Adversarial Training Collaborating Multi-Path Context Feature Aggregation Network for Maize Disease Density Prediction
Abstract
1. Introduction
- (1)
- We employ a multi-scale patch embedding module to extract multi-scale features from various types of maize images using multi-scale convolution with overlapping parts—thus adapting to different maize disease characteristics.
- (2)
- Our proposed multi-path context feature aggregation module uses a depth convolution and Transformer encoder to further extract detailed features and long-range features, and allows these two to interact in the same dimension in order for the multi-scale features to effectively improve the network’s ability to characterize features.
- (3)
- We use the adversarial training method to generate adversarial samples by adding noise perturbations to the input maize images; this disrupts the normal training of the network—thus improving the robustness of the network.
2. Materials and Methods
2.1. Dataset
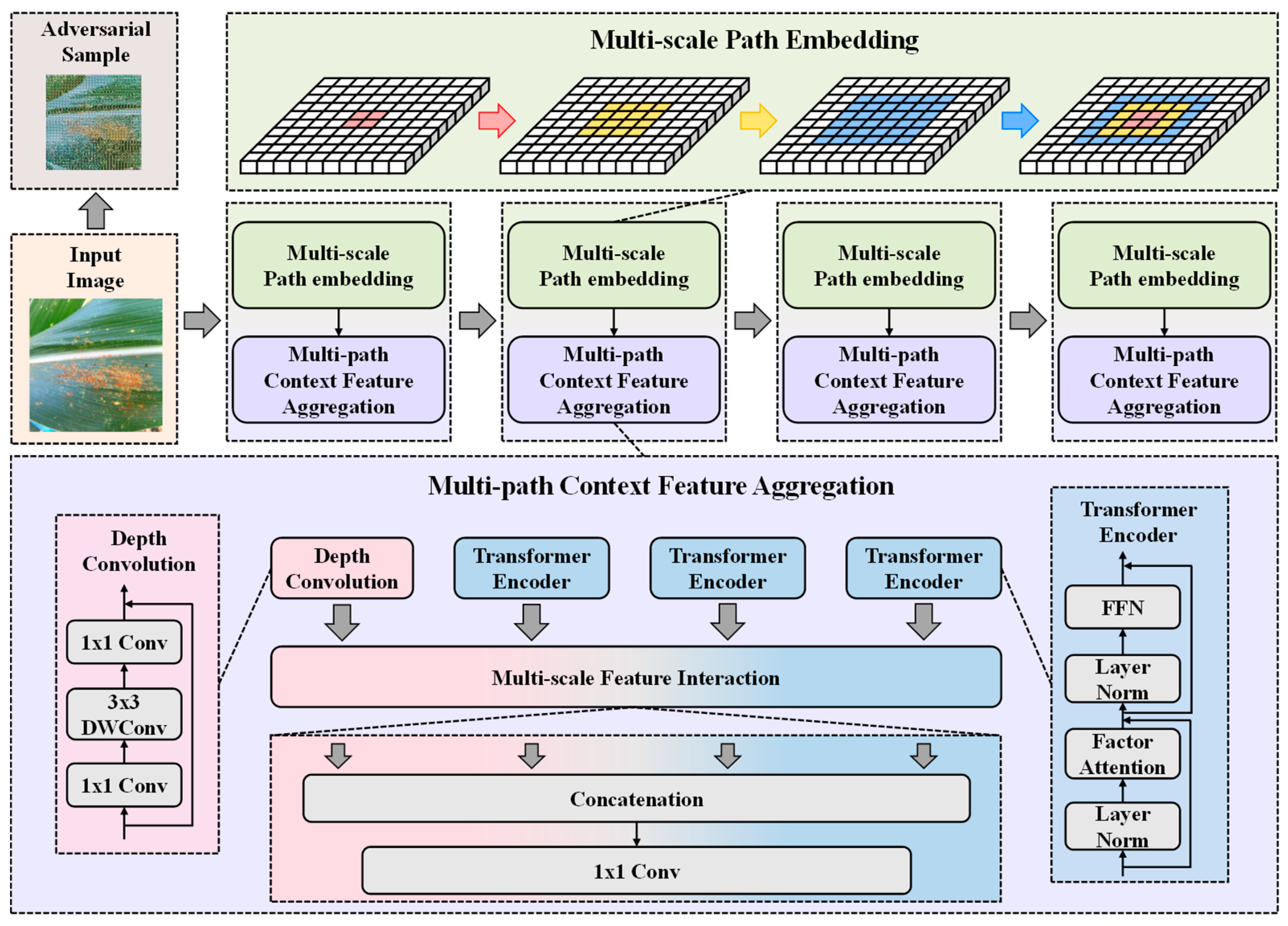
2.2. Overview of Network
2.3. Multi-Scale Patch Embedding
2.4. Multi-Path Context-Feature Aggregation
2.5. Loss Function
3. Experiments and Results
3.1. Experimental Settings
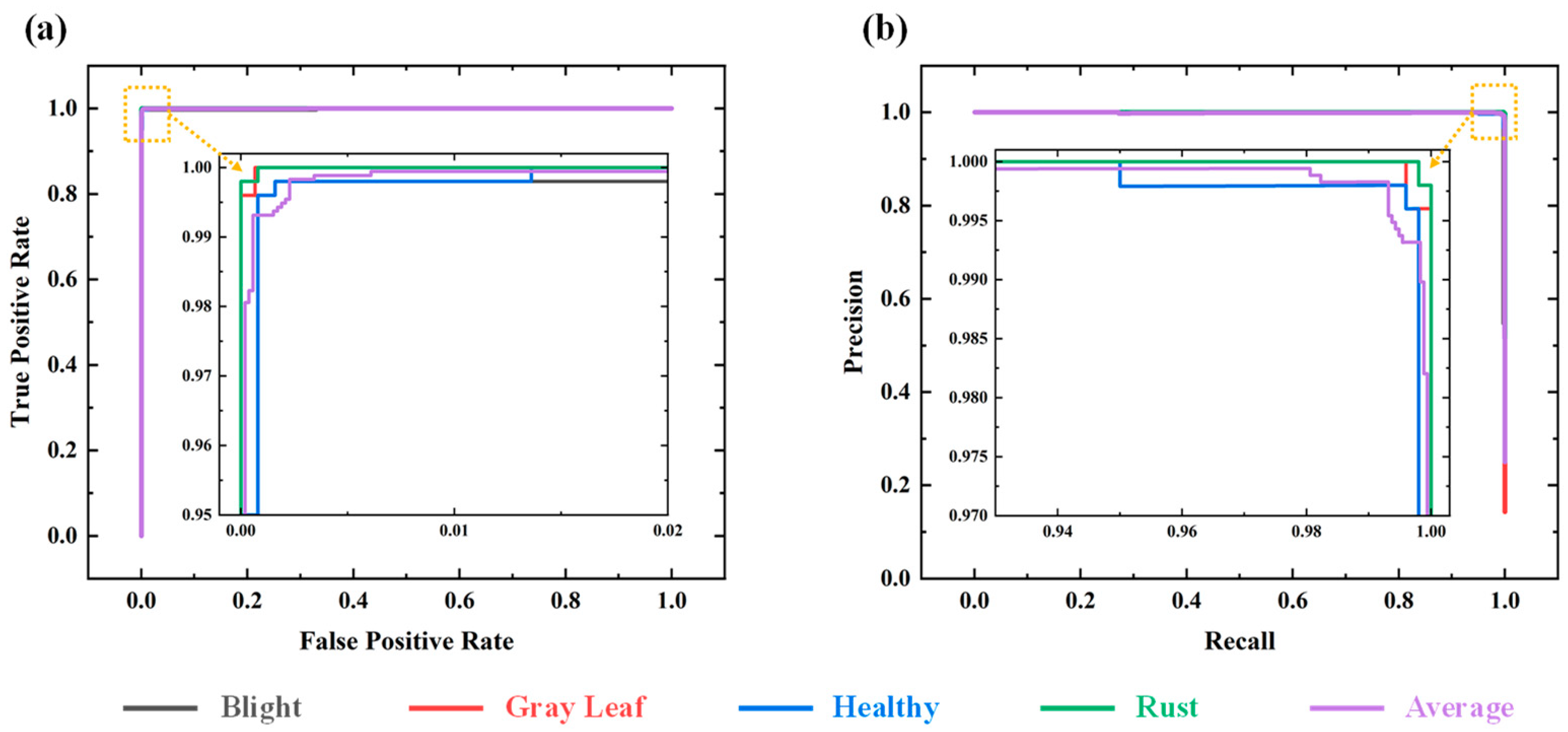
3.2. Evaluation Metrics
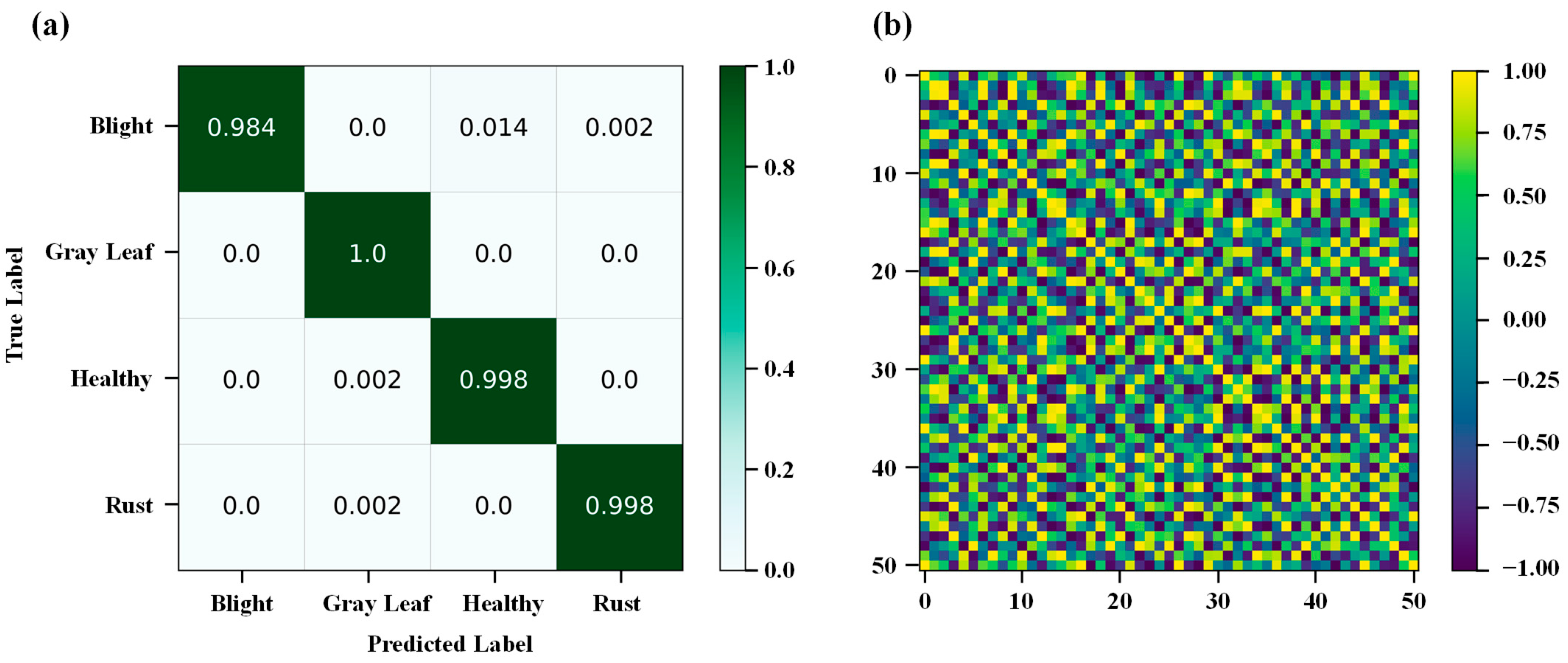
3.3. Quantitative Analysis
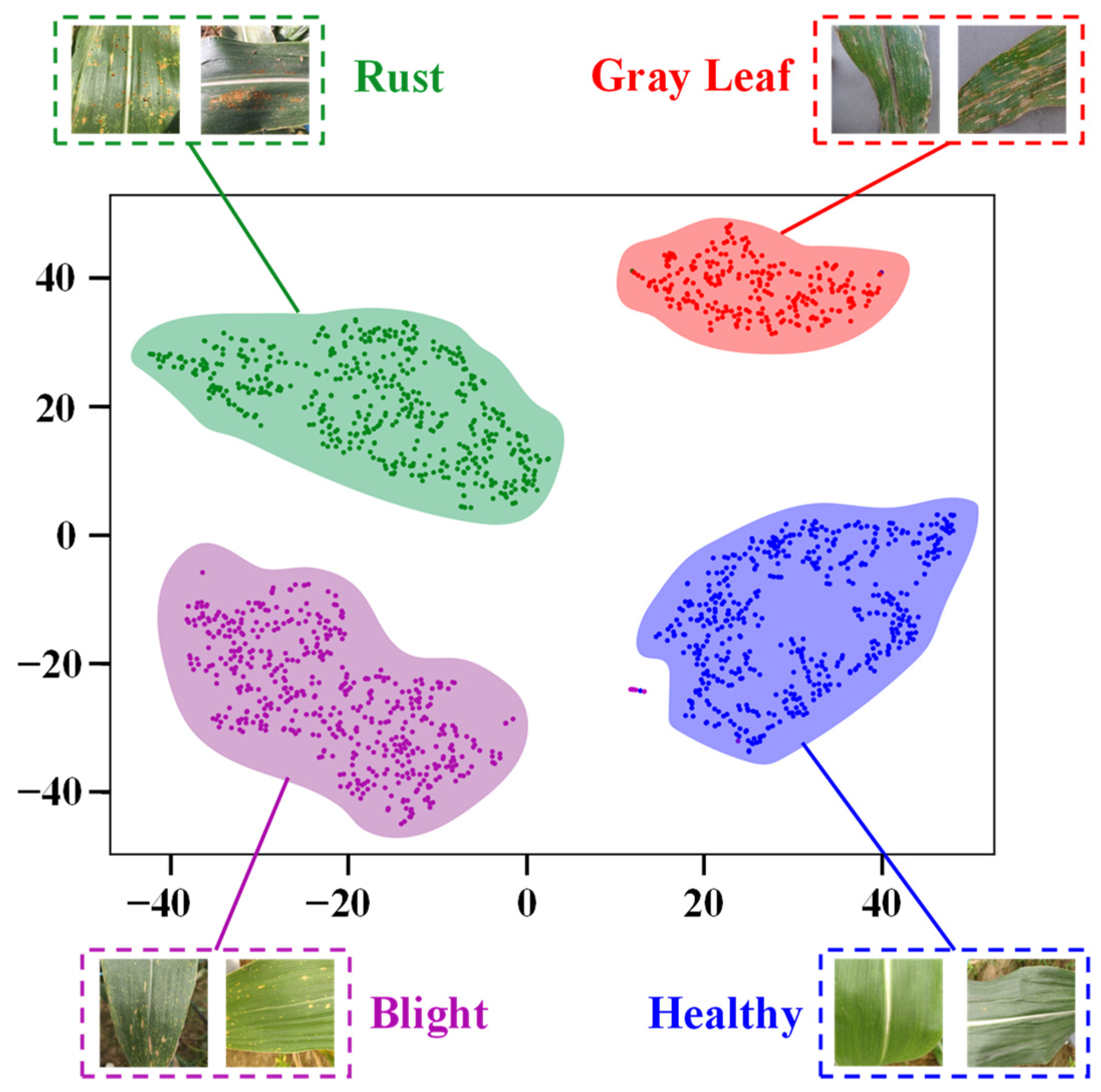
