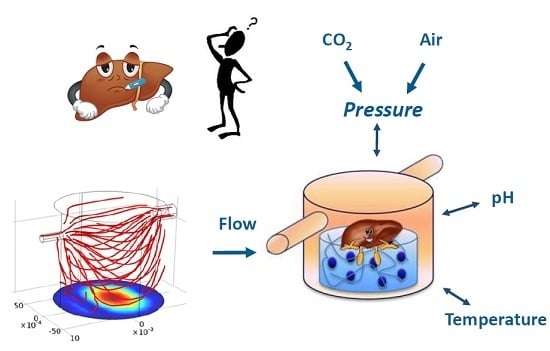
Environmental Control in Flow Bioreactors
Abstract
:1. Introduction
2. Materials and Methods
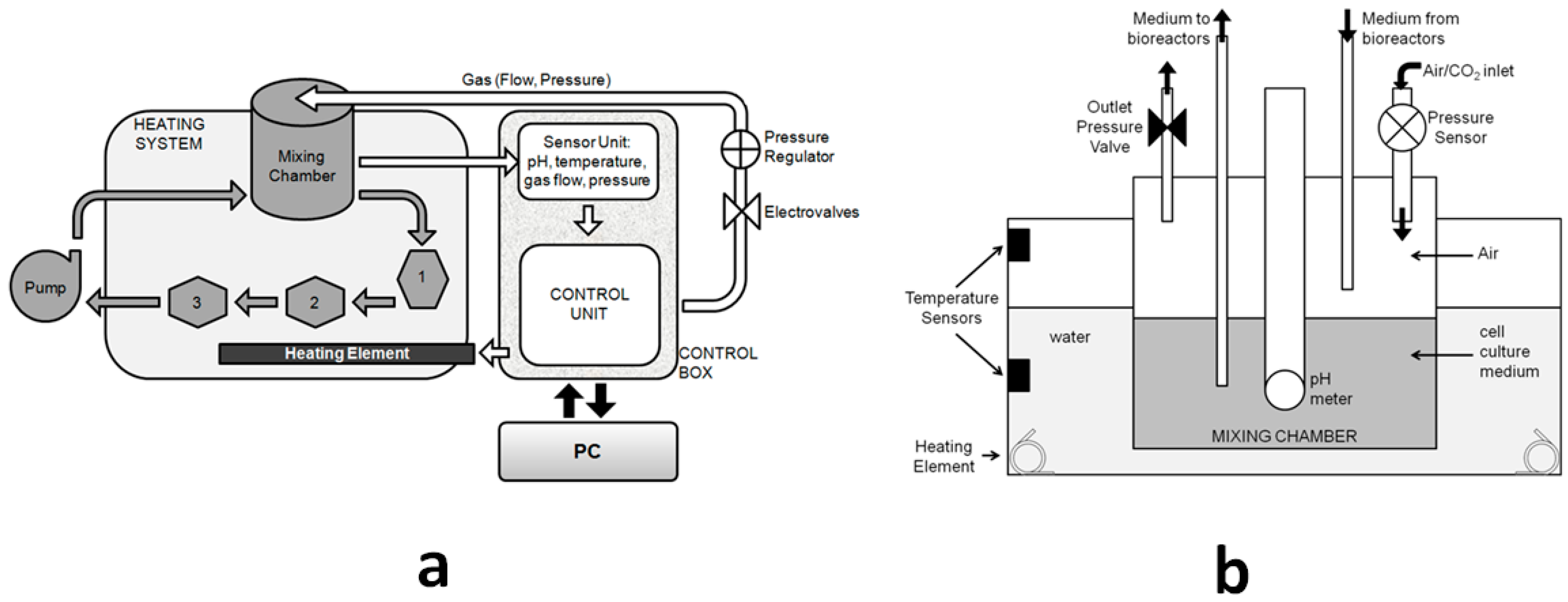
2.1. The SUITE System
2.1.1. Architecture of the SUITE System
- The software which reads, controls, and stores information from the sensors and actuators in the system;
- A control box linked to a PC, containing all the electronic circuits and electro-valves;
- A mixing chamber for pH regulation and media reservoir;
- A heating system;
- A peristaltic pump; and
- A series of modular bioreactors which can be combined in series or parallel to recapitulate physiological crosstalk between organs.
