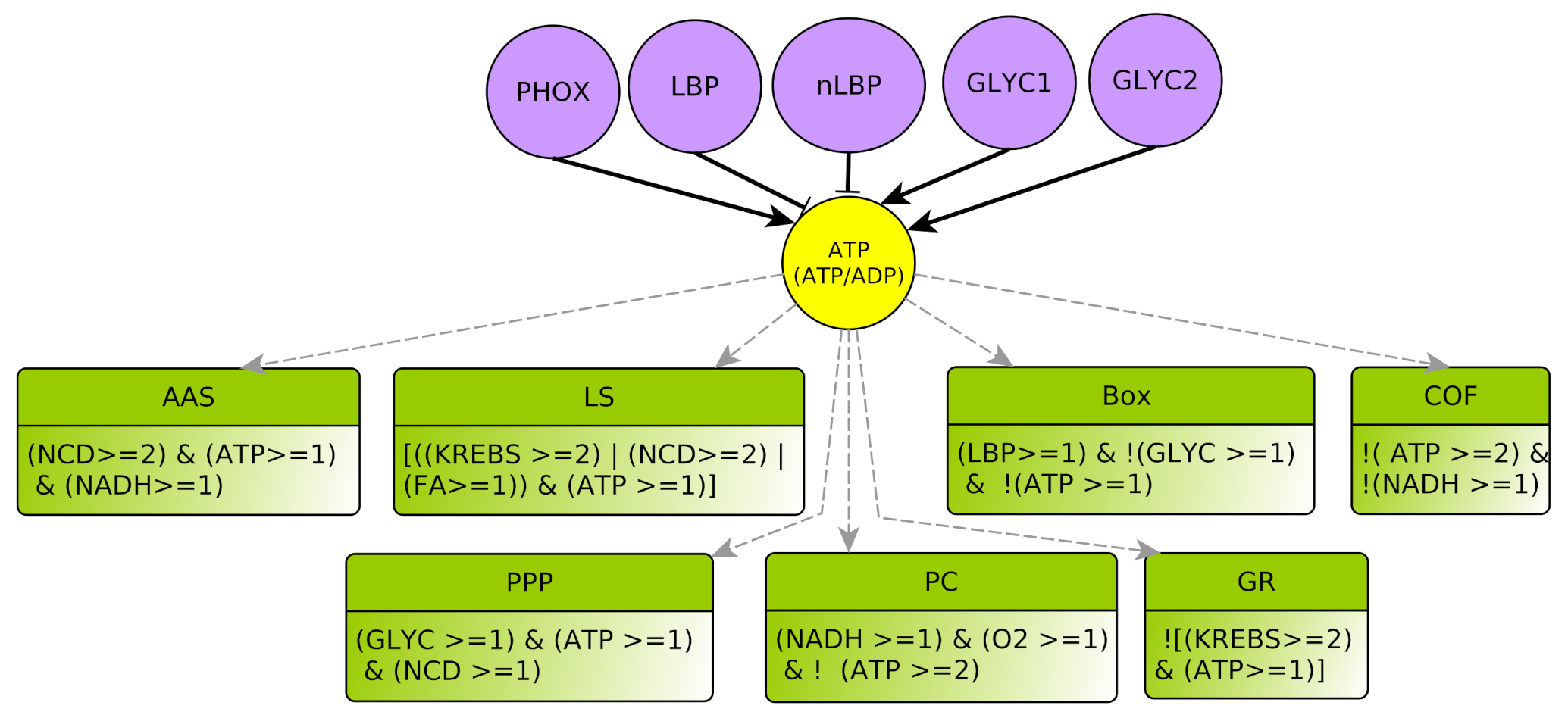
4.2.4. Biomass: LBP and nLBP
- ∘
- represents respectively the Production and storage of Lipid Biomass (all complex lipids, such as phospholipids or glycolipids) and Non-Lipidic Biomass (proteins and DNA/RNA). They are useful for component turnover and cell proliferation. Biomass production consumes energy: LBP and nLBP inhibit ATP (Notice that lipid synthesis is in fact the process that consumes ATP, see Section 4.3.3. Technically, as LS is a multiplex, one must delegate the ATP consumption to the variable LBP. The same applies to nLBP). In addition, lipid biomass production LBP can participate in Krebs activation through Box. Whatever the kind of biomass, there is either noticeable production and storage or not, so these variables are boolean.
