Critical Analysis of Risk Factors and Machine-Learning-Based Gastric Cancer Risk Prediction Models: A Systematic Review
Abstract
:1. Introduction
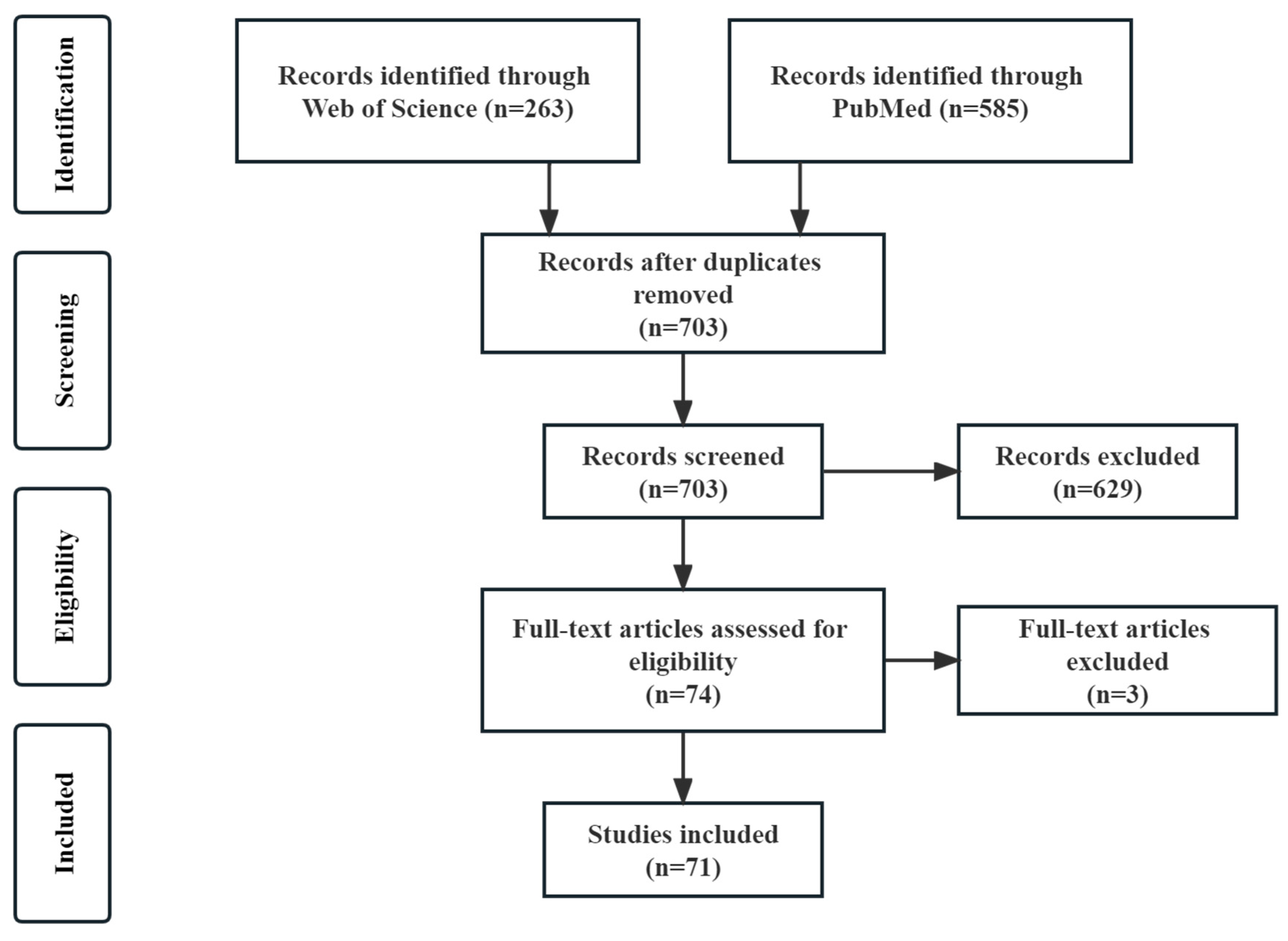
2. Materials and Methods
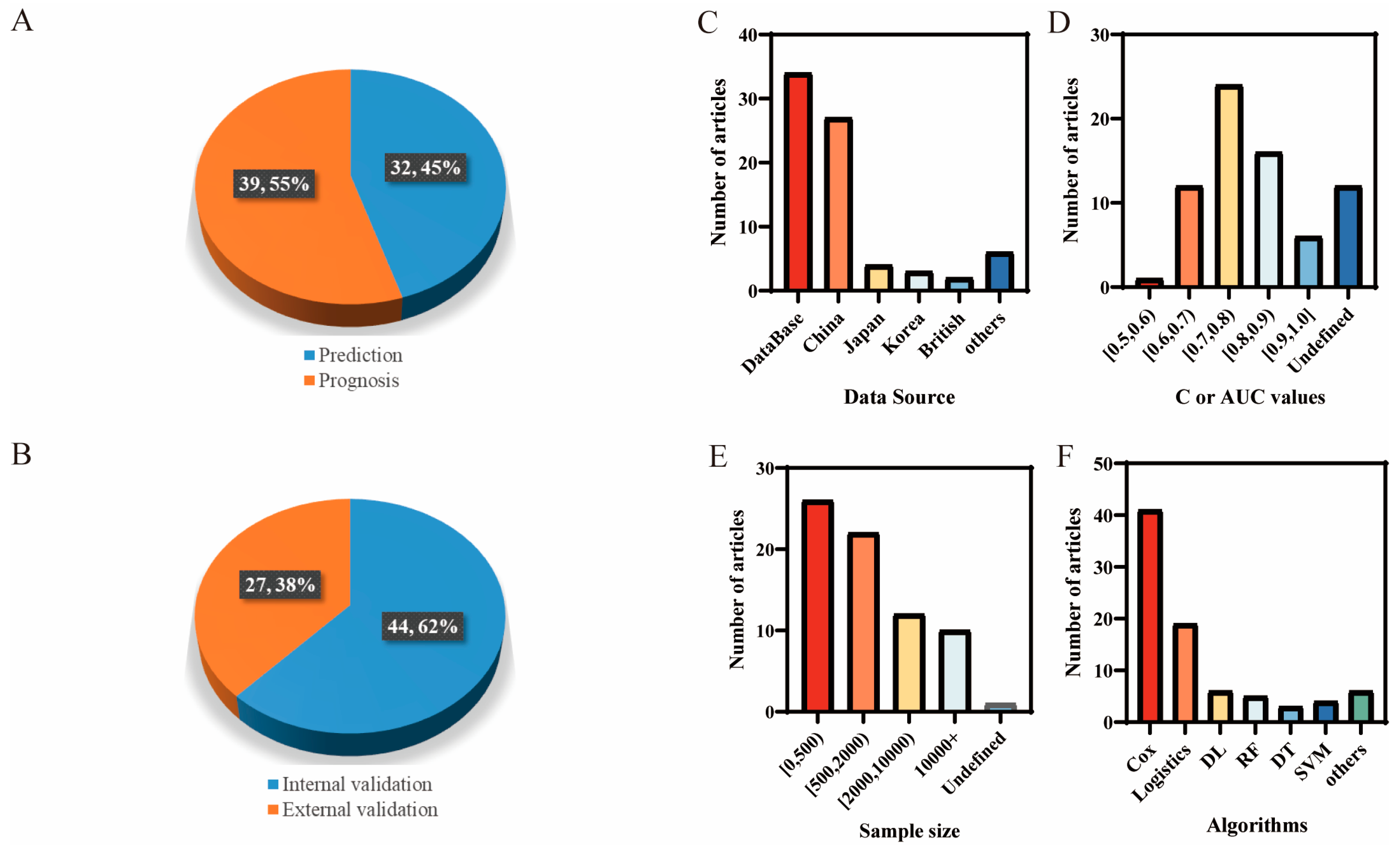
3. Results
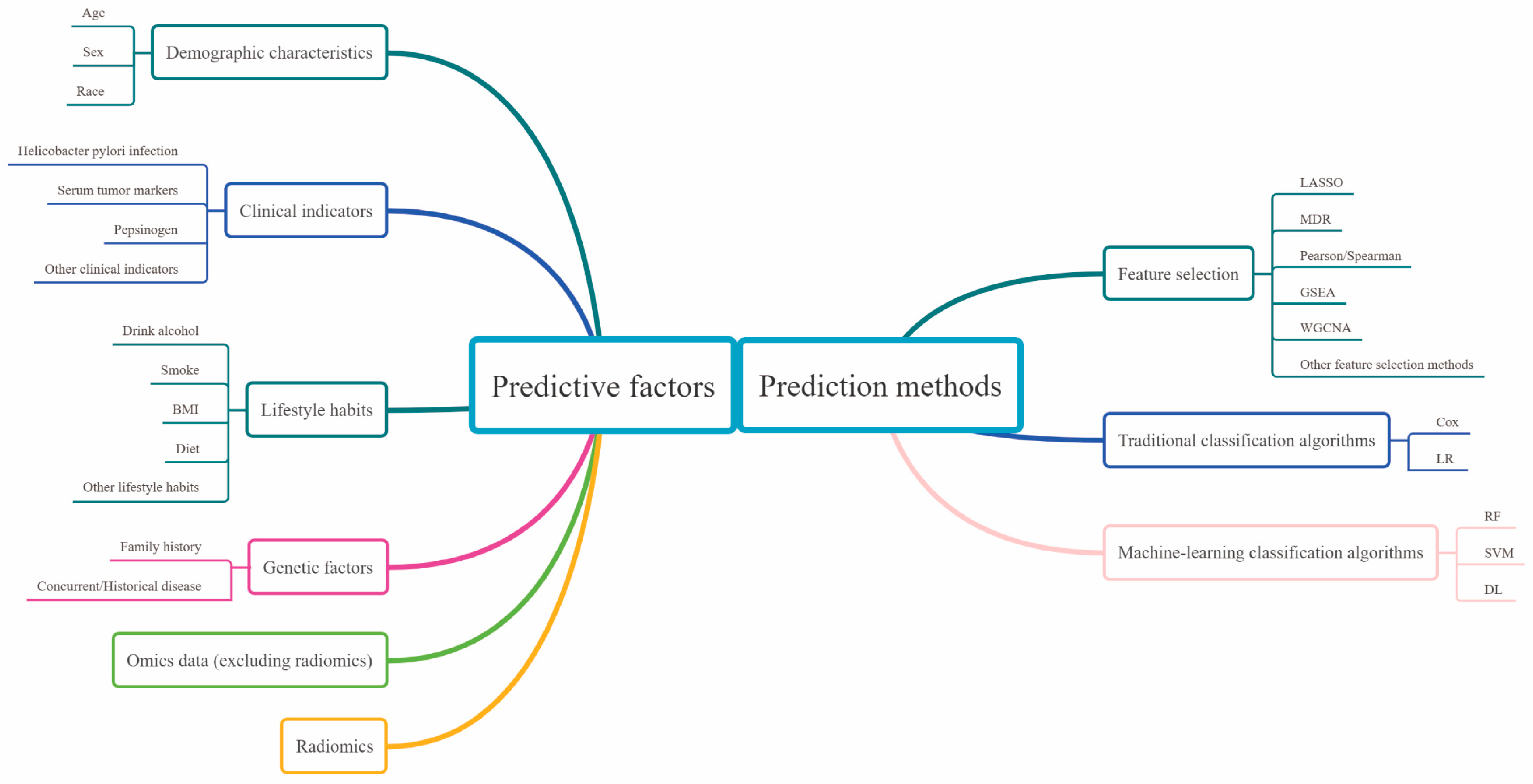
3.1. Predictive Factors
3.1.1. Demographic Characteristics
3.1.2. Clinical Indicators
3.1.3. Lifestyle Habits
3.1.4. Genetic Factors
3.1.5. Omics Data (Excluding Radiomics)
3.1.6. Radiomics
3.2. Analysis of the Prediction Methods
3.2.1. Feature Selection
3.2.2. Traditional Classification Algorithms
3.2.3. Mainstream of Machine-Learning Classification Algorithms
4. Discussion
4.1. Selection of Predictive Factors
4.2. Selection of Prediction Methods
4.3. Collection and Partition of Datasets
4.4. Criteria for Evaluating the Model Performance
4.5. Ethical Limitations of Model Application
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.Y.; Guo, Y.X.; Gu, X.R.; Huang, R.R.; Miao, W.J. Development and validation of an artificial neural network model for non-invasive gastric cancer screening and diagnosis. Sci. Rep. 2022, 12, 21795. [Google Scholar] [CrossRef]
- Pan, Y.Y.; Fu, H.; Jiang, X.W.; Zhu, M. A systematic review of the main predictors of gastric cancer risk prediction models. Jiangsu J. Prev. Med. 2021, 32, 689–692. [Google Scholar] [CrossRef]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, Nccn Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 2022, 20, 167–192. [Google Scholar] [CrossRef]
- Seeneevassen, L.; Bessède, E.; Mégraud, F.; Lehours, P.; Dubus, P.; Varon, C. Gastric Cancer: Advances in Carcinogenesis Research and New Therapeutic Strategies. Int. J. Mol. Sci. 2021, 22, 3418. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Shen, Z.; Zhang, X.; Sang, J.; Ding, Y.; Yang, X.; Li, J.; Chen, M.; Jin, C.; et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer 2020, 23, 126–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdbrook, D.A.; Singh, M.; Choudhury, Y.; Kalaw, E.M.; Koh, V.; Tan, H.S.; Kanesvaran, R.; Tan, P.H.; Peng, J.Y.S.; Tan, M.H.; et al. Automated Renal Cancer Grading Using Nuclear Pleomorphic Patterns. JCO Clin. Cancer Inform. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Hashimoto, D.A.; Rosman, G.; Rus, D.; Meireles, O.R. Artificial Intelligence in Surgery: Promises and Perils. Ann. Surg. 2018, 268, 70–76. [Google Scholar] [CrossRef]
- Wong, P.K.; Chan, I.N.; Yan, H.M.; Gao, S.; Wong, C.H.; Yan, T.; Yao, L.; Hu, Y.; Wang, Z.R.; Yu, H.H. Deep learning based radiomics for gastrointestinal cancer diagnosis and treatment: A minireview. World J. Gastroenterol. 2022, 28, 6363–6379. [Google Scholar] [CrossRef]
- Jin, P.; Ji, X.Y.; Kang, W.Z.; Li, Y.; Liu, H.; Ma, F.H.; Ma, S.; Hu, H.T.; Li, W.K.; Tian, Y.T. Artificial intelligence in gastric cancer: A systematic review. J. Cancer Res. Clin. Oncol. 2020, 146, 2339–2350. [Google Scholar] [CrossRef]
- Sgourakis, G.; Gockel, I.; Lang, H. Endoscopic and surgical resection of T1a/T1b esophageal neoplasms: A systematic review. World J. Gastroenterol. 2013, 19, 1424–1437. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ (Clin. Res. Ed.) 2021, 372, n160. [Google Scholar] [CrossRef]
- Iida, M.; Ikeda, F.; Hata, J.; Hirakawa, Y.; Ohara, T.; Mukai, N.; Yoshida, D.; Yonemoto, K.; Esaki, M.; Kitazono, T.; et al. Development and validation of a risk assessment tool for gastric cancer in a general Japanese population. Gastric Cancer 2018, 21, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Zhi, S.; Yang, B.; Zhou, S.; Tan, J.; Zhong, G.; Han, F. Immune-Related LncRNAs to Construct a Prognosis Risk-Assessment Model for Gastric Cancer. Curr. Oncol. 2022, 29, 4923–4935. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ma, Q.; Wang, L.; He, C.; Lu, S.; Ni, Z.; Hua, Z.; Zhu, Z.; Yang, Z.; Zheng, Y.; et al. Prediction Model of Tumor Regression Grade for Advanced Gastric Cancer After Preoperative Chemotherapy. Front. Oncol. 2021, 11, 607640. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, D.; Deng, X.; Lin, J.; Zheng, X.; Peng, F.; Meng, F.; Niu, W. Prediction of presurgical metabolic syndrome for gastric cancer-specific mortality is more evident in smokers: The FIESTA study. Cancer Med. 2023, 12, 3419–3432. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xu, Y.; Li, Y.; Chen, R.; Cai, W. Integrative Radiogenomics Approach for Risk Assessment of Postoperative and Adjuvant Chemotherapy Benefits for Gastric Cancer Patients. Front. Oncol. 2021, 11, 755271. [Google Scholar] [CrossRef]
- Yang, Y.; Long, Z.; Zhong, Z.M.; Liu, Q.; Yang, X. Construction and Evaluation of Gastric Cancer Risk Prediction Model. Indian J. Pharm. Sci. 2021, 83, 112–118. [Google Scholar] [CrossRef]
- Duan, F.; Song, C.; Wang, P.; Ye, H.; Dai, L.; Zhang, J.; Wang, K. Polygenic Risk Scores for Prediction of Gastric Cancer Based on Bioinformatics Screening and Validation of Functional lncRNA SNPs. Clin. Transl. Gastroenterol. 2021, 12, e00430. [Google Scholar] [CrossRef]
- Ishikura, N.; Ito, H.; Oze, I.; Koyanagi, Y.N.; Kasugai, Y.; Taniyama, Y.; Kawakatsu, Y.; Tanaka, T.; Ito, S.; Tajika, M.; et al. Risk Prediction for Gastric Cancer Using GWAS-Identifie Polymorphisms, Helicobacter pylori Infection and Lifestyle-Related Risk Factors in a Japanese Population. Cancers 2021, 13, 5525. [Google Scholar] [CrossRef]