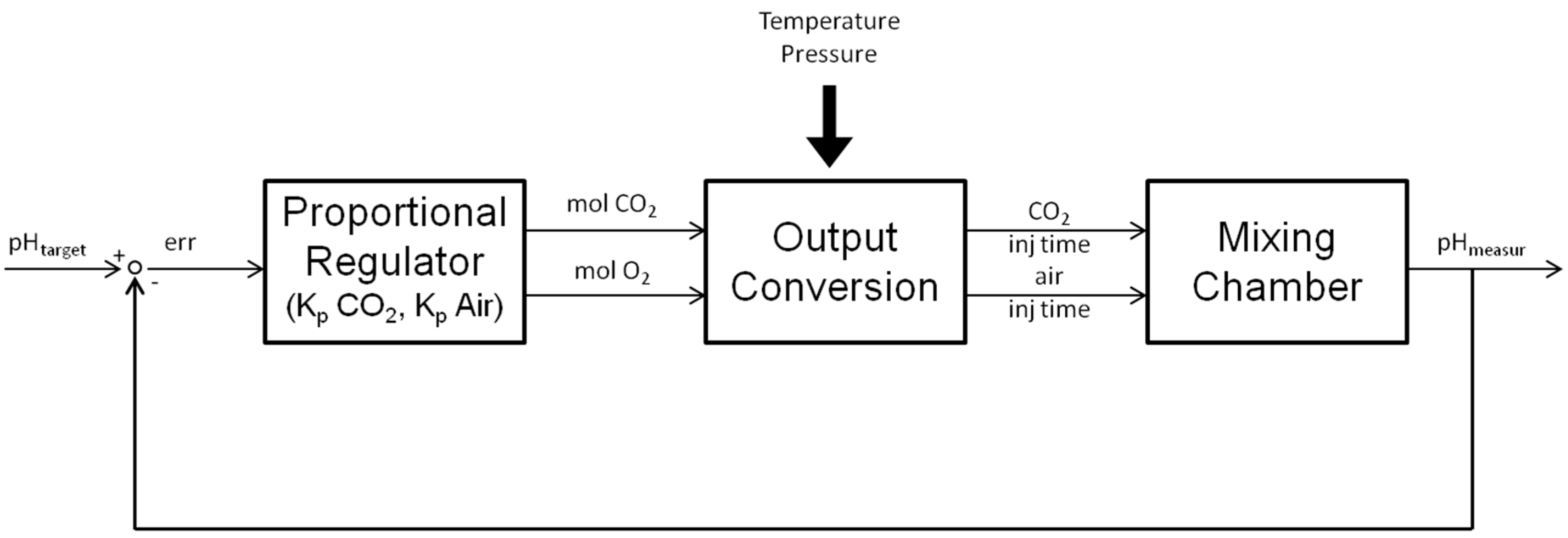
2.1.2. Control Algorithm
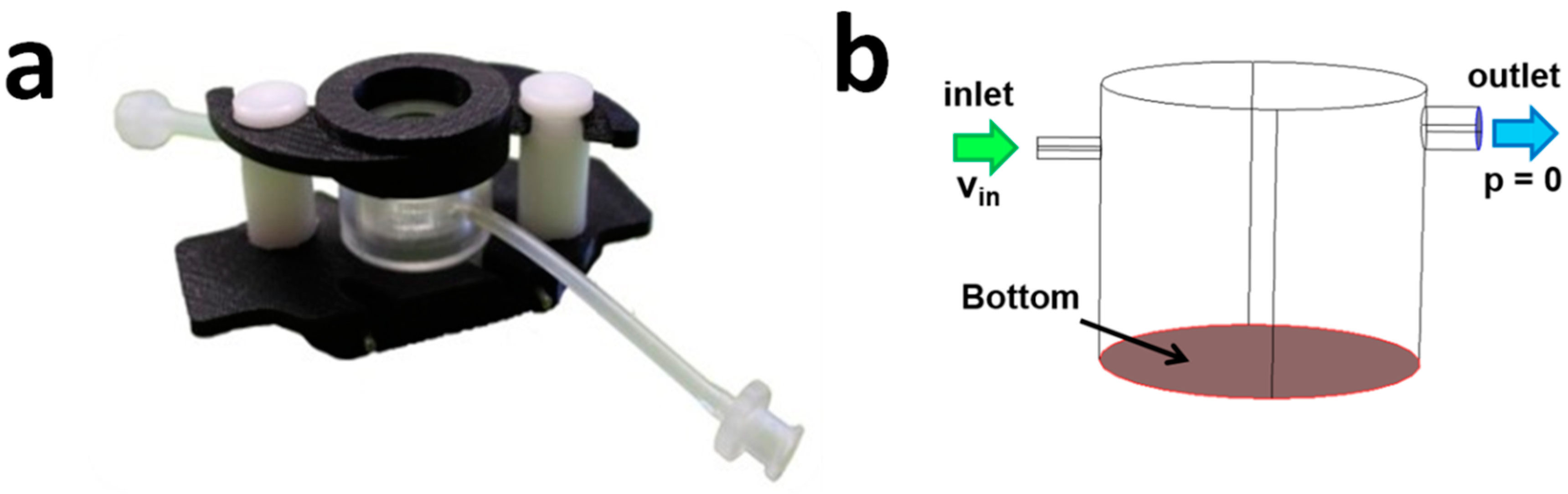
2.2. The LiveBox1 Bioreactor
Hydrodynamic Pressure in the LiveBox1
2.3. In Vitro Model of Portal Hypertension
2.3.1. Cell Culture
2.3.2. Cell Assay and Staining
2.3.3. Data Analysis
3. Results
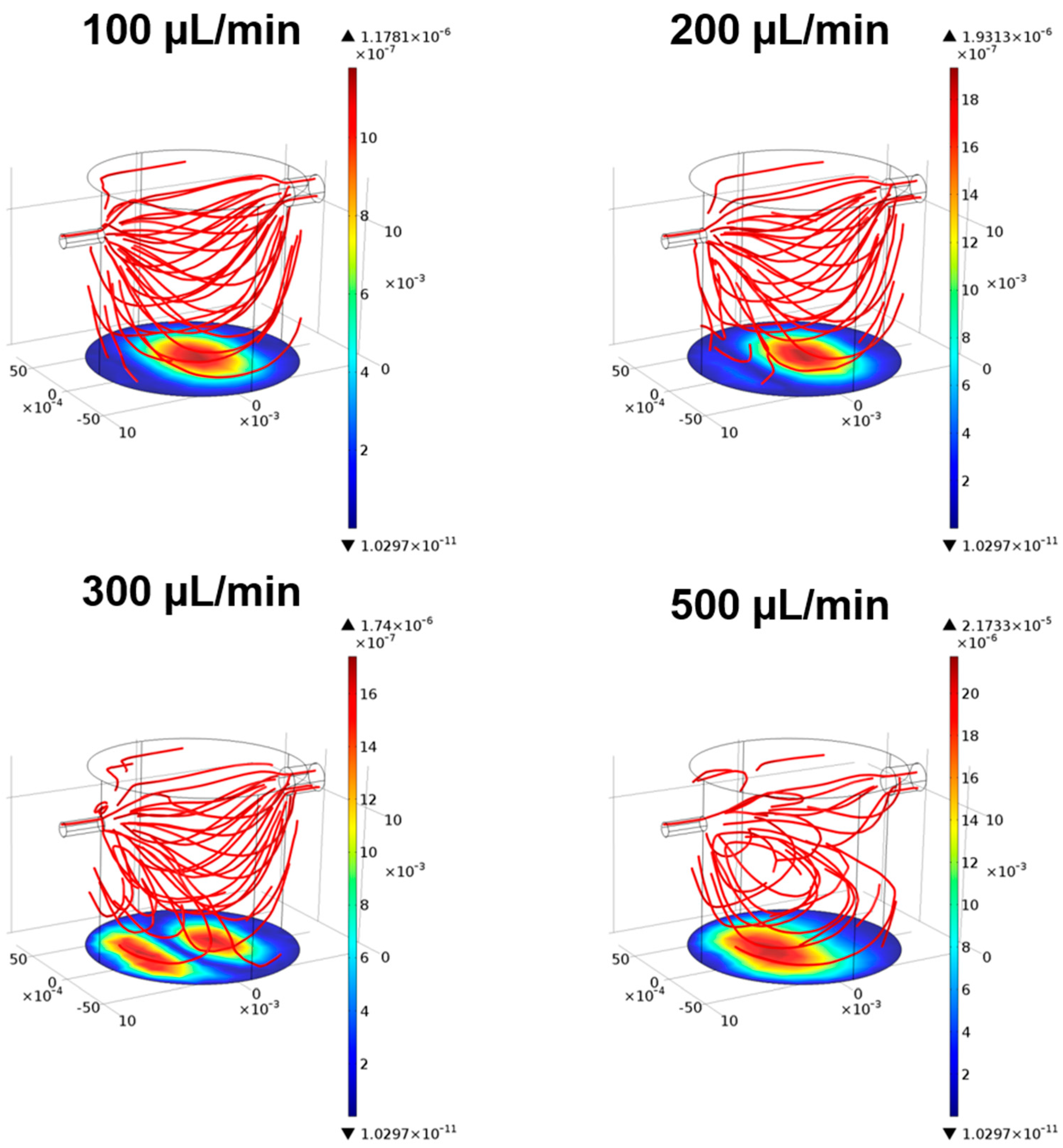
3.1. Fluid-Dynamics in the LB1
3.1.1. CFD Models
3.1.2. Hydrostatic and Hydrodynamic Pressure
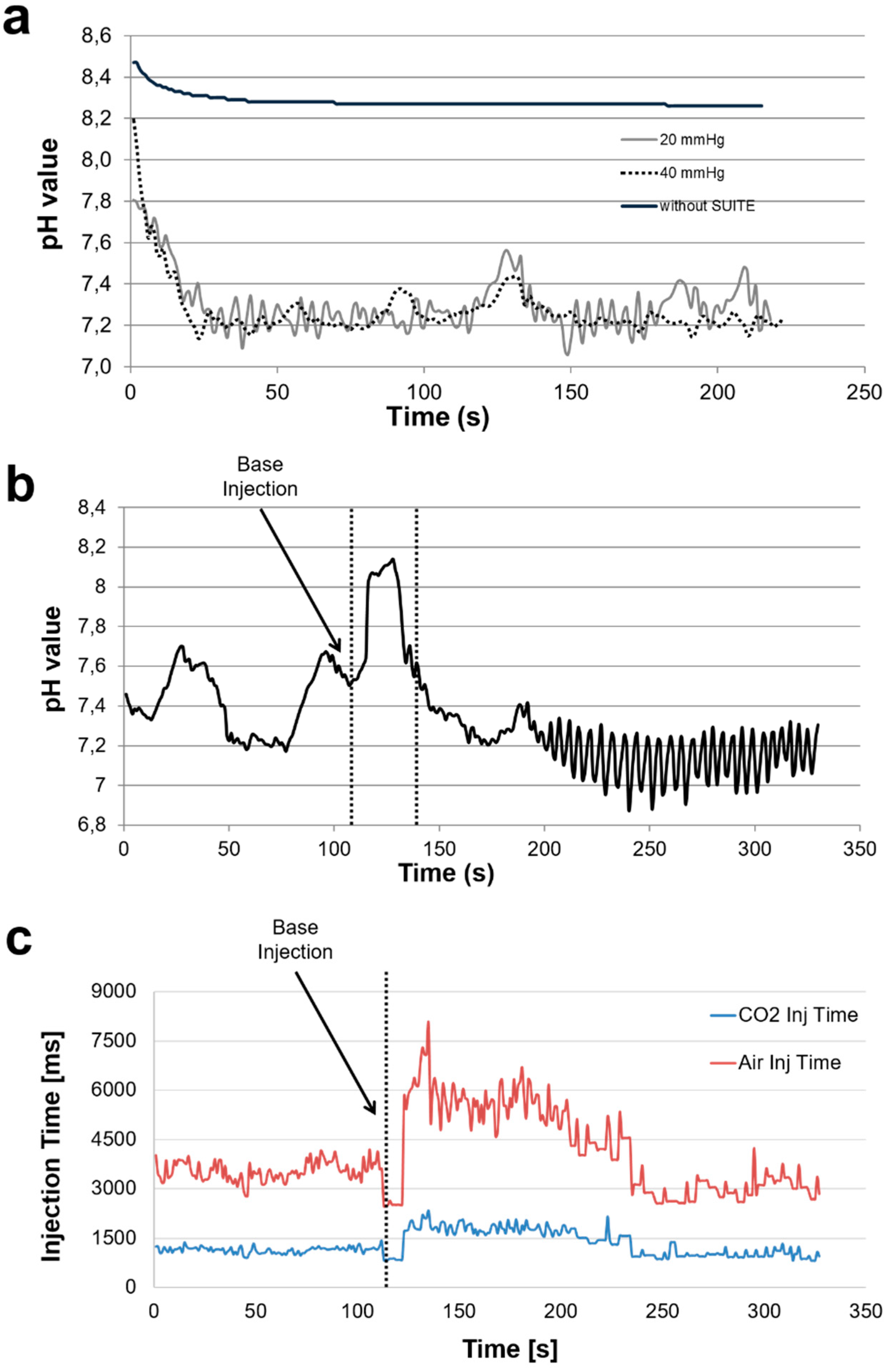
3.2. Validation of Control Algorithm
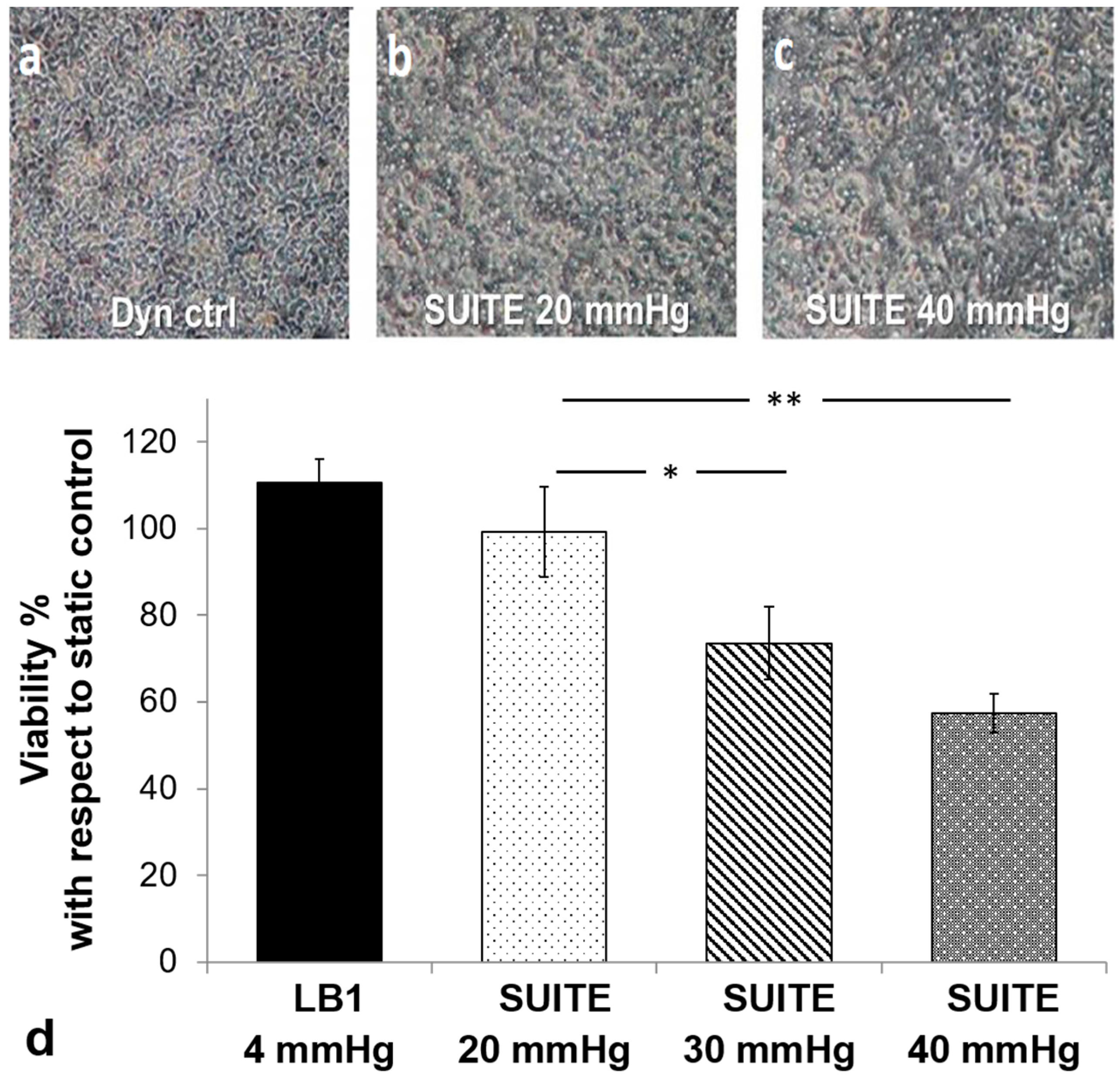
3.3. Cell Assays
4. Discussion
Supplementary Files
Supplementary File 1Author Contributions
Conflicts of Interest
Abbreviations
| SUITE | Supervising Unit for In Vitro TEsting |
| LB1 | LiveBox 1 Bioreactor |
| HUVEC | Human Umbilical Vein Endothelial Cells |
| C3A | human hepatocellular carcinoma cells |
| vWF | von Willebrand Factor |
| EMEM | Eagles Minimal Essential Medium |
References
- Di Nardo, P.; Minieri, M.; Ahluwalia, A. Engineering the Stem Cell Niche and the Differentiative Micro-and Macroenvironment: Technologies and Tools for Applying Biochemical, Physical and Structural Stimuli and Their Effects on Stem Cells. In Stem Cell Engineering; Springer: Berlin/Heidelberg, Germany, 2011; pp. 41–59. [Google Scholar] [CrossRef]