4.3. Multiplexes and Their Biological Meaning
4.3.1. Multiplexes Regulating Metabolites
NADH Regulator
- □
- AAS represents the processes for anabolic production of amino acids (Amino Acid Synthesis) which favor non-lipidic biomass production. They are mostly synthesized from other amino acids collected outside the cell: The multiplex AAS summarizes the necessary elements to produce new amino acids, such as nitrogen and carbon given off by the products of degradation of amino acids outside the cell , a large amount of , and at least for some of the amino acid synthesis reactions . The conjunction of these three conditions yields to the formula of this multiplex [31,32]. Moreover, being consumed, AAS inhibits NADH: this is encoded by the “!” (negation) on the outgoing arrow from AAS to NADH.
4.3.2. Multiplexes Regulating Pathways
GLYC Regulators
- □
- COF represents the cofactors needs to a correct course of glycolysis: and must not be limiting. This means that and , as already explained in Section 4.2.2. Therefore, the COF formula properly formalizes this regulation of GLYC.
- □
- GR represents metabolic glycolytic flow inhibitors such as an absence of the enzyme PFK (Phospho-fructokinase) and an accumulation of citrate. Enzyme PFK of the glycolysis is allosterically inhibited by [33], Thus, ATP participates to inhibition of GLYC via PFK. Moreover, pyruvate (the final product of glycolysis) fuels the TCA cycle and is transformed into citrate, which, if in excess, inhibits glycolysis. Reminding that the TCA cycle is abstracted by KREBS, an excess of citrate is produced when , which also participates in the inhibition of glycolysis. Here, we consider that GLYC is inhibited when both conditions are satisfied, so GLYC is inhibited if . The negation “!” at the beginning of the formula indicates the inhibition.
KREBS Regulators
- □
- □
- Box represents -oxidation, the fatty acid degradation pathway. It converts fatty acids to acetyl-CoA [35]. -oxidation can be performed when the energy level of the cell is relatively low , when the pool of stored lipids is large enough and when glycolysis is not efficient enough to produce energy [29]. The multiplex formula is the conjunction of these conditions.
- □
- -KG represents an accumulation of the -Ketoglutarate metabolite. This intermediate of a reaction of the Krebs cycle shows a significant flow only in the presence of oxygen [36]. Then, the accumulation can come from a saturation issued by glycolysis with a very high flow rate . Moreover, with a lower flow rate , the transformation of glutamine to -KG by glutaminolysis [23,37] can accumulate, provided that the amount of amino acids in the form of nitrogen and carbon is important .
FERM Regulator
- □
- EP represents an Excess of Pyruvate, which activates the fermentation flow. Fermentation always needs as a cofactor . Then, fermentation can be activated by an overproduction of pyruvate via glycolysis , or if glycolysis produces pyruvate at a standard rate together with an intracellular hypoxia .
PHOX Regulator
- □
- PC represents the PHOX Control. Oxidative phosphorylation requires a sufficient supply of oxygen as precursor of the chain. It also requires . Moreover, this pathway is only activated when the energy pool of the cell is not too high [38], i.e., .
4.3.3. Multiplexes Regulating Biomass
nLBP Regulators
- □
- PPP represents the production of nucleotides via the Pentose Phosphate Pathway that favors non-lipidic biomass production. It produces nucleotides, thus PPP activates non-lipidic biomass (nLBP). This pathway also produces , which is directly consumed to produce non-lipidic biomass, so that the end production result is neutral for the NADH ratio. Consequently, we do not put an activation arrow from PPP to NADH in the regulation graph (contrarily to AAS). Glycolysis is required for the activation of PPP because Glyceraldehyde-3-phosphate (G3P) is an intermediate reaction of glycolysis and the precursor of the Pentose Phosphate Pathway. In addition, for these endergonic reactions it needs a carbon input from the internalization of amino acids, as well as [39].
LBP regulators:
- □
- LS represents specifically the Synthesis of Lipids composed by fatty acids. Lipid creation uses energy and fatty acids can come directly from the cellular environment or via the fatty acid synthesis pathway, the main precursor of which is acetyl-CoA, which is in turn provided by Krebs cycle when it is over-functioning or by glutaminolysis directly derived from amino acids degradation [40].
5. Kinetic Parameters: How Local Reasoning Makes a Global Dynamics of the Mathematical Model Emerge
5.1. Kinetic Parameters and State Transitions
5.2. Local Identification of Parameters for Metabolic Regulations
GLYC Parameters
- EP if and ,
- KG if and ,
- over-activation of ATP (compared to “simple” activation performed at level 1).
- When the functioning of glycolysis will reach a level that is sufficient to activate EP even if and , as well as KG if and . Thus .