3.4. Interpretability Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Upadhyay, M.K.; Shukla, A.; Yadav, P.; Srivastava, S. A review of arsenic in crops, vegetables, animals and food products. Food Chem. 2019, 276, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Liu, Y.; Liu, W.; Liu, G.; Xie, R.; Wang, K.; Ming, B.; Wang, Y.; Zhao, R.; Zhang, W. How to increase maize production without extra nitrogen input. Resour. Conserv. Recycl. 2020, 160, 104913. [Google Scholar] [CrossRef]
- Adisa, O.M.; Botai, J.O.; Adeola, A.M.; Hassen, A.; Botai, C.M.; Darkey, D.; Tesfamariam, E. Application of artificial neural network for predicting maize production in South Africa. Sustainability 2019, 11, 1145. [Google Scholar] [CrossRef]
- Kaur, N.; Vashist, K.K.; Brar, A. Energy and productivity analysis of maize based crop sequences compared to rice-wheat system under different moisture regimes. Energy 2021, 216, 119286. [Google Scholar] [CrossRef]
- Letsoin, S.M.A.; Purwestri, R.C.; Perdana, M.C.; Hnizdil, P.; Herak, D. Monitoring of Paddy and Maize Fields Using Sentinel-1 SAR Data and NGB Images: A Case Study in Papua, Indonesia. Processes 2023, 11, 647. [Google Scholar] [CrossRef]
- Zhou, L.; Gu, X.; Cheng, S.; Yang, G.; Shu, M.; Sun, Q. Analysis of plant height changes of lodged maize using UAV-LiDAR data. Agriculture 2020, 10, 146. [Google Scholar] [CrossRef]
- Zhang, Y.; Wa, S.; Liu, Y.; Zhou, X.; Sun, P.; Ma, Q. High-accuracy detection of maize leaf diseases CNN based on multi-pathway activation function module. Remote Sens. 2021, 13, 4218. [Google Scholar] [CrossRef]