- Charvat, H.; Sasazuki, S.; Inoue, M.; Iwasaki, M.; Sawada, N.; Shimazu, T.; Yamaji, T.; Tsugane, S.; JPHC Study Group. Prediction of the 10-year probability of gastric cancer occurrence in the Japanese population: The JPHC study cohort II. Int. J. Cancer 2016, 138, 320–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wu, X.; Chen, Y. Stromal-Immune Score-Based Gene Signature: A Prognosis Stratification Tool in Gastric Cancer. Front. Oncol. 2019, 9, 1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.Y.; Wang, L.L.; Liang, S.Z.; Yang, C.; Xu, L.; Yu, M.C.; Wang, Y.X.; Dong, Q.J. Prediction of gastric cancer risk by a polygenic risk score of Helicobacter pylori. World J. Gastrointest. Oncol. 2022, 14, 1844–1855. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.Y.; Qiu, J.P.; Zhai, H.Z.; Wang, Y.Q.; Ma, P.P.; Li, M.Y.; Chen, B. Prognostic Implications of Novel Gene Signatures in Gastric Cancer Microenvironment. Med. Sci. Monit. 2020, 26, e924604. [Google Scholar] [CrossRef] [PubMed]
- Briggs, E.; de Kamps, M.; Hamilton, W.; Johnson, O.; McInerney, C.D.; Neal, R.D. Machine Learning for Risk Prediction of Oesophago-Gastric Cancer in Primary Care: Comparison with Existing Risk-Assessment Tools. Cancers 2022, 14, 5023. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.M.; Li, H.; Sun, D.Q.; Lei, L.; Ren, J.S.; Shi, J.F.; Li, N.; Peng, J.; Chen, W.Q. Classifying risk level of gastric cancer: Evaluation of questionnaire-based prediction model. Chin. J. Cancer Res. 2020, 32, 605–613. [Google Scholar] [CrossRef]
- Park, B.; Yang, S.; Lee, J.; Choi, I.J.; Kim, Y.-I.; Kim, J. Gastric Cancer Risk Prediction Using an Epidemiological Risk Assessment Model and Polygenic Risk Score. Cancers 2021, 13, 876. [Google Scholar] [CrossRef]
- In, H.; Langdon-Embry, M.; Gordon, L.; Schechter, C.B.; Wylie-Rosett, J.; Castle, P.E.; Margaret Kemeny, M.; Rapkin, B.D. Can a gastric cancer risk survey identify high-risk patients for endoscopic screening? A pilot study. J. Surg. Res. 2018, 227, 246–256. [Google Scholar] [CrossRef] [Green Version]
- Mahmoodi, S.A.; Mirzaie, K.; Mahmoodi, M.S.; Mahmoudi, S.M. A Medical Decision Support System to Assess Risk Factors for Gastric Cancer Based on Fuzzy Cognitive Map. Comput. Math. Methods Med. 2020, 2020, 1016284. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, D.; Zhang, W.; Hao, X.; Fang, M.; Wang, S.; Li, W.; Liu, Z.; Wang, R.; Zhou, J.; et al. A deep learning risk prediction model for overall survival in patients with gastric cancer: A multicenter study. Radiother. Oncol. 2020, 150, 73–80. [Google Scholar] [CrossRef]
- Kumar, S.; Huang, J.; Abbassi-Ghadi, N.; Mackenzie, H.A.; Veselkov, K.A.; Hoare, J.M.; Lovat, L.B.; Španel, P.; Smith, D.; Hanna, G.B. Mass Spectrometric Analysis of Exhaled Breath for the Identification of Volatile Organic Compound Biomarkers in Esophageal and Gastric Adenocarcinoma. Ann. Surg. 2015, 262, 981–990. [Google Scholar] [CrossRef]
- Yang, J.C.; Bo, L.M.; Han, T.; Ding, D.; Nie, M.M.; Yin, K. Pathway- and clinical-factor-based risk model predicts the prognosis of patients with gastric cancer. Mol. Med. Rep. 2018, 17, 6345–6356. [Google Scholar] [CrossRef]
- Li, T.D.; Chen, X.; Gu, M.L.; Deng, A.M.; Qian, C. Identification of the subtypes of gastric cancer based on DNA methylation and the prediction of prognosis. Clin. Epigenetics 2020, 12, 161. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.H.; Lagergren, J. A model for predicting individuals’ absolute risk of esophageal adenocarcinoma: Moving toward tailored screening and prevention. Int. J. Cancer 2016, 138, 2813–2819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, C.; Yin, F.X.; Liu, Y.P. Developing and validating nomograms for predicting the survival in patients with clinical local-advanced gastric cancer. Front. Oncol. 2022, 12, 1039498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.X.; Zhou, B.C.; Luo, P.Q.; Xu, A.; Han, W.X.; Wei, Z.J. A model established using marital status and other factors from the Surveillance, Epidemiology, and End Results database for early stage gastric cancer. J. Investig. Med. 2022, 70, 1373–1380. [Google Scholar] [CrossRef]
- Liu, Q.F.; Li, J.R.; Xin, B.W.; Sun, Y.Y.; Feng, D.G.; Fulham, M.J.J.; Wang, X.Y.; Song, S.L. F-18-FDG PET/CT Radiomics for Preoperative Prediction of Lymph Node Metastases and Nodal Staging in Gastric Cancer. Front. Oncol. 2021, 11, 723345. [Google Scholar] [CrossRef]
- Haga, Y.; Hato, S.; Ikenaga, M.; Yamamoto, K.; Tsuburaya, A.; Doi, K.; Ikejiri, K.; Hirata, T.; Yamamoto, M.; Ishikawa, S.; et al. Validation of an assessment tool: Estimation of Postoperative Overall Survival for Gastric Cancer. Eur. J. Surg. Oncol. 2018, 44, 515–523. [Google Scholar] [CrossRef]
- Gao, L.; Xue, J.; Liu, X.M.; Cao, L.; Wang, R.F.; Lei, L.L. A risk model based on autophagy-related lncRNAs for predicting prognosis and efficacy of immunotherapy and chemotherapy in gastric cancer patients. Aging 2021, 13, 25453–25465. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Z.F.; Zhou, L.S.; Wu, G.Z.; Ma, X.M.; Zheng, Y.; Liu, M.; Wang, Y.P.; Ji, R.; Guo, Q.H.; et al. In silico development and validation of a novel glucose and lipid metabolism-related gene signature in gastric cancer. Transl. Cancer Res. 2022, 11, 1977–1993. [Google Scholar] [CrossRef]
- Lee, T.Y.; Wang, C.B.; Chen, T.T.; Kuo, K.N.; Wu, M.S.; Lin, J.T.; Wu, C.Y.; Taiwan Gastrointestinal, D.; Heli. A Tool to Predict Risk for Gastric Cancer in Patients With Peptic Ulcer Disease on the Basis of a Nationwide Cohort. Clin. Gastroenterol. Hepatol. 2015, 13, 287-U108. [Google Scholar] [CrossRef]