- When the functioning of glycolysis will allow the activation of its targets (if the other conditions are satisfied), but neither EP if and , nor KG if and . Thus .
KREBS Parameters
FERM and PHOX Parameters
nLBP Parameters
LBP Parameters
NCD Parameters
O2 Parameters
NADH Parameters
- In the cell, PHOX is the principal consumer of NADH, KREBS is its principal producer, and the metabolic processes they abstract are nested, therefore they balance on a long-term basis.
- GLYC is a weak producer of NADH, FERM is an average consumer, and AAS is a weak consumer (but we do not know if GLYC and AAS balance on a long-term basis).
ATP Parameters
6. Validation Matrix: Encoding Global Biological Knowledge Using Temporal Logic
- The environmental conditions are defined by the four environmental variables described above (Section 4.2): presence of glucose, availability of fatty acids, etc.
- and the markers of the global functioning are the regulated variables: biomass production, the activity level of the Krebs cycle, concentration ratio , etc.
6.1. A Logic for Asymptotic Behaviors
- The pattern “osc” depicts oscillations of the considered marker. If one knows the highest and lowest boundaries of this oscillation, one can specify these boundaries, e.g., “osc(1,2)” indicates that the variable under consideration oscillates between 1 and 2. However, if such boundaries are unknown, “osc” simply specifies that the oscillation takes place without any constraints on the boundaries.
- The pattern “td(n)” specifies that the variable under consideration tends toward the value n. It specifies that the marker tends toward a global equilibrium.
6.2. Validation Matrix for the Regulation of Metabolism
6.2.1. Without Lipid Intake and without Oxygen Supply
- ⇝
- 1—FA = 0, exO2 = 0, GLC = 0, AA = 0: Leads to cell death. Cells require sufficient nutrients to supply the metabolic demands for cell growth and division. When all inputs are absent, no production of any component of the cell is possible and thus each variable tends toward 0. This would lead to cell death.
- ⇝
- 2 & 3—FA = 0, exO2 = 0, GLC = 0, AA = (1 or 2): The cell has only amino acids as nutrient supply. There is no general knowledge to assert whether amino acid intake alone can sustain carbon-dependent metabolic activity of the cell even if there are culture media without glucose for certain cancer cell lines [20,22]. Thus, we consider that cell fate is unknown, except that with no input of oxygen, the cell becomes in hypoxia and O2 tends toward 0.
- ⇝
- 4—FA = 0, exO2 = 0, GLC = 1, AA = 0: Leads to aerobic glycolysis. Without oxygen and with glucose as a unique carbon source, mitochondrial respiration cannot occur and the only metabolic pathway is fermentation. Even if ATP is consumed for cell maintenance, it at least does not tend to zero as the cell can survive with only glucose. is consumed during glycolysis and regenerated during fermentation; Glucose intake allows the production of lactate, which reduces to , which is recycled back to by glycolysis, so NADH oscillates. By the absence of oxygen supply, O2 tends toward 0 and mitochondrial activity is shunted, so PHOX tends toward 0. Lastly, GLC=1 denotes a glucose intake insufficient to make glycolysis reach its higher rate, so GLYC oscillates between 0 and 1.
- ⇝
- 5—FA = 0, exO2 = 0, GLC = 1, AA = 1: Leads to aerobic glycolysis. An additional supply of amino acids allows for non-lipidic biomass production, so nLBP does not tend toward 0. This anabolic process consumes , and as it is also produced, should oscillate. In addition, as in ⇝4, the cell is in an anaerobic process, and cytosolic metabolism and mitochondrial activity act similarly on the other markers.
- ⇝
- 6—FA = 0, exO2 = 0, GLC = 1, AA = 2: Leads to aerobic glycolysis. A huge supply on amino acids favors the reductive phase of Krebs to provide precursors for lipid synthesis, so LBP does not tend toward 0. The metabolic processes are the same as ⇝5, except that a large intake of amino acids activates glutaminolysis. It creates -ketoglutarate that can be converted with the reductive Krebs cycle in pyruvate. This accumulation of pyruvate could also be due to high activity of GLYC, therefore GLYC could sometimes reach its highest level. Thus, we prefer to relax its oscillatory behavior (“osc” without knowledge of the boundaries instead of “osc (0,1)”).
- ⇝
- 7—FA = 0, exO2 = 0, GLC = 2, AA = 0): Leads to aerobic glycolysis. With high glucose intake, glycolysis can sometimes reach its highest level. So, GLYC could possibly oscillate from its lowest to its highest level (“osc” instead of “osc (0,1)”). Moreover, the same processes as ⇝4 are impacted, and thus the behavior of all other markers remain identical.
