Menthol and Fatty Acid-Based Hydrophobic Deep Eutectic Solvents as Media for Enzyme Activation
Abstract
:1. Introduction
2. Results and Discussion
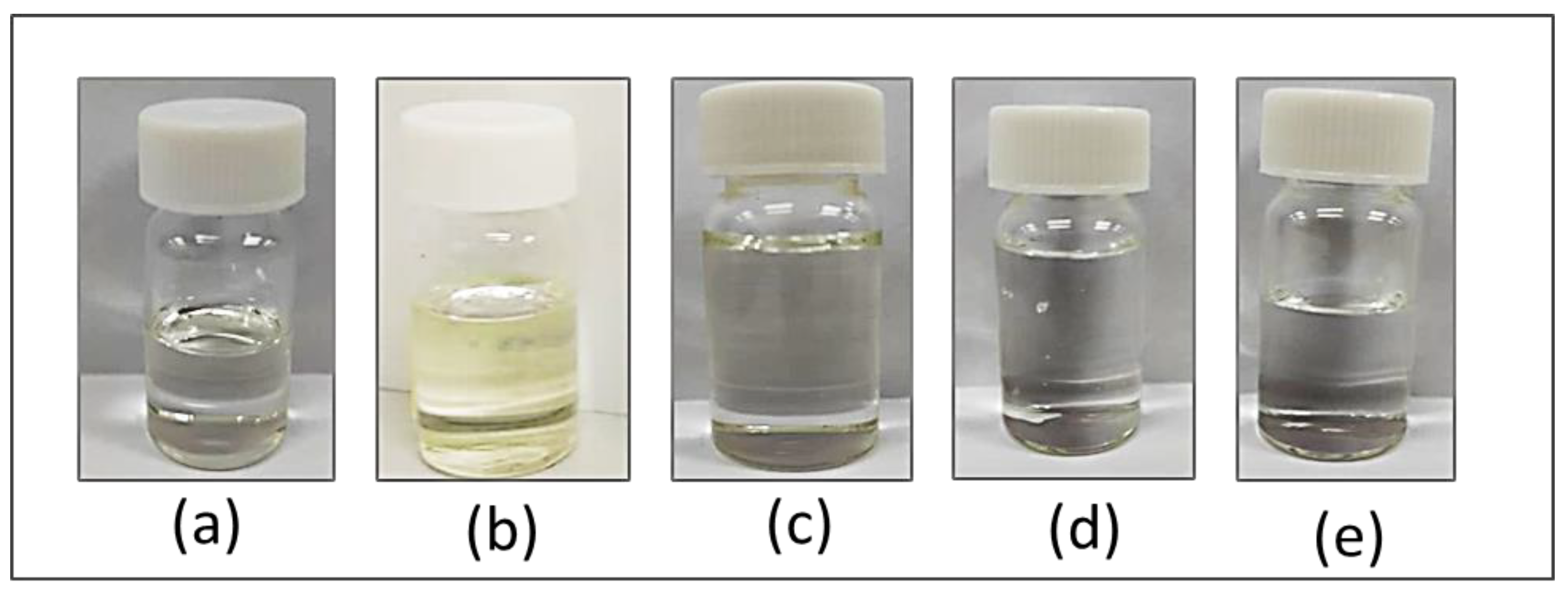
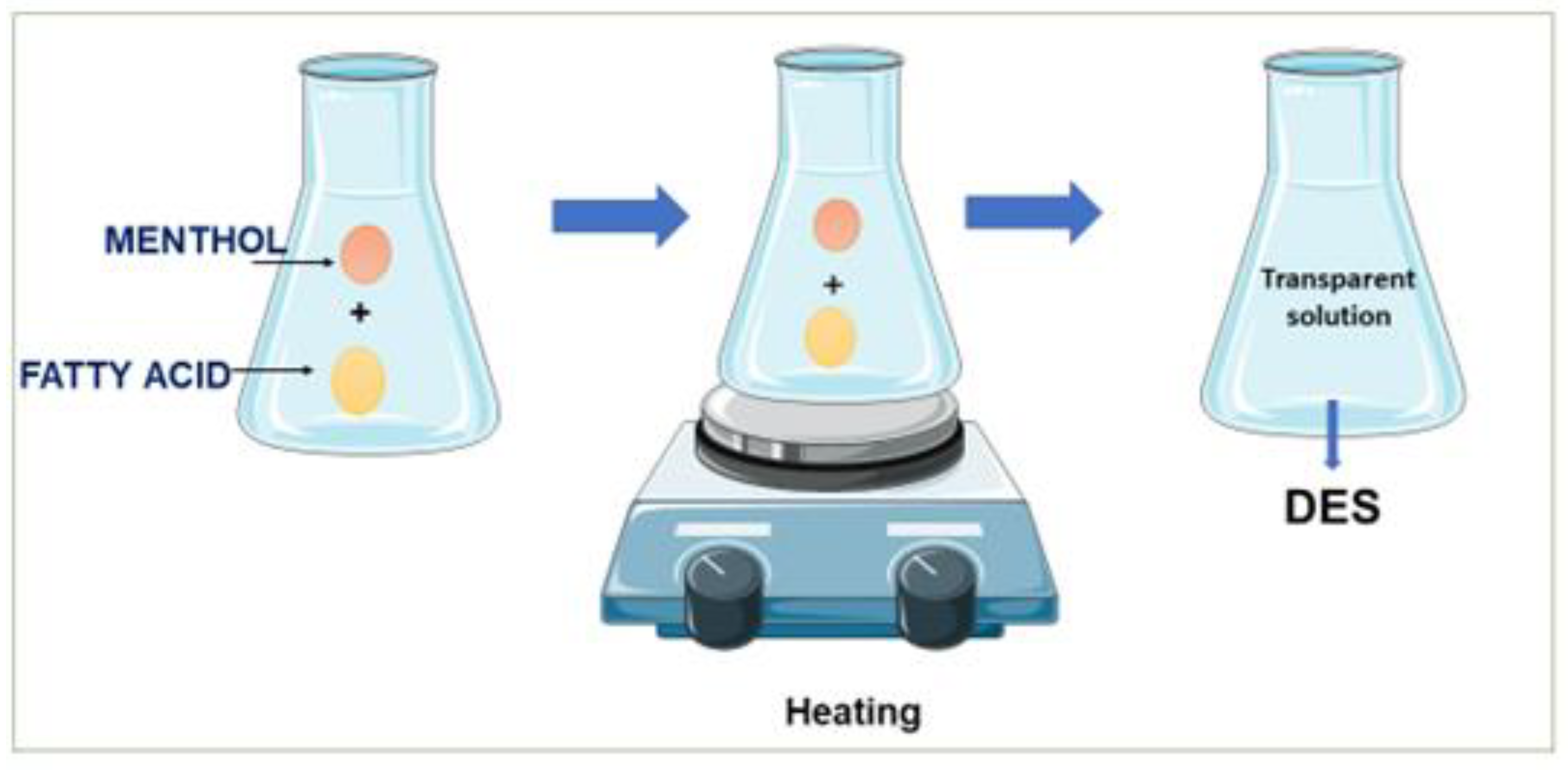
2.1. DES Preparation and the Visual Aspect
2.1.1. Solvent Miscibility Test Analyses