- Liu, Y.F.; Wu, J.H.; Huang, W.W.; Weng, S.W.; Wang, B.C.; Chen, Y.M.; Wang, H. Development and validation of a hypoxia-immune-based microenvironment gene signature for risk stratification in gastric cancer. J. Transl. Med. 2020, 18, 201. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Xiao, K. Prediction of gastric cancer risk: Association between ZBTB20 genetic variance and gastric cancer risk in Chinese Han population. Biosci. Rep. 2020, 40, BSR20202102. [Google Scholar] [CrossRef]
- Wang, P.; Shi, B.; Wen, Y.; Tang, X. Establishment of Combination of Syndrome and Disease Risk Predicting Model for Precancerous Lesion of Gastric Cancer. Chin. J. Integr. Tradit. West. Med. 2018, 38, 773–778. [Google Scholar]
- Wang, X.; Chen, Y.; Gao, Y.; Zhang, H.; Guan, Z.; Dong, Z.; Zheng, Y.; Jiang, J.; Yang, H.; Wang, L.; et al. Predicting gastric cancer outcome from resected lymph node histopathology images using deep learning. Nat. Commun. 2021, 12, 1637. [Google Scholar] [CrossRef]
- D’Journo, X.B.; Boulate, D.; Fourdrain, A.; Loundou, A.; van Berge Henegouwen, M.I.; Gisbertz, S.S.; O’Neill, J.R.; Hoelscher, A.; Piessen, G.; van Lanschot, J.; et al. Risk Prediction Model of 90-Day Mortality After Esophagectomy for Cancer. JAMA Surg. 2021, 156, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Ma, Z.; Zhang, X.; Hang, D.; Yin, R.; Feng, J.; Xu, L.; Shen, H. C-reactive protein and cancer risk: A pan-cancer study of prospective cohort and Mendelian randomization analysis. BMC Med. 2022, 20, 301. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, B.; Zhou, B.; Chen, J.; Qi, J.; Shi, L.; Yu, S.; Chen, G.; Kang, M.; Jin, X.; et al. A novel immune-related lncRNA pair signature for prognostic prediction and immune response evaluation in gastric cancer: A bioinformatics and biological validation study. Cancer Cell Int. 2022, 22, 69. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Jiang, F.; Shen, Y.; Li, X.; Hu, X.; Wei, P.; Shen, X. Prognostic Prediction Using a Stemness Index-Related Signature in a Cohort of Gastric Cancer. Front. Mol. Biosci. 2020, 7, 570702. [Google Scholar] [CrossRef]
- Zheng, H.; Liu, H.; Li, H.; Dou, W.; Wang, X. Weighted Gene Co-expression Network Analysis Identifies a Cancer-Associated Fibroblast Signature for Predicting Prognosis and Therapeutic Responses in Gastric Cancer. Front. Mol. Biosci. 2021, 8, 744677. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, R.; Zhou, Y.; Wang, Q.; Yang, C.Y.; Hao, B.C.; Ke, C.F. Prediction of distant metastasis and survival prediction of gastric cancer patients with metastasis to the liver, lung, bone, and brain: Research based on the SEER database. Ann. Transl. Med. 2022, 10, 16. [Google Scholar] [CrossRef]
- Praud, D.; Rota, M.; Pelucchi, C.; Bertuccio, P.; Rosso, T.; Galeone, C.; Zhang, Z.F.; Matsuo, K.; Ito, H.; Hu, J.; et al. Cigarette smoking and gastric cancer in the Stomach Cancer Pooling (StoP) Project. Eur. J. Cancer Prev. 2018, 27, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Miceli, R.; Raimondi, A.; Kim, Y.W.; Kang, W.K.; Langley, R.E.; Choi, Y.Y.; Kim, K.M.; Nankivell, M.G.; Morano, F.; et al. Individual Patient Data Meta-Analysis of the Value of Microsatellite Instability as a Biomarker in Gastric Cancer. J. Clin. Oncol. 2019, 37, 3392–3400. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Guo, Y.; Li, Z.W.; Shui, G.; Tian, H.; Li, B.W.; Kadeerhan, G.; Li, Z.X.; Li, X.; Zhang, Y.; et al. Identification and Validation of Plasma Metabolomic Signatures in Precancerous Gastric Lesions That Progress to Cancer. JAMA Netw. Open 2021, 4, e2114186. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Lee, W.J.J.; Chua, S.J.; Zhu, F.; Yeoh, K.G.; Zhang, Y. Mucosal microbiome associates with progression to gastric cancer. Theranostics 2022, 12, 48–58. [Google Scholar] [CrossRef] [PubMed]
- van den Boorn, H.G.; Abu-Hanna, A.; Ter Veer, E.; van Kleef, J.J.; Lordick, F.; Stahl, M.; Ajani, J.A.; Guimbaud, R.; Park, S.H.; Dutton, S.J.; et al. SOURCE: A Registry-Based Prediction Model for Overall Survival in Patients with Metastatic Oesophageal or Gastric Cancer. Cancers 2019, 11, 187. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Chen, X.; Shen, Y.; Shen, X. Identification and Validation of an m6A Modification of JAK-STAT Signaling Pathway-Related Prognostic Prediction Model in Gastric Cancer. Front. Genet. 2022, 13, 891744. [Google Scholar] [CrossRef]
- Lei, L.; Li, N.; Yuan, P.; Liu, D. A new risk model based on a 11-m(6)A-related lncRNA signature for predicting prognosis and monitoring immunotherapy for gastric cancer. BMC Cancer 2022, 22, 365. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, C.; Liu, Y.; Zhao, Y.; Lin, D.; Hu, Y.; Yu, J.; Li, G. A gastric cancer LncRNAs model for MSI and survival prediction based on support vector machine. BMC Genom. 2019, 20, 846. [Google Scholar] [CrossRef] [Green Version]
- Guan, K.; Liu, X.; Li, J.; Ding, Y.; Li, J.; Cui, G.; Cui, X.; Sun, R. Expression Status And Prognostic Value Of M6A-associated Genes in Gastric Cancer. J. Cancer 2020, 11, 3027–3040. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Zhang, J.; Li, Z.; Wei, K.; Lei, Y.; Ren, J.; Wu, L.; Shi, Z.; Meng, X.; Yang, X.; et al. A CT-based deep learning radiomics nomogram for predicting the response to neoadjuvant chemotherapy in patients with locally advanced gastric cancer: A multicenter cohort study. EClinicalMedicine 2022, 46, 101348. [Google Scholar] [CrossRef]
- Lee, I.S.; Lee, H.; Hur, H.; Kanda, M.; Yook, J.H.; Kim, B.S.; Woo, Y.; Kodera, Y.; Kim, K.; Goel, A. Transcriptomic Profiling Identifies a Risk Stratification Signature for Predicting Peritoneal Recurrence and Micrometastasis in Gastric Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 2292–2300. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.H.; Hwang, J.E.; Jang, H.J.; Lee, H.S.; Oh, S.C.; Shim, J.J.; Lee, K.W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef] [Green Version]