- ⇝
- 8—FA = 0, exO2 = 0, GLC = 2, AA = 1): Leads to aerobic glycolysis. Here, the catabolic activity (glycolysis, fermentation, oxidative respiration and Krebs cycle) is similar to ⇝7, so that NADH behaves similarly. The more precise knowledge comes from the endergonic production of non-lipidic biomass, so that nLBP does not tend toward 0 and ATP can temporarily decrease to 0, so it oscillates.
- ⇝
- 9—FA = 0, exO2 = 0, GLC = 2, AA = 2: Leads to aerobic glycolysis. For the same reasons as ⇝6, this context allows for the production of lipid biomass, so LBP does not tend toward 0. Other markers behave similarly as in ⇝8.
6.2.2. Without Lipid Intake and with Oxygen Supply
- ⇝
- 10—FA = 0, exO2 = 1, GLC = 0, AA = 0: Leads to cell death. The cell is now in normoxia, thus O2 does not tend toward 0. All other markers tend toward 0 because, without glucose and amino acids entries, there is no glucose metabolism, thus no carbon and cofactors sources for anabolism, so we expect no pyruvate available for the Krebs cycle, and thus no oxidative phosphorylation; and lastly this also affects the production of ATP.
- ⇝
- 11 & 12—FA = 0, exO2 = 1, GLC = 0, AA = (1 or 2): No consensus phenotype. With oxygen and amino acids (glutamine) but no glucose, the general knowledge does not allow us to decide if amino acids intake is sufficient to sustain carbon dependent metabolic activity of the cell (grey boxes): Some cells survive, and some others do not. Only oxygen makes no doubt. Indeed, we prefer to take a cautious approach so as not to restrict the genericity of our study of the Warburg effect.
- ⇝
- 13—FA = 0, exO2 = 1, GLC = 1, AA = 0: Aerobic respiration survival. With normal intake, except amino acids and lipids, respiration can operate the processes involved in creating : GLYC, KREBS and PHOX oscillate. In response, O2 (consumed by oxidative phosphorylation) and NADH (produced by the Krebs cycle) also, oscillate. is produced but there is not enough general knowledge about its consumption to assert that ATP tends toward 1 or that it oscillates, so we only assert that it does not tend toward 0. Lastly, according to normal aerobic metabolism, FERM tend toward 0 (as fermentation is less efficient than oxidative phosphorylation).
- ⇝
- 14—FA = 0, exO2 = 1, GLC = 1, AA = 1: Cell culture conditions. This line can be considered to be the context representing a healthy cell. It adds amino acid inputs to ⇝13, so that non-lipid biomass can be produced (nLBP does not tend toward 0). Aerobic processes follow the behavior of ⇝13 but we can be more specific about ATP: there is now an consumption by biomass production, so that ATP oscillates.
- ⇝
- 15—FA = 0, exO2 = 1, GLC = 1, AA = 2: Aerobic respiration. This line is similar to ⇝14 except that the large input of amino acids enables a lipid biomass production.
- ⇝
- 16—FA = 0, exO2 = 1, GLC = 2 and AA = 0: Warburg/Crabtree effect. GLYC oscillates as in ⇝14, but the high glucose uptake provokes Warburg/Crabtree phenotype and leads to high anaerobic glycolysis, even in the presence of oxygen. From the general knowledge, we only assert that FERM does not tend toward 0. Glycolysis activity suffices to regenerate : as explained in ⇝4, NADH oscillates. As with ⇝13 ATP does not tend toward 0. On the opposite, oxidative phosphorylation might be present when the Warburg/Crabtree effect occurs, but for sure not constantly so PHOX does not tend toward 1. Therefore, oxygen could be partially consumed, but O2 does not tend toward 0 because its consumption by oxidative phosphorylation cannot counterbalance the external intake.
- ⇝
- 17 & 18—FA = 0, exO2 = 1, GLC = 2, AA = (1 or 2): Warburg/Crabtree effect. The Warburg/Crabtree occurs as in ⇝16. Additionally, here, the presence of amino acids intake allows biomass productions, so that nLBP and BLP do not tend toward 0, and thus ATP oscillates.
6.2.3. With Lipid Intake
- ⇝
- 19 and 28—FA = 1, exO2 = (0 or 1), GLC = 0, AA = 0: Lead to cell death. Lipid intake alone is unable to sustain all carbon-dependent metabolic activity of the cell, so, as in ⇝1 and ⇝10 respectively, all markers tend toward 0, except O2 for ⇝28.