2.1.2. Thermal Analysis
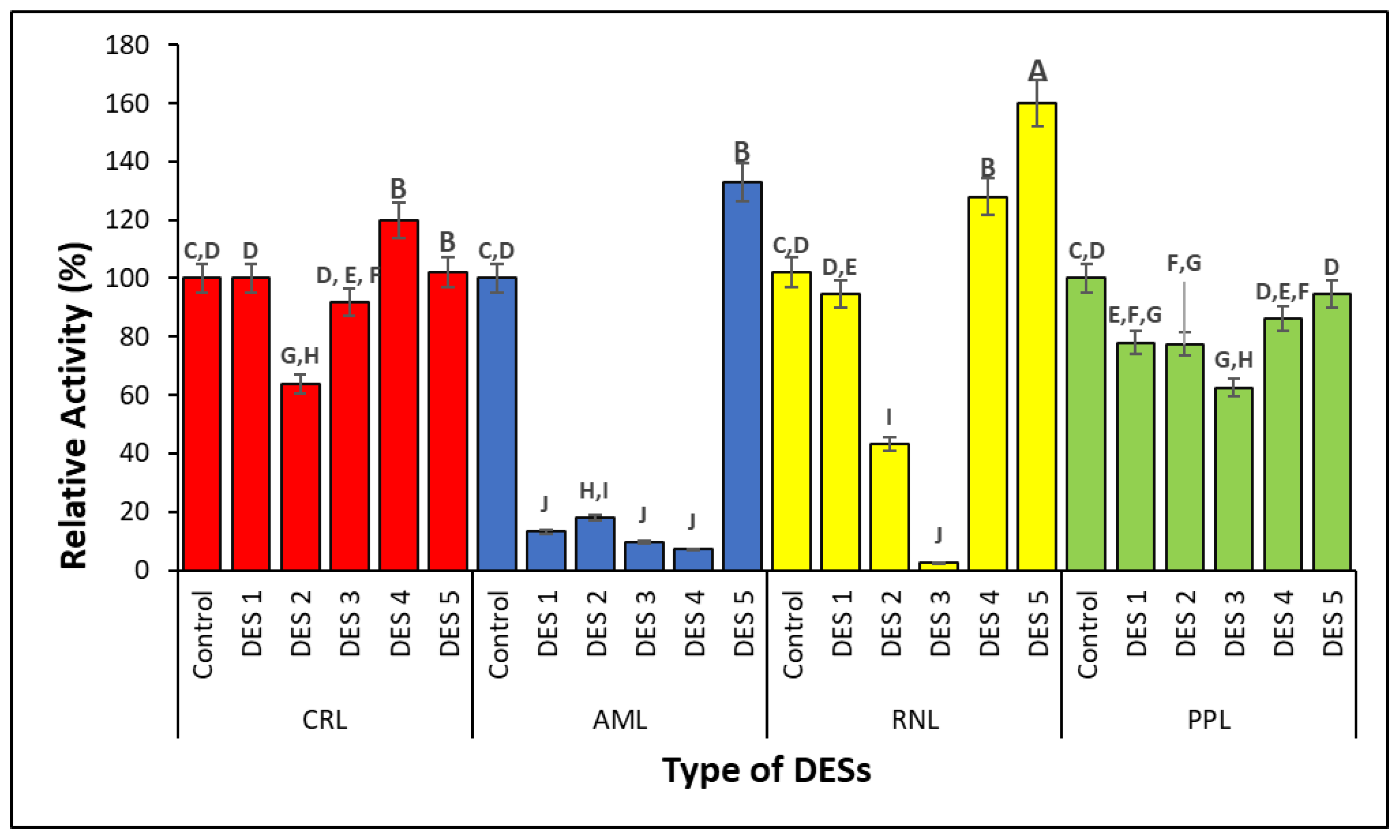
2.2. Screening of Lipases in Different Types of DESs
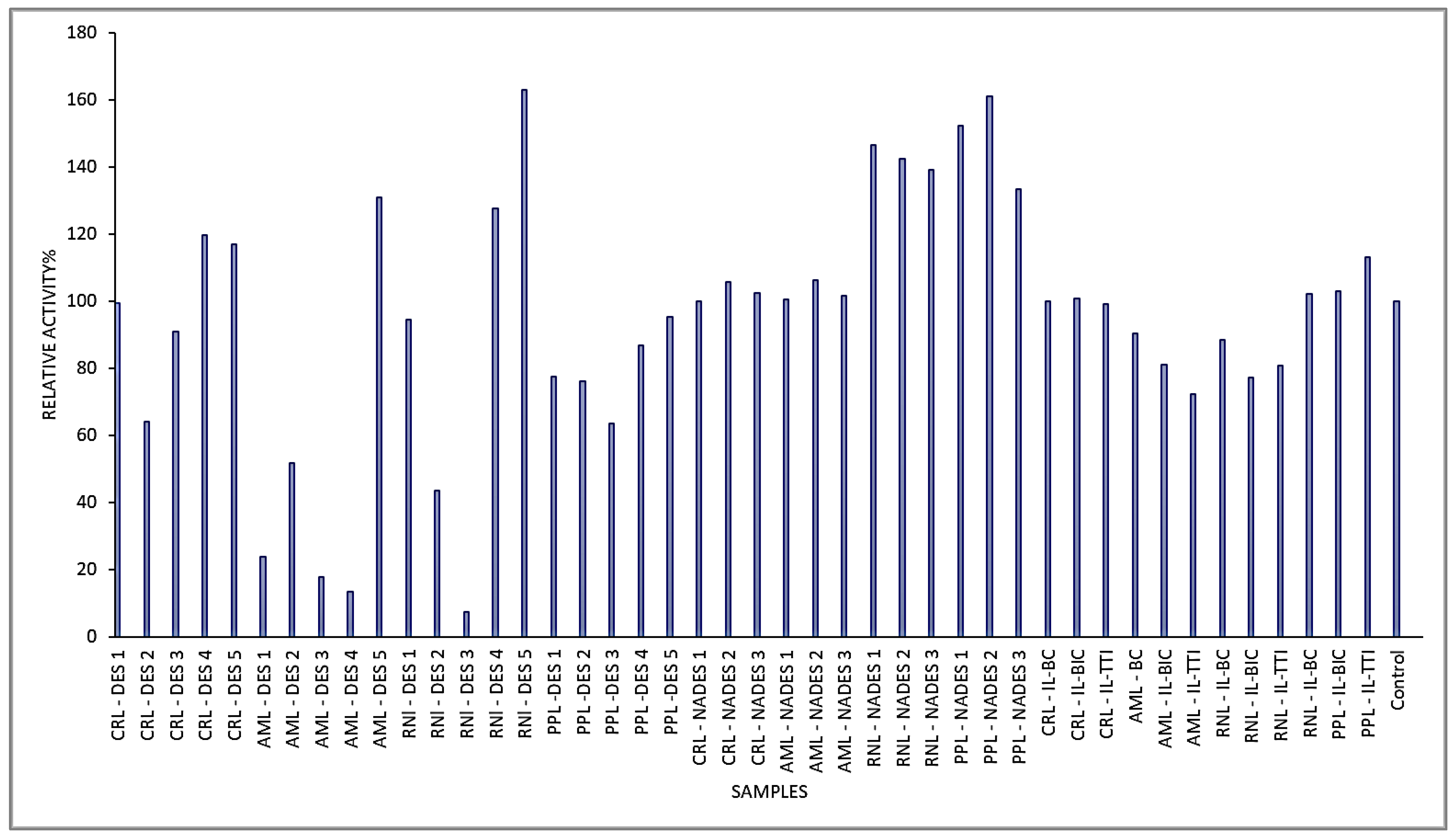