- Cai, W.Y.; Dong, Z.N.; Fu, X.T.; Lin, L.Y.; Wang, L.; Ye, G.D.; Luo, Q.C.; Chen, Y.C. Identification of a Tumor Microenvironment-relevant Gene set-based Prognostic Signature and Related Therapy Targets in Gastric Cancer. Theranostics 2020, 10, 8633–8647. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wei, C.; Zhong, Y.; Zhang, Y.; Long, J.; Huang, S.; Xie, F.; Tian, Y.; Wang, X.; Zhao, H. Development and Validation of a Prognostic Nomogram for Gastric Cancer Based on DNA Methylation-Driven Differentially Expressed Genes. Int. J. Biol. Sci. 2020, 16, 1153–1165. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.G.; Hu, C.; Li, M.; Fan, Y.; Otter, N.; Sam, I.; Gou, H.; Hu, Y.; Kwok, T.; et al. Cell graph neural networks enable the precise prediction of patient survival in gastric cancer. NPJ Precis. Oncol. 2022, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, D.; Yu, R.; Li, C.; Song, Y.; Chen, X.; Fan, Y.; Liu, Y.; Qu, X. Immune Landscape of Gastric Carcinoma Tumor Microenvironment Identifies a Peritoneal Relapse Relevant Immune Signature. Front. Immunol. 2021, 12, 651033. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Zheng, T.; Yan, J.; Ma, J.; Lin, C.; Dong, S.; Wei, C.; Li, T.; Zhang, X.; Li, G. Identification of a 3-Gene Model as Prognostic Biomarker in Patients With Gastric Cancer. Front. Oncol. 2022, 12, 930586. [Google Scholar] [CrossRef]
- Zhou, L.; Li, S.H.; Wu, Y.; Xin, L. Establishment of a prognostic model of four genes in gastric cancer based on multiple data sets. Cancer Med. 2021, 10, 3309–3322. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, C.; Liu, Y. Establishing a cancer driver gene signature-based risk model for predicting the prognoses of gastric cancer patients. Aging 2022, 14, 2383–2399. [Google Scholar] [CrossRef]
- Feng, B.; Huang, L.; Liu, Y.; Chen, Y.; Zhou, H.; Yu, T.; Xue, H.; Chen, Q.; Zhou, T.; Kuang, Q.; et al. A Transfer Learning Radiomics Nomogram for Preoperative Prediction of Borrmann Type IV Gastric Cancer From Primary Gastric Lymphoma. Front. Oncol. 2021, 11, 802205. [Google Scholar] [CrossRef]
- Wang, S.; Ye, F.; Sheng, Y.; Yu, W.; Liu, Y.; Liu, D.; Zhang, K. Development and Validation of Nomograms to Predict Operative Link for Gastritis Assessment Any-Stage and Stages III-IV in the Chinese High-Risk Gastric Cancer Population. Front. Med. 2021, 8, 724566. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pu, K.; Li, C.; Wang, Y.; Zhou, Y. A Novel Six-Gene-Based Prognostic Model Predicts Survival and Clinical Risk Score for Gastric Cancer. Front. Genet. 2021, 12, 615834. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Zhang, W.; Che, Y.; Bai, W.; Huang, G. LASSO-based Cox-PH model identifies an 11-lncRNA signature for prognosis prediction in gastric cancer. Mol. Med. Rep. 2018, 18, 5579–5593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, L.; Qu, X.; He, J.; Cheng, L.; Zhang, R.; Sun, M.; Yang, Y.; Wang, J.; Wang, M.; Zhu, X.; et al. Predictive model for risk of gastric cancer using genetic variants from genome-wide association studies and high-evidence meta-analysis. Cancer Med. 2020, 9, 7310–7316. [Google Scholar] [CrossRef]
- Gao, B.; Feng, C.; Chai, F.; Wei, S.; Hong, N.; Ye, Y.; Wang, Y.; Cheng, J. CT-detected extramural venous invasion-related gene signature for the overall survival prediction in patients with gastric cancer. Cancer Med. 2021, 10, 7816–7830. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Yang, Q.; Wang, H.; Tan, M.; Zou, Y.; Liu, J. A predictive model for assessing prognostic risks in gastric cancer patients using gene expression and methylation data. BMC Med. Genom. 2021, 14, 14. [Google Scholar] [CrossRef]
- Han, Z.; Lan, J.; Wang, T.; Hu, Z.; Huang, Y.; Deng, Y.; Zhang, H.; Wang, J.; Chen, M.; Jiang, H.; et al. A Deep Learning Quantification Algorithm for HER2 Scoring of Gastric Cancer. Front. Neurosci. 2022, 16, 877229. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, C.; Xie, J.; Wang, W.; Zha, X.; Lv, W.; Chen, H.; Hu, Y.; Li, T.; Yu, J.; et al. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine 2018, 36, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Chen, H.; Han, Z.; Huang, W.; Hu, Y.; Zhao, M.; Lin, T.; Yu, J.; Liu, H.; Jiang, Y.; et al. Genomics Score Based on Genome-Wide Network Analysis for Prediction of Survival in Gastric Cancer: A Novel Prognostic Signature. Front. Genet. 2020, 11, 835. [Google Scholar] [CrossRef]
- Gu, C.; Xie, L.; Li, B.; Zhang, L.; Li, F.; Wang, W.; Su, J.; Xu, Z. Quantification of Tumor Abnormal Proteins in the Diagnosis and Postoperative Prognostic Evaluation of Gastric Cancer. Clin. Med. Insights Oncol. 2022, 16, 11795549221104440. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Lv, J.; Zhu, M.; Yan, C.W.; Deng, B.; Yu, C.Q.; Guo, Y.; Ni, J.; She, Q.; Wang, T.P.; et al. Development, validation, and evaluation of a risk assessment tool for personalized screening of gastric cancer in Chinese populations. BMC Med. 2023, 21, 159. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.D.; Epplein, M.; Lin, F.-C.; Troester, M.A.; Nichols, H.B.; Butt, J.; Pan, K.; You, W.; Olshan, A. Discrimination between Precancerous Gastric Lesions and Gastritis Using a Gastric Cancer Risk Stratification Model. Asian Pac. J. Cancer Prev. APJCP 2023, 24, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhang, Y. Development and validation of prognostic nomogram for young patients with gastric cancer. Ann. Transl. Med. 2019, 7, 641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, C. Development and validation of a Surveillance, Epidemiology, and End Results (SEER)-based prognostic nomogram for predicting survival in elderly patients with gastric cancer after surgery. J. Gastrointest. Oncol. 2021, 12, 278–296. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, W.; Gluth, A.; Hinz, U.; Koliogiannis, D.; Strobel, O.; Hackert, T.; Werner, J.; Büchler, M.W. Outcomes after extended pancreatectomy in patients with borderline resectable and locally advanced pancreatic cancer. Br. J. Surg. 2016, 103, 1683–1694. [Google Scholar] [CrossRef]