- ⇝
- 20 & 21—FA = 1, exO2 = 0, GLC = 0, AA = (1 or 2): No consensus phenotype. As with ⇝2&3, there is no general knowledge to assert whether lipid intake alone can sustain carbon-dependent metabolic activity of the cell: we consider that the future phenotype of the cell is unknown, except for hypoxia.
- ⇝
- 22–27—FA = 1, exO2 = 0, GLC = (1 or 2), AA = (0, 1 or 2): Leads to aerobic glycolysis. With respect to ⇝4 to ⇝9, fatty acid intake only sustains lipidic biomass production, and consequently the consumption. Lipids synthesis can be done as soon as is available for the cell even in absence of amino acids and thus LBP does not tend toward 0, and the consumption makes ATP oscillate. Other markers keep the behavior already described in ⇝4 to ⇝9.
- ⇝
- 29 & 30—FA = 1, exO2 = 1, GLC = 0, AA = (1 or 2): No consensus phenotype. As with ⇝11&12, the general knowledge does not allow us to decide if oxygen and lipid intake are sufficient to sustain carbon-dependent metabolic activity of the cell. Only normoxia makes no doubt.
- ⇝
- 31—FA = 1, exO2 = 1, GLC = 1, AA = 0: Aerobic respiration survival. Compared to ⇝13, the same reasoning as ⇝22-27 applies.
- ⇝
- 32—FA = 1, exO2 = 1, GLC = 1, AA = 1: Compared to the cell culture conditions of ⇝14, fatty acid intake only sustains lipidic biomass production, thus LBP does not tend toward 0 and the other markers keep the same behavior.
- ⇝
- 33—FA = 1, exO2 = 1, GLC = 1, AA = 2: Aerobic respiration. With respect to ⇝32, glutaminolysis feeds Krebs (anaplerotic reactions) and this can fuel lipid synthesis through citrate export from mitochondria. This excess of citrate opens the possibility to fuel fermentation in addition to lipid production. Thus, as a precaution, we leave the behavior of FERM unknown.
- ⇝
- 34–36—FA=1, exO2=1, GLC=0, AA=(0,1 or 2): Warburg/Crabtree effect. Compared to lines from ⇝16 to ⇝18, the same reasoning as ⇝22–27 applies: LBP does not tend toward 0, and ATP oscillates.
7. Computer-Aided Validation of the Model Dynamics
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hammad, N.; Rosas-Lemus, M.; Uribe-Carvajal, S.; Rigoulet, M.; Devin, A. The Crabtree and Warburg effects: Do metabolite-induced regulations participate in their induction? Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, D.; van Berlo, R.; de Ridder, D.; Teusink, B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 2009, 5, 323. [Google Scholar] [CrossRef] [PubMed]
- Simeonidis, E.; Murabito, E.; Smallbone, K.; Westerhoff, H.V. Why does yeast ferment? A flux balance analysis study. Biochem. Soc. Trans. 2010, 38, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Wortel, M.T.; Bosdriesz, E.; Teusink, B.; Bruggeman, F.J. Evolutionary pressures on microbial metabolic strategies in the chemostat. Sci. Rep. 2016, 6, 29503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Veiga Moreira, J.; Hamraz, M.; Abolhassani, M.; Schwartz, L.; Jolicœur, M.; Peres, S. Metabolic therapies inhibit tumor growth in vivo and in silico. Sci. Rep. 2019, 9, 3153. [Google Scholar] [CrossRef] [Green Version]
- Bernot, G.; Comet, J.P.; Richard, A.; Guespin, J. Application of formal methods to biological regulatory networks: Extending Thomas’ asynchronous logical approach with temporal logic. J. Theor. Biol. 2004, 229, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Bernot, G.; Comet, J.P.; Khalis, Z.; Richard, A.; Roux, O.F. A Genetically Modified Hoare Logic. Theor. Comput. Sci. 2019, 765, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R. (Ed.) Kinetic logic: A boolean approach to the analysis of complex regulatory systems. In Lecture Notes in Biomathematics; Springer: Berlin/Heidelberg, Germany, 1979; Volume 29. [Google Scholar]