2.3. Comparison of Lipase Activity in Menthol-Based DESs and Other Solvents
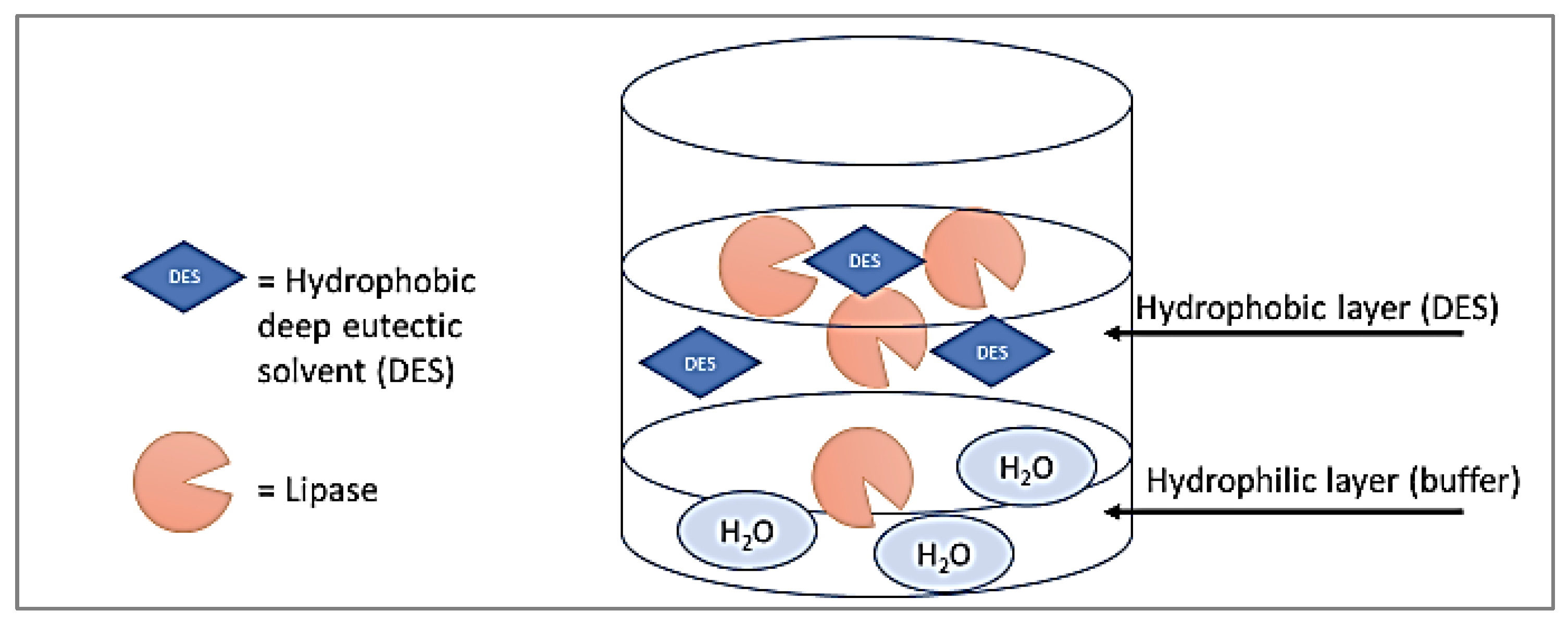
2.4. Activated Lipase Enzymatic Activity and Adsorption