- Li, Z.; Lin, Y.; Cheng, B.; Zhang, Q.; Cai, Y. Prognostic Model for Predicting Overall and Cancer-Specific Survival Among Patients With Cervical Squamous Cell Carcinoma: A SEER Based Study. Front. Oncol. 2021, 11, 651975. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, S.; Cai, W.; Hong, Z.N.; Yang, C.; Lin, Y.; Zhu, J.; Lin, Z.; Christoph, D.C.; Bohnenberger, H.; et al. A nomogram to predict the recurrence-free survival and analyze the utility of chemotherapy in stage IB non-small cell lung cancer. Transl. Lung Cancer Res. 2022, 11, 75–86. [Google Scholar] [CrossRef]
- Zheng, Y.Z.; Qin, H.B.; Li, Z.Z.; Jiang, H.S.; Zhang, G.; Yang, S.W.; Wang, X.M.; Xu, Y.C.; Deng, Z.H.; Liu, G.W. Prognostic Factors and a Nomogram Predicting Survival in Patients with Breast Ductal Carcinoma in situ with Microinvasion: A Population-Based Study. Clin. Epidemiol. 2021, 13, 1095–1108. [Google Scholar] [CrossRef]
- Han, Y.; Wang, J.; Sun, Y.; Yu, P.; Yuan, P.; Ma, F.; Fan, Y.; Luo, Y.; Zhang, P.; Li, Q.; et al. Prognostic Model and Nomogram for Estimating Survival of Small Breast Cancer: A SEER-based Analysis. Clin. Breast Cancer 2021, 21, e497–e505. [Google Scholar] [CrossRef]
- Liang, Y.X.; Deng, J.Y.; Guo, H.H.; Ding, X.W.; Wang, X.N.; Wang, B.G.; Zhang, L.; Liang, H. Characteristics and prognosis of gastric cancer in patients aged ≥ 70 years. World J. Gastroenterol. 2013, 19, 6568–6578. [Google Scholar] [CrossRef]
- Crew, K.D.; Neugut, A.I. Epidemiology of gastric cancer. World J. Gastroenterol. 2006, 12, 354–362. [Google Scholar] [CrossRef]
- Karimi, P.; Islami, F.; Anandasabapathy, S.; Freedman, N.D.; Kamangar, F. Gastric Cancer: Descriptive Epidemiology, Risk Factors, Screening, and Prevention. Cancer Epidemiol. Biomark. Prev. 2014, 23, 700–713. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, Z.; Zhang, Z.; Wu, J.; Ren, D.; Yan, X.; Wang, Q.; Wang, Y.; Wang, H.; Zhang, J.; et al. A rising trend of gastric cardia cancer in Gansu Province of China. Cancer Lett. 2008, 269, 18–25. [Google Scholar] [CrossRef]
- Hu, K.; Wang, S.; Wang, Z.; Li, L.; Huang, Z.; Yu, W.; Chen, Z.; Wu, Q.-F. Clinicopathological risk factors for gastric cancer: A retrospective cohort study in China. BMJ Open 2019, 9, e030639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsu, Y.; Hiki, N.; Nunobe, S.; Ohashi, M.; Honda, M.; Yamaguchi, T.; Nakajima, T.; Sano, T. Clinicopathological features of gastric cancer in young patients. Gastric Cancer 2016, 19, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, J.; Shi, Z.; Shen, X.; Luo, T.; Bi, J.; Nie, M. Clinicopathologic characteristics and prognostic of gastric cancer in young patients. Scand. J. Gastroenterol. 2016, 51, 1043–1049. [Google Scholar] [CrossRef]
- Lui, F.H.; Tuan, B.; Swenson, S.L.; Wong, R.J. Ethnic disparities in gastric cancer incidence and survival in the USA: An updated analysis of 1992-2009 SEER data. Dig. Dis. Sci. 2014, 59, 3027–3034. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, V.W.; Andrews, P.A.; Ruiz, B.; Correa, P. Incidence of esophageal and gastric cancers among Hispanics, non-Hispanic whites and non-Hispanic blacks in the United States: Subsite and histology differences. Cancer Causes Control CCC 2007, 18, 585–593. [Google Scholar] [CrossRef]
- Kamineni, A.; Williams, M.A.; Schwartz, S.M.; Cook, L.S.; Weiss, N.S. The incidence of gastric carcinoma in Asian migrants to the United States and their descendants. Cancer Causes Control CCC 1999, 10, 77–83. [Google Scholar] [CrossRef]
- Lee, J.; Demissie, K.; Lu, S.E.; Rhoads, G.G. Cancer incidence among Korean-American immigrants in the United States and native Koreans in South Korea. Cancer Control J. Moffitt Cancer Cent. 2007, 14, 78–85. [Google Scholar] [CrossRef]
- Pinheiro, P.S.; Sherman, R.L.; Trapido, E.J.; Fleming, L.E.; Huang, Y.; Gomez-Marin, O.; Lee, D. Cancer incidence in first generation U.S. Hispanics: Cubans, Mexicans, Puerto Ricans, and new Latinos. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2162–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byfield, S.A.; Earle, C.C.; Ayanian, J.Z.; McCarthy, E.P. Treatment and outcomes of gastric cancer among United States-born and foreign-born Asians and Pacific Islanders. Cancer 2009, 115, 4595–4605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Koshiol, J.; Wei, W.Q.; Kreimer, A.R.; Ren, J.S.; Gravitt, P.; Chen, W.; Kim, E.; Abnet, C.C.; Zhang, Y.; Kamangar, F.; et al. The gastric cardia is not a target for human papillomavirus-induced carcinogenesis. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1137–1139. [Google Scholar] [CrossRef] [Green Version]
- Kamangar, F.; Dawsey, S.M.; Blaser, M.J.; Perez-Perez, G.I.; Pietinen, P.; Newschaffer, C.J.; Abnet, C.C.; Albanes, D.; Virtamo, J.; Taylor, P.R. Opposing risks of gastric cardia and noncardia gastric adenocarcinomas associated with Helicobacter pylori seropositivity. J. Natl. Cancer Inst. 2006, 98, 1445–1452. [Google Scholar] [CrossRef] [Green Version]