- Thomas, R. Regulatory networks seen as asynchronous automata: A logical description. J. Theor. Biol. 1991, 153, 1–23. [Google Scholar] [CrossRef]
- Khalis, Z.; Comet, J.P.; Richard, A.; Bernot, G. The SMBioNet Method for Discovering Models of Gene Regulatory Networks. Genes Genomes Genom. 2009, 3, 15–22. [Google Scholar]
- Boyenval, D.; Bernot, G.; Collavizza, H.; Comet, J.P. What is a cell cycle checkpoint? The TotemBioNet answer. In Proceedings of the 18th International Conference on Computational Methods in Systems Biology (CMSB), Online, 23–25 September 2020; Volume 12314, pp. 362–372. [Google Scholar]
- Gibart, L.; Bernot, G.; Collavizza, H.; Comet, J.P. TotemBioNet Enrichment Methodology: Application to the Qualitative Regulatory Network of the Cell Metabolism. In Proceedings of the 12th International Conference on Bioinformatics Models, Methods and Algorithms, Online, 11–13 February 2021. [Google Scholar]
- Khoodeeram, R. Discrete Coarse-Grained Modelling of Energy Metabolism Using Formal Approach: A Study of the Dynamics in Cell Proliferation. Ph.D. Thesis, Université Côte d’Azur, Nice, France, 2020. [Google Scholar]
- Khoodeeram, R.; Bernot, G.; Trosset, J.Y. An Ockham Razor model of energy metabolism. In Proceedings of the Thematic Research School on Advances in Systems and Synthetic Biology, Modelling Complex Biological Systems in the Context of Genomics; EDP Sciences: Les Ulis, France, 2016; pp. 81–101. ISBN 978-2-7598-2116-7. [Google Scholar]
- Thomas, R.; D’Ari, R. Biological Feedback; CRC Press: Boca Raton, FL, USA, 1990. [Google Scholar]
- Clarke, E.; Emerson, E. Design and syntheses of synchronization skeletons using branching time temporal logic. Workshop Log. Programs 1981, 131, 52–71. [Google Scholar]
- Yin, C.; Qie, S.; Sang, N. Carbon Source Metabolism and Its Regulation in Cancer Cells. Crit. Rev. Eukaryot. Gene Expr. 2012, 22, 17–35. [Google Scholar] [CrossRef] [Green Version]
- de Alteriis, E.; Cartenì, F.; Parascandola, P.; Serpa, J.; Mazzoleni, S. Revisiting the Crabtree/Warburg effect in a dynamic perspective: A fitness advantage against sugar-induced cell death. Cell Cycle 2018, 17, 688–701. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell. Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef]
- Rigoulet, M.; Bouchez, C.L.; Paumard, P.; Ransac, S.; Cuvellier, S.; Duvezin-Caubet, S.; Mazat, J.P.; Devin, A. Cell energy metabolism: An update. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148276. [Google Scholar] [CrossRef]
- Murray, D.B.; Beckmann, M.; Kitano, H. Regulation of yeast oscillatory dynamics. Proc. Natl. Acad. Sci. USA 2007, 104, 2241–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, C.V. Links between metabolism and cancer. Genes Dev. 2012, 26, 877–890. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, J.d.V.; Hamraz, M.; Abolhassani, M.; Bigan, E.; Pérès, S.; Paulevé, L.; Nogueira, M.L.; Steyaert, J.M.; Schwartz, L. The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect. Metabolites 2016, 6, 33. [Google Scholar] [CrossRef]
- Sober, E. Ockham’s Razors: A User’s Manual; Cambridge University Press: Cambridge, UK, 2015. [Google Scholar] [CrossRef]
- Axelrod, B. Chapter 3—Glycolysis. In Metabolic Pathways, 3rd ed.; Greenberg, D.M., Ed.; Academic Press: Cambridge, MA, USA, 1967; pp. 112–145. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Lowenstein, J.M. Chapter 4—The Tricarboxylic Acid Cycle. In Metabolic Pathways, 3rd ed.; Greenberg, D.M., Ed.; Academic Press: Cambridge, MA, USA, 1967; pp. 146–270. [Google Scholar] [CrossRef]
- Green, D.E.; MacLennan, D.H. Chapter 2—The Mitochondrial System of Enzymes. In Metabolic Pathways, 3rd ed.; Greenberg, D.M., Ed.; Academic Press: Cambridge, MA, USA, 1967; pp. 47–111. [Google Scholar] [CrossRef]