2.5. Half-Life (t1/2) and Stability of Free and Activated Lipase in the Presence of DES
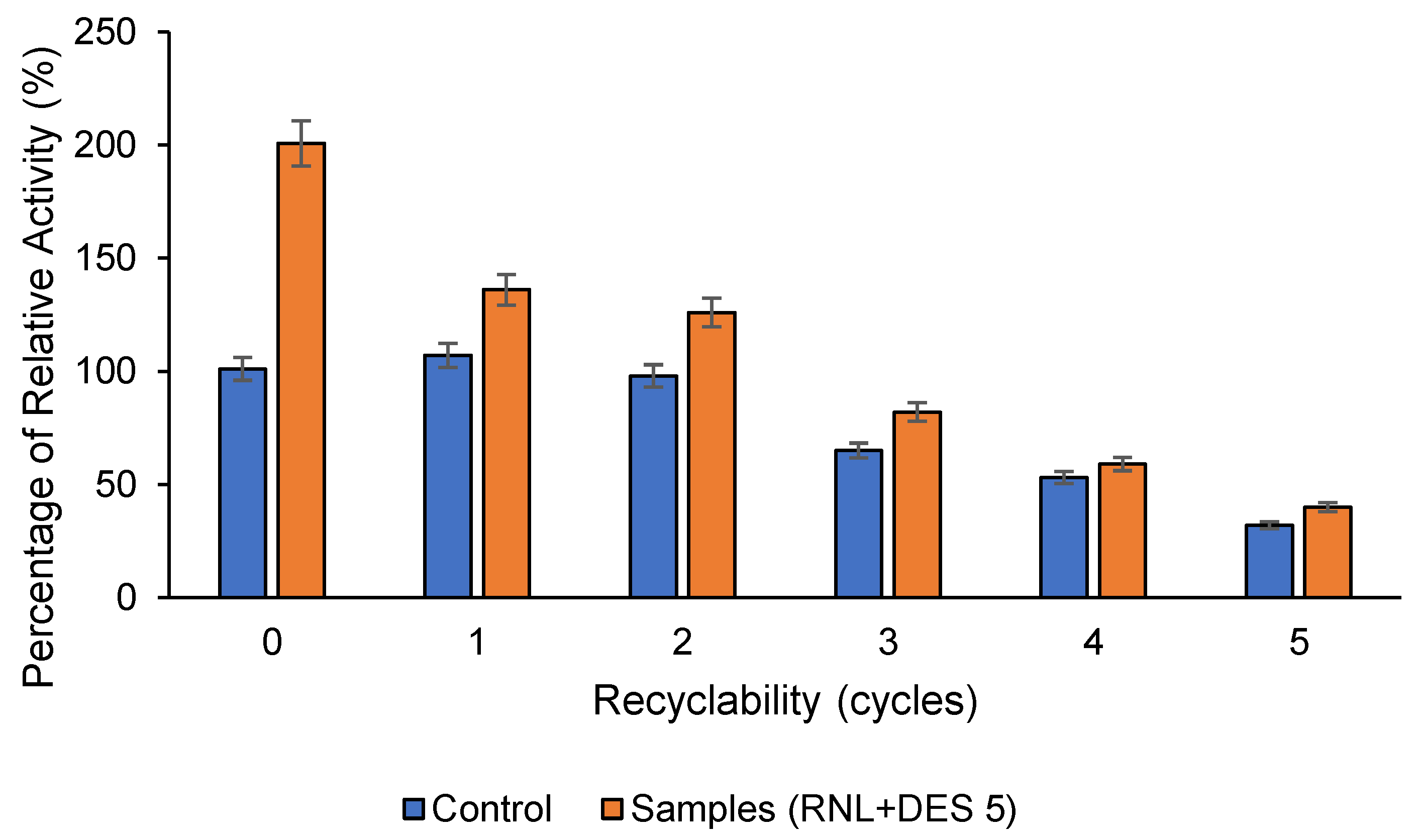