- Arora, J.; Agrawal, U. Classification of Maize leaf diseases from healthy leaves using Deep Forest. J. Artif. Intell. Syst. 2020, 2, 14–26. [Google Scholar] [CrossRef]
- Ali, I.; HUO, X.-x.; Khan, I.; Ali, H.; Khan, B.; Khan, S.U. Technical efficiency of hybrid maize growers: A stochastic frontier model approach. J. Integr. Agric. 2019, 18, 2408–2421. [Google Scholar] [CrossRef]
- Alemu, G.T.; Nigussie, Z.; Haregeweyn, N.; Berhanie, Z.; Wondimagegnehu, B.A.; Ayalew, Z.; Molla, D.; Okoyo, E.N.; Baributsa, D. Cost-benefit analysis of on-farm grain storage hermetic bags among small-scale maize growers in northwestern Ethiopia. Crop Prot. 2021, 143, 105478. [Google Scholar] [CrossRef]
- Lv, M.; Zhou, G.; He, M.; Chen, A.; Zhang, W.; Hu, Y. Maize leaf disease identification based on feature enhancement and DMS-robust alexnet. IEEE Access 2020, 8, 57952–57966. [Google Scholar] [CrossRef]
- Waldamichael, F.G.; Debelee, T.G.; Schwenker, F.; Ayano, Y.M.; Kebede, S.R. Machine learning in cereal crops disease detection: A review. Algorithms 2022, 15, 75. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Attention embedded lightweight network for maize disease recognition. Plant Pathol. 2021, 70, 630–642. [Google Scholar] [CrossRef]
- Orchi, H.; Sadik, M.; Khaldoun, M. On using artificial intelligence and the internet of things for crop disease detection: A contemporary survey. Agriculture 2022, 12, 9. [Google Scholar] [CrossRef]
- Fenu, G.; Malloci, F.M. Forecasting plant and crop disease: An explorative study on current algorithms. Big Data Cogn. Comput. 2021, 5, 2. [Google Scholar] [CrossRef]