- Bakhti, S.Z.; Latifi-Navid, S.; Safaralizadeh, R. Helicobacter pylori-related risk predictors of gastric cancer: The latest models, challenges, and future prospects. Cancer Med. 2020, 9, 4808–4822. [Google Scholar] [CrossRef]
- Yang, L.; Kartsonaki, C.; Yao, P.; de Martel, C.; Plummer, M.; Chapman, D.; Guo, Y.; Clark, S.; Walters, R.G.; Chen, Y.; et al. The relative and attributable risks of cardia and non-cardia gastric cancer associated with Helicobacter pylori infection in China: A case-cohort study. Lancet. Public Health 2021, 6, e888–e896. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zhao, Y.; Shan, Z.; Zhang, J. CA724 predicts overall survival in locally advanced gastric cancer patients with neoadjuvant chemotherapy. BMC Cancer 2021, 21, 4. [Google Scholar] [CrossRef]
- Reim, D.; Choi, Y.S.; Yoon, H.M.; Park, B.; Eom, B.W.; Kook, M.C.; Ryu, K.W.; Choi, I.J.; Joo, J.; Kim, Y.W. Alpha-fetoprotein is a significant prognostic factor for gastric cancer: Results from a propensity score matching analysis after curative resection. Eur. J. Surg. Oncol. EJSO 2017, 43, 1542–1549. [Google Scholar] [CrossRef]
- Xiao, J.; Ye, Z.S.; Wei, S.H.; Zeng, Y.; Lin, Z.M.; Wang, Y.; Teng, W.H.; Chen, L.C. Prognostic significance of pretreatment serum carcinoembryonic antigen levels in gastric cancer with pathological lymph node-negative: A large sample single-center retrospective study. World J. Gastroenterol. 2017, 23, 8562–8569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokunaga, R.; Imamura, Y.; Nakamura, K.; Uchihara, T.; Ishimoto, T.; Nakagawa, S.; Iwatsuki, M.; Baba, Y.; Sakamoto, Y.; Miyamoto, Y.; et al. Carbohydrate antigen 19-9 is a useful prognostic marker in esophagogastric junction adenocarcinoma. Cancer Med. 2015, 4, 1659–1666. [Google Scholar] [CrossRef] [PubMed]
- Emoto, S.; Ishigami, H.; Yamashita, H.; Yamaguchi, H.; Kaisaki, S.; Kitayama, J. Clinical significance of CA125 and CA72-4 in gastric cancer with peritoneal dissemination. Gastric Cancer 2012, 15, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Noie, T.; Ohashi, M.; Oba, K.; Takahashi, Y. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2014, 17, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Tian, Y.; Xu, G.; Liu, Z.; Liu, S.; Zheng, G.; Guo, M.; Lian, X.; Fan, D.; Zhang, H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 2017, 17, 737. [Google Scholar] [CrossRef]
- Zhao, S.; Bi, Y.; Wang, Z.; Zhang, F.; Zhang, Y.; Xu, Y. Accuracy evaluation of combining gastroscopy, multi-slice spiral CT, Her-2, and tumor markers in gastric cancer staging diagnosis. World J. Surg. Oncol. 2022, 20, 152. [Google Scholar] [CrossRef]
- In, H.; Sarkar, S.; Ward, J.; Friedmann, P.; Parides, M.; Yang, J.; Epplein, M. Serum Pepsinogen as a Biomarker for Gastric Cancer in the United States: A Nested Case-Control Study Using the PLCO Cancer Screening Trial Data. Cancer Epidemiol. Biomark. Prev. 2022, 31, 1426–1432. [Google Scholar] [CrossRef]
- Agkoc, M.; Dursun, H.; Albayrak, F.; Yilmaz, O.; Kiziltunc, A.; Yilmaz, A.; Gundogdu, C. Usefulness of serum pepsinogen levels as a screening test for atrophic gastritis and gastric cancer. Eurasian J. Med. 2010, 42, 15–18. [Google Scholar] [CrossRef]
- Miki, K. Gastric cancer screening by combined assay for serum anti-Helicobacter pylori IgG antibody and serum pepsinogen levels—”ABC method”. Proc. Jpn. Academy. Ser. B Phys. Biol. Sci. 2011, 87, 405–414. [Google Scholar] [CrossRef]
- Wang, R.; Chen, X.Z. Prevalence of atrophic gastritis in southwest China and predictive strength of serum gastrin-17: A cross-sectional study (SIGES). Sci. Rep. 2020, 10, 4523. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Liu, Z.; Zhang, K. Pepsinogen Serology and Gastritis OLGA Staging in Mucosal Atrophy Assessment: A Cross-Sectional Study Involving East China Endoscopy Population. Gastroenterol. Res. Pract. 2020, 2020, 2324505. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Wu, Y.; Song, Z.; Yu, Y.; Yu, X. The potential value of serum pepsinogen for the diagnosis of atrophic gastritis among the health check-up populations in China: A diagnostic clinical research. BMC Gastroenterol. 2017, 17, 88. [Google Scholar] [CrossRef]
- Kabat, G.C.; Ng, S.K.; Wynder, E.L. Tobacco, alcohol intake, and diet in relation to adenocarcinoma of the esophagus and gastric cardia. Cancer Causes Control CCC 1993, 4, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Ekström, A.M.; Hansson, L.E.; Bergström, R.; Nyrén, O. Tobacco, alcohol and the risk of gastric cancer by sub-site and histologic type. Int. J. Cancer 1999, 83, 223–229. [Google Scholar] [CrossRef]
- Freedman, N.D.; Abnet, C.C.; Leitzmann, M.F.; Mouw, T.; Subar, A.F.; Hollenbeck, A.R.; Schatzkin, A. A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. Am. J. Epidemiol. 2007, 165, 1424–1433. [Google Scholar] [CrossRef]
- Tramacere, I.; Pelucchi, C.; Bagnardi, V.; Rota, M.; Scotti, L.; Islami, F.; Corrao, G.; Boffetta, P.; La Vecchia, C.; Negri, E. A meta-analysis on alcohol drinking and esophageal and gastric cardia adenocarcinoma risk. Ann. Oncol. 2012, 23, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Hansson, L.E.; Baron, J.; Nyrén, O.; Bergström, R.; Wolk, A.; Adami, H.O. Tobacco, alcohol and the risk of gastric cancer. A population-based case-control study in Sweden. Int. J. Cancer 1994, 57, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Maguire, F.B.; Movsisyan, A.S.; Morris, C.R.; Parikh-Patel, A.; Keegan, T.H.M.; Tong, E.K. Evaluation of Cancer Deaths Attributable to Tobacco in California, 2014-2019. JAMA Netw. Open 2022, 5, e2246651. [Google Scholar] [CrossRef]