- Kumagai, H. Amino Acid Production. In The Prokaryotes: Applied Bacteriology and Biotechnology; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 169–177. [Google Scholar] [CrossRef]
- Reitzer, L. Amino Acid Synthesis. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 1–17. [Google Scholar] [CrossRef]
- Mansour, T.E. Studies on Heart Phosphofructokinase: Purification, Inhibition, and Activation. J. Biol. Chem. 1963, 238, 2285–2292. [Google Scholar] [CrossRef]
- Shi, L.; Tu, B.P. Acetyl-CoA and the Regulation of Metabolism: Mechanisms and Consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houten, S.M.; Wanders, R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Yang, M.; Gaur, U.; Xu, H.; Yao, Y.; Li, D. Alpha-Ketoglutarate: Physiological Functions and Applications. Biomol. Ther. 2016, 24, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKeehan, W.L. Glutaminolysis in Animal Cells. In Carbohydrate Metabolism in Cultured Cells; Morgan, M.J., Ed.; Springer: Berlin/Heidelberg, Germany, 1986; pp. 111–150. [Google Scholar] [CrossRef]
- Wilson, D.F. Oxidative phosphorylation: Regulation and role in cellular and tissue metabolism. J. Physiol. 2017, 595, 7023–7038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregg, X.T.; Prchal, J.T. Chapter 44—Red Blood Cell Enzymopathies. In Hematology, 7th ed.; Hoffman, R., Benz, E.J., Silberstein, L.E., Heslop, H.E., Weitz, J.I., Anastasi, J., Salama, M.E., Abutalib, S.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 616–625. [Google Scholar] [CrossRef]
- Stein, O.; Stein, Y. Lipid Synthesis, Intracellular Transport, Storage, and Secretion. J. Cell Biol. 1967, 33, 319–339. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry, 6th ed.; W.H. Freeman and Company: New York, NY, USA, 2012. [Google Scholar]
- Koolman, J.; Röhm, K.H. Color Atlas of Biochemistry, 3rd ed.; Revised and Updated Edition; Thieme Publishing Group: Stuttgart, Germany, 2012. [Google Scholar]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L.; Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W.H. Freeman and Company: New York, NY, USA, 2002. [Google Scholar]
- Goldfeder, J.; Kugler, H. BRE:IN—A Backend for Reasoning About Interaction Networks with Temporal Logic. In Computational Methods in Systems Biology; Lecture Notes in Computer, Science; Bortolussi, L., Sanguinetti, G., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 289–295. [Google Scholar] [CrossRef]
- Richard, A. Fair Paths in CTL. Available online: https://gitlab.com/totembionet/totembionet (accessed on 30 May 2021).
- Cimatti, A.; Clarke, E.; Giunchiglia, E.; Giunchiglia, F.; Pistore, M.; Roveri, M.; Sebastiani, R.; Tacchella, A. NuSMV 2: An OpenSource Tool for Symbolic Model Checking. In Proceedings of the CAV 2002: Computer Aided Verification, Copenhagen, Denmark, Copenhagen, Denmark, 27–31 July 2002; pp. 359–364. [Google Scholar]
- Gibart, L.; Collavizza, H.; Comet, J.P. Greening R. Thomas’ Framework with Environment Variables: A Divide and Conquer Approach. In Proceedings of the 19th International Conference on Computational Methods in Systems Biology (CMSB), Bordeaux, France, 22–24 September 2021. [Google Scholar]
- Damiani, C.; Colombo, R.; Gaglio, D.; Mastroianni, F.; Pescini, D.; Westerhoff, H.V.; Mauri, G.; Vanoni, M.; Alberghina, L. A metabolic core model elucidates how enhanced utilization of glucose and glutamine, with enhanced glutamine-dependent lactate production, promotes cancer cell growth: The WarburQ effect. PLoS Comput. Biol. 2017, 13, e1005758. [Google Scholar] [CrossRef] [Green Version]