- Zhang, Z.; He, X.; Sun, X.; Guo, L.; Wang, J.; Wang, F. Image recognition of maize leaf disease based on GA-SVM. Chem. Eng. Trans. 2015, 46, 199–204. [Google Scholar]
- Aravind, K.; Raja, P.; Mukesh, K.; Aniirudh, R.; Ashiwin, R.; Szczepanski, C. Disease classification in maize crop using bag of features and multiclass support vector machine. In Proceedings of the 2nd International Conference on Inventive Systems and Control, Coimbatore, India, 19–20 January 2018; pp. 1191–1196. [Google Scholar]
- Zhang, S.; Shang, Y.; Wang, L. Plant disease recognition based on plant leaf image. J. Anim. Plant Sci. 2015, 25, 42–45. [Google Scholar]
- Alehegn, E. Ethiopian maize diseases recognition and classification using support vector machine. Int. J. Comput. Vis. Robot. 2019, 9, 90–109. [Google Scholar] [CrossRef]
- Waheed, A.; Goyal, M.; Gupta, D.; Khanna, A.; Hassanien, A.E.; Pandey, H.M. An optimized dense convolutional neural network model for disease recognition and classification in corn leaf. Comput. Electron. Agric. 2020, 175, 105456. [Google Scholar] [CrossRef]
- Gui, P.; Dang, W.; Zhu, F.; Zhao, Q. Towards automatic field plant disease recognition. Comput. Electron. Agric. 2021, 191, 106523. [Google Scholar] [CrossRef]
- Qian, X.; Zhang, C.; Chen, L.; Li, K. Deep learning-based identification of maize leaf diseases is improved by an attention mechanism: Self-Attention. Front. Plant Sci. 2022, 13, 864486. [Google Scholar] [CrossRef] [PubMed]