- Shikata, K.; Doi, Y.; Yonemoto, K.; Arima, H.; Ninomiya, T.; Kubo, M.; Tanizaki, Y.; Matsumoto, T.; Iida, M.; Kiyohara, Y. Population-based prospective study of the combined influence of cigarette smoking and Helicobacter pylori infection on gastric cancer incidence: The Hisayama Study. Am. J. Epidemiol. 2008, 168, 1409–1415. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349.e315. [Google Scholar] [CrossRef]
- Camilleri, M.; Malhi, H.; Acosta, A. Gastrointestinal Complications of Obesity. Gastroenterology 2017, 152, 1656–1670. [Google Scholar] [CrossRef] [Green Version]
- Mukaisho, K.; Nakayama, T.; Hagiwara, T.; Hattori, T.; Sugihara, H. Two distinct etiologies of gastric cardia adenocarcinoma: Interactions among pH, Helicobacter pylori, and bile acids. Front. Microbiol. 2015, 6, 412. [Google Scholar] [CrossRef] [Green Version]
- Poorolajal, J.; Moradi, L.; Mohammadi, Y.; Cheraghi, Z.; Gohari-Ensaf, F. Risk factors for stomach cancer: A systematic review and meta-analysis. Epidemiol. Health 2020, 42, e2020004. [Google Scholar] [CrossRef] [PubMed]
- Naemi Kermanshahi, M.; Safaei, E.; Tutunchi, H.; Naghshi, S.; Mobarak, S.; Asadi, M.; Sadeghi, O. Fruit and vegetable intake in relation to gastric cancer risk: A comprehensive and updated systematic review and dose-response meta-analysis of cohort studies. Front. Nutr. 2023, 10, 973171. [Google Scholar] [CrossRef]
- Kurosawa, M.; Kikuchi, S.; Xu, J.; Inaba, Y. Highly salted food and mountain herbs elevate the risk for stomach cancer death in a rural area of Japan. J. Gastroenterol. Hepatol. 2006, 21, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- D’Elia, L.; Rossi, G.; Ippolito, R.; Cappuccio, F.P.; Strazzullo, P. Habitual salt intake and risk of gastric cancer: A meta-analysis of prospective studies. Clin. Nutr. 2012, 31, 489–498. [Google Scholar] [CrossRef]
- Wu, B.; Yang, D.; Yang, S.; Zhang, G. Dietary Salt Intake and Gastric Cancer Risk: A Systematic Review and Meta-Analysis. Front. Nutr. 2021, 8, 801228. [Google Scholar] [CrossRef]
- Tsugane, S. Salt, salted food intake, and risk of gastric cancer: Epidemiologic evidence. Cancer Sci. 2005, 96, 1–6. [Google Scholar] [CrossRef]
- Wong, B.C.; Lam, S.K. Epidemiology of gastric cancer in relation to diet and Helicobacter pylori infection. J. Gastroenterol. Hepatol. 1998, 13, S166–S172. [Google Scholar] [CrossRef] [PubMed]
- Yaghoobi, M.; Bijarchi, R.; Narod, S.A. Family history and the risk of gastric cancer. Br. J. Cancer 2010, 102, 237–242. [Google Scholar] [CrossRef]
- Lauwers, G.Y.; Mullen, J.T.; Chelcun Schreiber, K.E.; Chung, D.C. Familial Gastric Cancers: A Review with Focus on Hereditary Diffuse Gastric Cancer Syndrome. AJSP Rev. Rep. 2014, 19, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef]
- Correa, P. Human gastric carcinogenesis: A multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar] [PubMed]
- Correa, P.; Shiao, Y.H. Phenotypic and genotypic events in gastric carcinogenesis. Cancer Res. 1994, 54, 1941s–1943s. [Google Scholar] [PubMed]
- Tang, L.; Tang, B.; Lei, Y.; Yang, M.; Wang, S.; Hu, S.; Xie, Z.; Liu, Y.; Vlodavsky, I.; Yang, S. Helicobacter pylori-Induced Heparanase Promotes H. pylori Colonization and Gastritis. Front. Immunol. 2021, 12, 675747. [Google Scholar] [CrossRef] [PubMed]
- Toh, J.W.T.; Wilson, R.B. Pathways of Gastric Carcinogenesis, Helicobacter pylori Virulence and Interactions with Antioxidant Systems, Vitamin C and Phytochemicals. Int. J. Mol. Sci. 2020, 21, 6451. [Google Scholar] [CrossRef]
- Kinoshita, H.; Hayakawa, Y.; Koike, K. Metaplasia in the Stomach-Precursor of Gastric Cancer? Int. J. Mol. Sci. 2017, 18, 2063. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Wu, A.H.; Tseng, C.C.; Bernstein, L. Hiatal hernia, reflux symptoms, body size, and risk of esophageal and gastric adenocarcinoma. Cancer 2003, 98, 940–948. [Google Scholar] [CrossRef]
- Ye, W.; Chow, W.H.; Lagergren, J.; Yin, L.; Nyrén, O. Risk of adenocarcinomas of the esophagus and gastric cardia in patients with gastroesophageal reflux diseases and after antireflux surgery. Gastroenterology 2001, 121, 1286–1293. [Google Scholar] [CrossRef]
- Yuan, Q.; Deng, D.; Pan, C.; Ren, J.; Wei, T.; Wu, Z.; Zhang, B.; Li, S.; Yin, P.; Shang, D. Integration of transcriptomics, proteomics, and metabolomics data to reveal HER2-associated metabolic heterogeneity in gastric cancer with response to immunotherapy and neoadjuvant chemotherapy. Front. Immunol. 2022, 13, 951137. [Google Scholar] [CrossRef] [PubMed]
- Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.N.; Lambin, P.; Bottari, F.; Tsoutzidis, N.; Miraglio, B.; et al. A review in radiomics: Making personalized medicine a reality via routine imaging. Med. Res. Rev. 2022, 42, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019, 9, 1303–1322. [Google Scholar] [CrossRef]
- Zheng, B.H.; Liu, L.Z.; Zhang, Z.Z.; Shi, J.Y.; Dong, L.Q.; Tian, L.Y.; Ding, Z.B.; Ji, Y.; Rao, S.X.; Zhou, J.; et al. Radiomics score: A potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer 2018, 18, 1148. [Google Scholar] [CrossRef]