- Mattei, G.; Giusti, S.; Ahluwalia, A. Design Criteria for Generating Physiologically Relevant In Vitro Models in Bioreactors. Processes 2014, 2, 548–569. [Google Scholar] [CrossRef]
- Ahluwalia, A. Allometric Scaling in-Vitro. Sci. Rep. 2017, 7, 42113. [Google Scholar] [CrossRef] [PubMed]
- Bratt-Leal, A.M.; Carpenedo, R.L.; McDevitt, T.C. Engineering the Embryoid Body Microenvironment to Direct Embryonic Stem Cell Differentiation. Biotechnol. Prog. 2009, 25, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Pfau, J.C.; Schneider, J.C.; Archer, A.J.; Sentissi, J.; Leyva, F.J.; Cramton, J. Environmental Oxygen Tension Affects Phenotype in Cultured Bone Marrow-Derived Macrophages. AJP Lung Cell. Mol. Physiol. 2004, 286, L354–L362. [Google Scholar] [CrossRef] [PubMed]
- Biechele, P.; Busse, C.; Solle, D.; Scheper, T.; Reardon, K. Sensor Systems for Bioprocess Monitoring. Eng. Life Sci. 2015, 15, 469–488. [Google Scholar] [CrossRef]
- Abu-Absi, N.R.; Kenty, B.M.; Cuellar, M.E.; Borys, M.C.; Sakhamuri, S.; Strachan, D.J.; Hausladen, M.C.; Li, Z.J. Real Time Monitoring of Multiple Parameters in Mammalian Cell Culture Bioreactors Using an in-Line Raman Spectroscopy Probe. Biotechnol. Bioeng. 