| # Parameters for ATP | # Parameters for O2 | # Parameters for NADH |
|---|---|---|
| # Parameters for GLYC | ||
| # Parameters for nLBP | ||
| # Parameters for LBP | ||
| # Parameters for KREBS | ||
| # Parameters for NCD | ||
| # Parameters for PHOX | # Parameters for FERM | |
| Biological Context | FA | exO2 | GLC | AA | ATP (0–2) | O2 (0–1) | GLYC (0–2) | nLBP (0–1) | LBP (0–1) | FERM (0–1) | KREBS (0–2) | PHOX (0–1) | NADH (0–1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neither lipids nor oxygen supply | |||||||||||||
| 1 | 0 | 0 | 0 | 0 | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) |
| 2 & 3 | 0 | 0 | 0 | 1 & 2 | td(0) | ||||||||
| 4 | 0 | 0 | 1 | 0 | !td(0) | td(0) | osc(0-1) | !td(0) | td(0) | osc | |||
| 5 | 0 | 0 | 1 | 1 | osc | td(0) | osc(0-1) | !td(0) | !td(0) | td(0) | osc | ||
| 6 | 0 | 0 | 1 | 2 | osc | td(0) | osc | !td(0) | !td(0) | !td(0) | td(0) | osc | |
| 7 | 0 | 0 | 2 | 0 | !td(0) | td(0) | osc | !td(0) | td(0) | osc | |||
| 8 | 0 | 0 | 2 | 1 | osc | td(0) | osc | !td(0) | !td(0) | td(0) | osc | ||
| 9 | 0 | 0 | 2 | 2 | osc | td(0) | osc | !td(0) | !td(0) | !td(0) | td(0) | osc | |
| No lipids but oxygen supply | |||||||||||||
| 10 | 0 | 1 | 0 | 0 | td(0) | !td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) |
| 11 & 12 | 0 | 1 | 0 | 1 & 2 | !td(0) | ||||||||
| 13 | 0 | 1 | 1 | 0 | !td(0) | osc | osc | td(0) | osc | osc | osc | ||
| 14 | 0 | 1 | 1 | 1 | osc | osc | osc | !td(0) | td(0) | osc | osc | osc | |
| 15 | 0 | 1 | 1 | 2 | osc | osc | osc | !td(0) | !td(0) | td(0) | osc | osc | osc |
| 16 | 0 | 1 | 2 | 0 | !td(0) | !td(0) | osc | !td(0) | !td(1) | osc | |||
| 17 & 18 | 0 | 1 | 2 | 1&2 | osc | !td(0) | osc | !td(0) | !td(0) | !td(0) | !td(1) | osc | |
| Lipids but no oxygen supply | |||||||||||||
| 19 | 1 | 0 | 0 | 0 | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) |
| 20 & 21 | 1 | 0 | 0 | 1 & 2 | td(0) | ||||||||
| 22 | 1 | 0 | 1 | 0 | osc | td(0) | osc(0-1) | !td(0) | !td(0) | td(0) | osc | ||
| 23 | 1 | 0 | 1 | 1 | osc | td(0) | osc(0-1) | !td(0) | !td(0) | !td(0) | td(0) | osc | |
| 24 | 1 | 0 | 1 | 2 | osc | td(0) | osc | !td(0) | !td(0) | !td(0) | td(0) | osc | |
| 25 | 1 | 0 | 2 | 0 | osc | td(0) | osc | !td(0) | !td(0) | td(0) | osc | ||
| 26 & 27 | 1 | 0 | 2 | 1 & 2 | osc | td(0) | osc | !td(0) | !td(0) | !td(0) | td(0) | osc | |
| Lipids and oxygen supply | |||||||||||||
| 28 | 1 | 1 | 0 | 0 | td(0) | !td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) | td(0) |
| 29 & 30 | 1 | 1 | 0 | 1 & 2 | !td(0) | ||||||||
| 31 | 1 | 1 | 1 | 0 | osc | osc | osc | !td(0) | td(0) | osc | osc | osc | |
| 32 | 1 | 1 | 1 | 1 | osc | osc | osc | !td(0) | !td(0) | td(0) | osc | osc | osc |
| 33 | 1 | 1 | 1 | 2 | osc | osc | osc | !td(0) | !td(0) | osc | osc | osc | |
| 34 | 1 | 1 | 2 | 0 | osc | !td(0) | osc | !td(0) | !td(0) | !td(1) | osc | ||
| 35 & 36 | 1 | 1 | 2 | 1 & 2 | osc | !td(0) | osc | !td(0) | !td(0) | !td(0) | !td(1) | osc | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibart, L.; Khoodeeram, R.; Bernot, G.; Comet, J.-P.; Trosset, J.-Y. Regulation of Eukaryote Metabolism: An Abstract Model Explaining the Warburg/Crabtree Effect. Processes 2021, 9, 1496. https://doi.org/10.3390/pr9091496
Gibart L, Khoodeeram R, Bernot G, Comet J-P, Trosset J-Y. Regulation of Eukaryote Metabolism: An Abstract Model Explaining the Warburg/Crabtree Effect. Processes. 2021; 9(9):1496. https://doi.org/10.3390/pr9091496
Chicago/Turabian StyleGibart, Laetitia, Rajeev Khoodeeram, Gilles Bernot, Jean-Paul Comet, and Jean-Yves Trosset. 2021. "Regulation of Eukaryote Metabolism: An Abstract Model Explaining the Warburg/Crabtree Effect" Processes 9, no. 9: 1496. https://doi.org/10.3390/pr9091496