- Ba, W.; Wang, S.; Shang, M.; Zhang, Z.; Wu, H.; Yu, C.; Xing, R.; Wang, W.; Wang, L.; Liu, C.; et al. Assessment of deep learning assistance for the pathological diagnosis of gastric cancer. Mod. Pathol. 2022, 35, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Awad, E.; Levine, S.; Anderson, M.; Anderson, S.L.; Conitzer, V.; Crockett, M.J.; Everett, J.A.C.; Evgeniou, T.; Gopnik, A.; Jamison, J.C.; et al. Computational ethics. Trends Cogn. Sci. 2022, 26, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Hall, M.A. Correlation-Based Feature Selection for Machine Learning; Morgan Kaufmann Publishers Inc.: Burlington, MA, USA, 2000. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B-Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Farsal, W.; Anter, S.; Ramdani, M. Deep Learning: An Overview. In Proceedings of the 12th International Conference on Intelligent Systems: Theories and Applications, Rabat, Morocco, 24–25 October 2018; p. 38. [Google Scholar]
- Chen, W.; Wang, X.; Duan, H.; Zhang, X.; Dong, T.; Nie, S. Application of deep learning in cancer prognosis prediction model. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi = J. Biomed. Eng. = Shengwu Yixue Gongchengxue Zazhi 2020, 37, 918–929. [Google Scholar] [CrossRef]
- Wen, F.; Huang, J.; Lu, X.; Huang, W.; Wang, Y.; Bai, Y.; Ruan, S.; Gu, S.; Chen, X.; Shu, P. Identification and prognostic value of metabolism-related genes in gastric cancer. Aging 2020, 12, 17647–17661. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Taninaga, J.; Nishiyama, Y.; Fujibayashi, K.; Gunji, T.; Sasabe, N.; Iijima, K.; Naito, T. Prediction of future gastric cancer risk using a machine learning algorithm and comprehensive medical check-up data: A case-control study. Sci. Rep. 2019, 9, 12384. [Google Scholar] [CrossRef] [Green Version]
- Basheer, I.A.; Hajmeer, M. Artificial neural networks: Fundamentals, computing, design, and application. J. Microbiol. Methods 2000, 43, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Zhang, J. PalmHash Code vs. PalmPhasor Code. Neurocomputing 2013, 108, 1–12. [Google Scholar] [CrossRef]
- Leng, L.; Li, M.; Kim, C.; Bi, X. Dual-source discrimination power analysis for multi-instance contactless palmprint recognition. Multimed. Tools Appl. 2017, 76, 333–354. [Google Scholar] [CrossRef]
- Cantarero, G.G.; Jarabo, R.M. The area under the ROC curve. Med. Clin. 1996, 106, 355–356. [Google Scholar]
- Bradley, A.P. The use of the area under the roc curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef] [Green Version]
- Hong, C. Confusion plot for the confusion matrix. J. Korean Data Inf. Sci. Sociaty 2021, 32, 427–437. [Google Scholar] [CrossRef]
- Vickers, A.J. Prediction models: Revolutionary in principle, but do they do more good than harm? J. Clin. Oncol. 2011, 29, 2951–2952. [Google Scholar] [CrossRef]



| The Relationship between Each Factor and GC | ||
|---|---|---|
| Demographic characteristics | Age | GC is highly correlated with age with a gradually increasing incidence observed after 40 years. |
| Sex | Men are twice women in both incidence cases and mortality cases. | |
| Race | The incidence of GC varies greatly among different races. | |
| Clinical indicators | Hp infection | Chronic Hp infection is considered the leading cause of NCGC but is not associated with CGC. |
| Serum tumor markers | Serum tumor markers are relatively high in advanced GC. | |
| Pepsinogen | PG is a marker of atrophic gastritis, associated with GC indirectly. | |
| Lifestyle habits | Drink alcohol | Drinking alcohol has been identified as one of the CGC risk factors. |
| Smoke | The duration of cigarette or pipe smoking is positively associated with GC risk. | |
| BMI | Some studies have linked GC to obesity, but others disagree. | |
| Diet | A higher intake of total fruit and vegetable are associated with a lower risk of GC. Fried/pickled/smoked and high-salt diet may increase the risk of GC. | |
| Genetic factors | Family history | 10% of GC cases show familial aggregation. |
| Concurrent/Historical disease | Some diseases are closely related to the development of GC but need to be considered along with other factors. | |
| Omics data (excluding radiomics) | The novel-integrated multi-omics strategy may facilitate the development of a more tailored approach to GC therapy. | |
| Radiomics | Radiomics has certain intuition and high accuracy in the examination of GC. | |
| Included Factors | Quantity |
|---|---|
| Demographic characteristics | |
| Age | 51 |
| Gender | 44 |
| Race | 6 |
| Clinical indicators | |
| Helicobacter pylori | 12 |
| Tumor markers | 6 |
| Pepsinogen (PG I/PG II/PGR) | 5 |
| G-17 | 2 |
| Hemoglobin | 2 |
| Lifestyle habits | |
| Drink alcohol | 11 |
| Smoke | 14 |
| Body weight/Body Mass Index (BMI) | 12 |
| Exercise | 4 |
| Diet | |
| Intake of fruits and vegetables | 6 |
| Fried/pickled/smoked | 4 |
| High-salt diet | 6 |
| Regularity and speed of eating Genetic factors | 3 |
| Family history of GC | 12 |
| Concurrent/Historical disease | |
| Atrophic gastritis | 6 |
| Gastric ulcer | 3 |
| Dyspepsia | 3 |
| Omics data (excluding radiomics) | 32 |
| Radiomics | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Z.; He, Z.; Miao, W.; Huang, R. Critical Analysis of Risk Factors and Machine-Learning-Based Gastric Cancer Risk Prediction Models: A Systematic Review. Processes 2023, 11, 2324. https://doi.org/10.3390/pr11082324
Fan Z, He Z, Miao W, Huang R. Critical Analysis of Risk Factors and Machine-Learning-Based Gastric Cancer Risk Prediction Models: A Systematic Review. Processes. 2023; 11(8):2324. https://doi.org/10.3390/pr11082324
Chicago/Turabian StyleFan, Zeyu, Ziju He, Wenjun Miao, and Rongrong Huang. 2023. "Critical Analysis of Risk Factors and Machine-Learning-Based Gastric Cancer Risk Prediction Models: A Systematic Review" Processes 11, no. 8: 2324. https://doi.org/10.3390/pr11082324