2011, 108, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Trummer, E.; Fauland, K.; Seidinger, S.; Schriebl, K.; Lattenmayer, C.; Kunert, R.; Vorauer-Uhl, K.; Weik, R.; Borth, N.; Katinger, H.; et al. Process Parameter Shifting: Part I. Effect of DOT, pH, and Temperature on the Performance of Epo-Fc Expressing CHO Cells Cultivated in Controlled Batch Bioreactors. Biotechnol. Bioeng. 2006, 94, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Olmer, R.; Lange, A.; Selzer, S.; Kasper, C.; Haverich, A.; Martin, U.; Zweigerdt, R. Suspension Culture of Human Pluripotent Stem Cells in Controlled, Stirred Bioreactors. Tissue Eng. Part C Methods 2012, 18, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Maharbiz, M.M.; Holtz, W.J.; Howe, R.T.; Keasling, J.D. Microbioreactor Arrays with Parametric Control for High-Throughput Experimentation. Biotechnol. Bioeng. 2004, 85, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Mehta, G.; Mehta, K.; Sud, D.; Song, J.W.; Bersano-Begey, T.; Futai, N.; Heo, Y.S.; Mycek, M.A.; Linderman, J.J.; Takayama, S. Quantitative Measurement and Control of Oxygen Levels in Microfluidic Poly (dimethylsiloxane) Bioreactors during Cell Culture. Biomed. Microdevices 2007, 9, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Vinci, B.; Murphy, E.; Iori, E.; Meduri, F.; Fattori, S.; Marescotti, M.C.; Castagna, M.; Avogaro, A.; Ahluwalia, A. An