- DeChant, C.; Wiesner-Hanks, T.; Chen, S.; Stewart, E.L.; Yosinski, J.; Gore, M.A.; Nelson, R.J.; Lipson, H. Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology 2017, 107, 1426–1432. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, B.; Zhai, Y.; Chen, Q.; Zhou, Y. Maize diseases identification method based on multi-scale convolutional global pooling neural network. IEEE Access 2021, 9, 27959–27970. [Google Scholar] [CrossRef]
- Ahila Priyadharshini, R.; Arivazhagan, S.; Arun, M.; Mirnalini, A. Maize leaf disease classification using deep convolutional neural networks. Neural Comput. Appl. 2019, 31, 8887–8895. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, J.; Willette, J.; Hwang, S.J. Mpvit: Multi-path vision transformer for dense prediction. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, Louisiana, USA, 19–24 June 2022; pp. 7287–7296. [Google Scholar]
- Aregbesola, E.; Ortega-Beltran, A.; Falade, T.; Jonathan, G.; Hearne, S.; Bandyopadhyay, R. A detached leaf assay to rapidly screen for resistance of maize to Bipolaris maydis, the causal agent of southern corn leaf blight. Eur. J. Plant Pathol. 2020, 156, 133–145. [Google Scholar] [CrossRef]
- Saito, B.C.; Silva, L.Q.; Andrade, J.A.d.C.; Goodman, M.M. Adaptability and stability of corn inbred lines regarding resistance to gray leaf spot and northern leaf blight. Crop Breed. Appl. Biotechnol. 2018, 18, 148–154. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Z.; Tian, L.; Ding, Y.; Zhang, J.; Zhou, J.; Liu, P.; Chen, Y.; Wu, L. Comparative proteomics combined with analyses of transgenic plants reveal Zm REM 1.3 mediates maize resistance to southern corn rust. Plant Biotechnol. J. 2019, 17, 2153–2168. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Linderman, G.C.; Steinerberger, S. Clustering with t-SNE, provably. SIAM J. Math. Data Sci. 2019, 1, 313–332. [Google Scholar] [CrossRef] [PubMed]
- Thakur, P.S.; Sheorey, T.; Ojha, A. VGG-ICNN: A Lightweight CNN model for crop disease identification. Multimed. Tools Appl. 2023, 82, 497–520. [Google Scholar] [CrossRef]
- Li, E.; Wang, L.; Xie, Q.; Gao, R.; Su, Z.; Li, Y. A novel deep learning method for maize disease identification based on small sample-size and complex background datasets. Ecol. Inform. 2023, 75, 102011. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Yang, J.; Li, S. Transformer helps identify kiwifruit diseases in complex natural environments. Comput. Electron. Agric. 2022, 200, 107258. [Google Scholar] [CrossRef]


| Type | Leaf Blight | Gray Leaf | Healthy Leaf | Leaf Rust |
|---|---|---|---|---|
| Training Set | 1743 | 750 | 1935 | 1523 |
| Testing Set | 500 | 250 | 500 | 500 |
| Total | 2243 | 1000 | 2435 | 2023 |
| Metrics | VGG11 | EfficientNet | Inception-v3 | MobileNet | ResNet50 | ViT | Improved ViT | Our Proposed Method |
|---|---|---|---|---|---|---|---|---|
| Accuracy | 97.9% | 91.6% | 97.2% | 90.2% | 96.6% | 93.9% | 98.7% | 99.5% |
| Precision | ||||||||
| Leaf Blight | 99% | 90% | 97% | 88% | 99% | 92% | 99% | 98.4% |
| Gray Leaf | 100% | 97% | 99% | 99% | 100% | 96% | 100% | 99.6% |