in Vitro Model of Glucose and Lipid Metabolism in a Multicompartmental Bioreactor. Biotechnol. J. 2012, 7, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Vinci, B.; Murphy, E.; Iori, E.; Marescotti, M.C.; Avogaro, A.; Ahluwalia, A. Flow-Regulated Glucose and Lipid Metabolism in Adipose Tissue, Endothelial Cell and Hepatocyte Cultures in a Modular Bioreactor. Biotechnol. J. 2010, 5, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Iori, E.; Vinci, B.; Murphy, E.; Marescotti, M.C.; Avogaro, A.; Ahluwalia, A. Glucose and Fatty Acid Metabolism in a 3 Tissue In Vitro Model Challenged with Normo-and Hyperglycaemia. PLoS ONE 2012, 7, e34704. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.K.; Chen, L.; Groszmann, R.J. Pathophysiology of Portal Hypertension. Clin. Liver Dis. 1997, 1, 1–12. [Google Scholar] [CrossRef]
- Lee, S.S.; Hadengue, A.; Girod, C.; Braillon, A.; Lebrec, D. Reduction of Intrahepatic Vascular Space in the Pathogenesis of Portal Hypertension: In Vitro and in Vivo Studies in the Rat. Gastroenterology 1987, 9, 157–161. [Google Scholar] [CrossRef]
- Liu, S.; Premont, R.T.; Kontos, C.D.; Zhu, S.; Rockey, D.C. A Crucial Role for GRK2 in Regulation of Endothelial Cell Nitric Oxide Synthase Function in Portal Hypertension. Nat. Med. 2005, 11, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Van der Worp, H.B.; Howells, D.W.; Sena, E.S.; Porritt, M.J.; Rewell, S.; O’Collins, V.; Macleod, M.R. Can Animal Models of Disease Reliably Inform Human Studies? PLoS Med. 2010, 7, e1000245. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, D.; Vozzi, F.; Cisternino, A.; Vozzi, G.; Ahluwalia, A. A High-Throughput Bioreactor System for Simulating Physiological Environments. IEEE Trans. Ind. Electron. 2008, 55, 232–241. [Google Scholar] [CrossRef]
- Abts, H.; Arain, S. Process Monitoring in Suspension–Adapted CHO Cell Cultures. BioProcess Int. 2008, 6, 64–66. [Google Scholar]
- Vinci, B.; Cavallone, D.; Vozzi, G.; Mazzei, D.; Domenici, C.; Brunetto, M.; Ahluwalia, A. In Vitro Liver Model Using Microfabricated Scaffolds in a Modular Bioreactor. Biotechnol. J. 2010, 5, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, D.; Guzzardi, M.A.; Giusti, S.; Ahluwalia, A. A Low Shear Stress Modular Bioreactor for Connected Cell Culture under High Flow Rates. Biotechnol. Bioeng. 2010, 106, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Giusti, S.; Sbrana, T.; La Marca, M.; Di Patria, V.; Martinucci, V.; Tirella, A.; Domenici, C.; Ahluwalia, A. A Novel Dual-Flow Bioreactor Simulates Increased Fluorescein Permeability in