| Healthy Leaf | 96% | 88% | 96% | 88% | 94% | 91% | 97% | 98.6% |
| Leaf Rust | 98% | 94% | 98% | 92% | 96% | 98% | 99% | 99.8% |
| Recall | ||||||||
| Leaf Blight | 96% | 86% | 94% | 86% | 91% | 90% | 97% | 98.4% |
| Gray Leaf | 97% | 89% | 98% | 85% | 97% | 92% | 99% | 100% |
| Healthy Leaf | 100% | 92% | 98% | 93% | 99% | 95% | 99% | 99.8% |
| Leaf Rust | 99% | 98% | 100% | 96% | 99% | 97% | 100% | 99.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Shen, P.; Ye, Z.; Zhu, Z.; Xu, C.; Liu, Y.; Mei, L. Adversarial Training Collaborating Multi-Path Context Feature Aggregation Network for Maize Disease Density Prediction. Processes 2023, 11, 1132. https://doi.org/10.3390/pr11041132
Yang W, Shen P, Ye Z, Zhu Z, Xu C, Liu Y, Mei L. Adversarial Training Collaborating Multi-Path Context Feature Aggregation Network for Maize Disease Density Prediction. Processes. 2023; 11(4):1132. https://doi.org/10.3390/pr11041132
Chicago/Turabian StyleYang, Wei, Peiquan Shen, Zhaoyi Ye, Zhongmin Zhu, Chuan Xu, Yi Liu, and Liye Mei. 2023. "Adversarial Training Collaborating Multi-Path Context Feature Aggregation Network for Maize Disease Density Prediction" Processes 11, no. 4: 1132. https://doi.org/10.3390/pr11041132
APA StyleYang, W., Shen, P., Ye, Z., Zhu, Z., Xu, C., Liu, Y., & Mei, L. (2023). Adversarial Training Collaborating Multi-Path Context Feature Aggregation Network for Maize Disease Density Prediction. Processes, 11(4), 1132. https://doi.org/10.3390/pr11041132