Epithelial Tissue Barriers. Biotechnol. J. 2014, 9, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Baudin, B.; Bruneel, A.; Bosselut, N.; Vaubourdolle, M. A Protocol for Isolation and Culture of Human Umbilical Vein Endothelial Cells. Nat. Protoc. 2007, 2, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Vozzi, F.; Bianchi, F.; Ahluwalia, A.; Domenici, C. Hydrostatic Pressure and Shear Stress Affect Endothelin-1 and Nitric Oxide Release by Endothelial Cells in Bioreactors. Biotechnol. J. 2014, 9, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Galbusera, M.; Zoja, C.; Donadelli, R.; Paris, S.; Morigi, M.; Benigni, A.; Figliuzzi, M.; Remuzzi, G.; Remuzzi, A. Fluid Shear Stress Modulates von Willebrand Factor Release From Human Vascular Endothelium. Blood 1997, 90, 1558–1564. [Google Scholar] [PubMed]
- Blendis, L.M.; Orrego, H.; Crossley, I.R.; Blake, J.E.; Medline, A.; Israel, Y. The Role of Hepatocyte Enlargement in Hepatic Pressure in Cirrhotic and Noncirrhotic Alcoholic Liver Disease. Hepatology 2007, 2, 539S–546S. [Google Scholar] [CrossRef]
- Mahmoodi, S.; Poshtan, J.; Jahed-Motlagh, M.R.; Montazeri, A. Nonlinear Model Predictive Control of a pH Neutralization Process Based on Wiener–Laguerre Model. Chem. Eng. J. 2009, 146, 328–337. [Google Scholar] [CrossRef]
- Henson, M.; Seborg, D.E. Adaptive Nonlinear Control of a pH Neutralization Process. IEEE Trans. Control Syst. Technol. 1994, 2, 169–182. [Google Scholar] [CrossRef]

| Pressure (mmHg) | Air Inject Time (s) | CO2 Inject Time (s) |
|---|---|---|
| 15 | 1.94 ± 0.22% | 4.06 ± 0.18% |
| 20 | 1.54 ± 0.23% | 3.15 ± 0.28% |
| 25 | 1.46 ± 0.21% | 2.88 ± 0.24% |
| 30 | 1.10 ± 0.09% | 2.11 ± 0.47% |
| 40 | 0.78 ± 0.06% | 2.03 ± 0.23% |
| 50 | 0.83 ± 0.18% | 1.57 ± 0.16% |
| Static Ctrl (0 mmHg) | LB1 (4 mm Hg) | SUITE 30 mmHg | SUITE 40 mmHg | SUITE 50 mmHg |
|---|---|---|---|---|
| 18.14 ± 2.86% | 28.44 ± 5.02% | 29.11 ± 5.98% | 37.28 ± 4.67% | 41.84 ± 6.01% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giusti, S.; Mazzei, D.; Cacopardo, L.; Mattei, G.; Domenici, C.; Ahluwalia, A. Environmental Control in Flow Bioreactors. Processes 2017, 5, 16. https://doi.org/10.3390/pr5020016
Giusti S, Mazzei D, Cacopardo L, Mattei G, Domenici C, Ahluwalia A. Environmental Control in Flow Bioreactors. Processes. 2017; 5(2):16. https://doi.org/10.3390/pr5020016
Chicago/Turabian StyleGiusti, Serena, Daniele Mazzei, Ludovica Cacopardo, Giorgio Mattei, Claudio Domenici, and Arti Ahluwalia. 2017. "Environmental Control in Flow Bioreactors" Processes 5, no. 2: 16. https://doi.org/10.3390/pr5020016