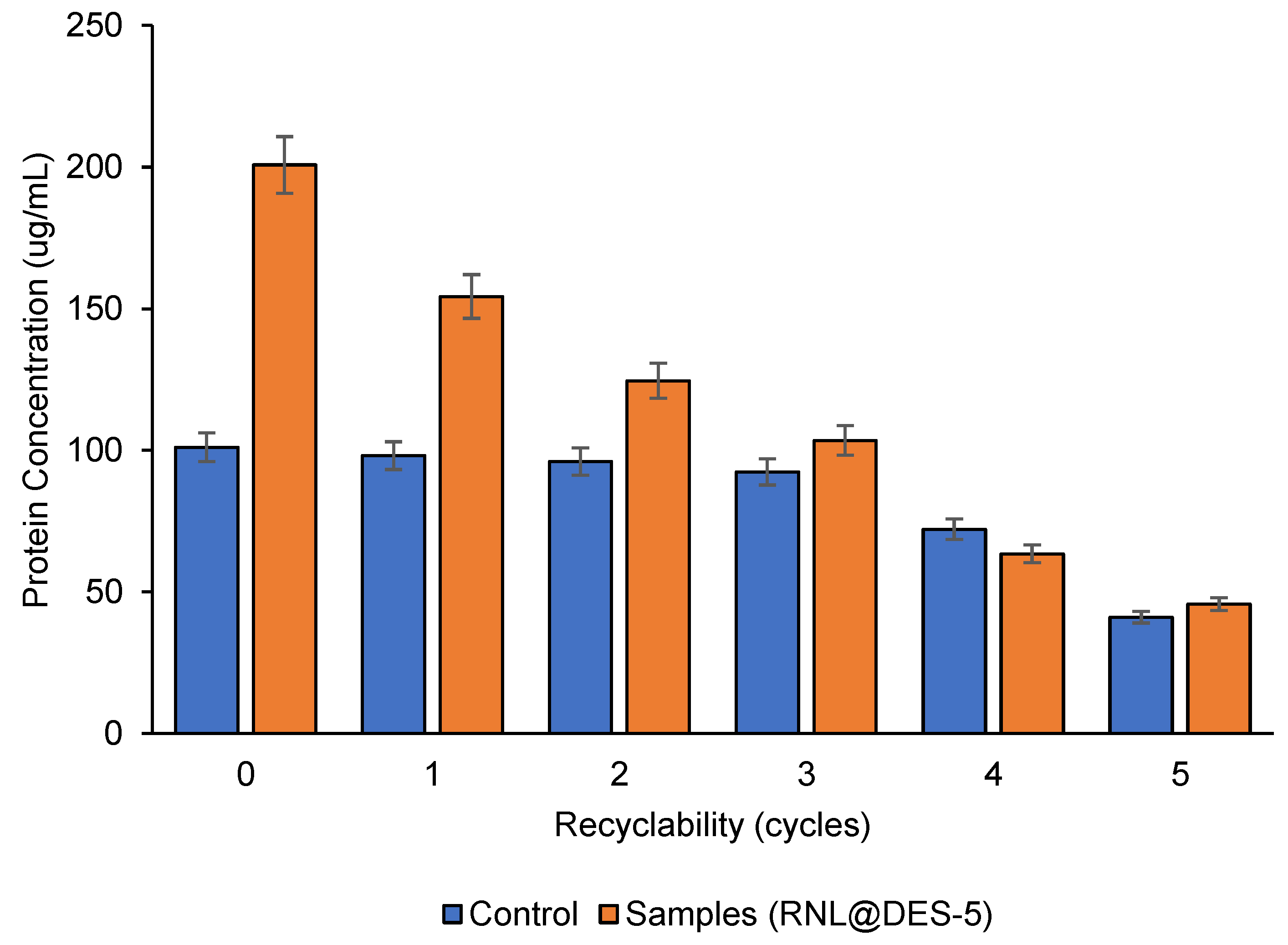
2.6. Recyclability of Activated Lipase and Total Protein Recyclability
2.7. Kinetic Parameters of the Enhanced Lipase
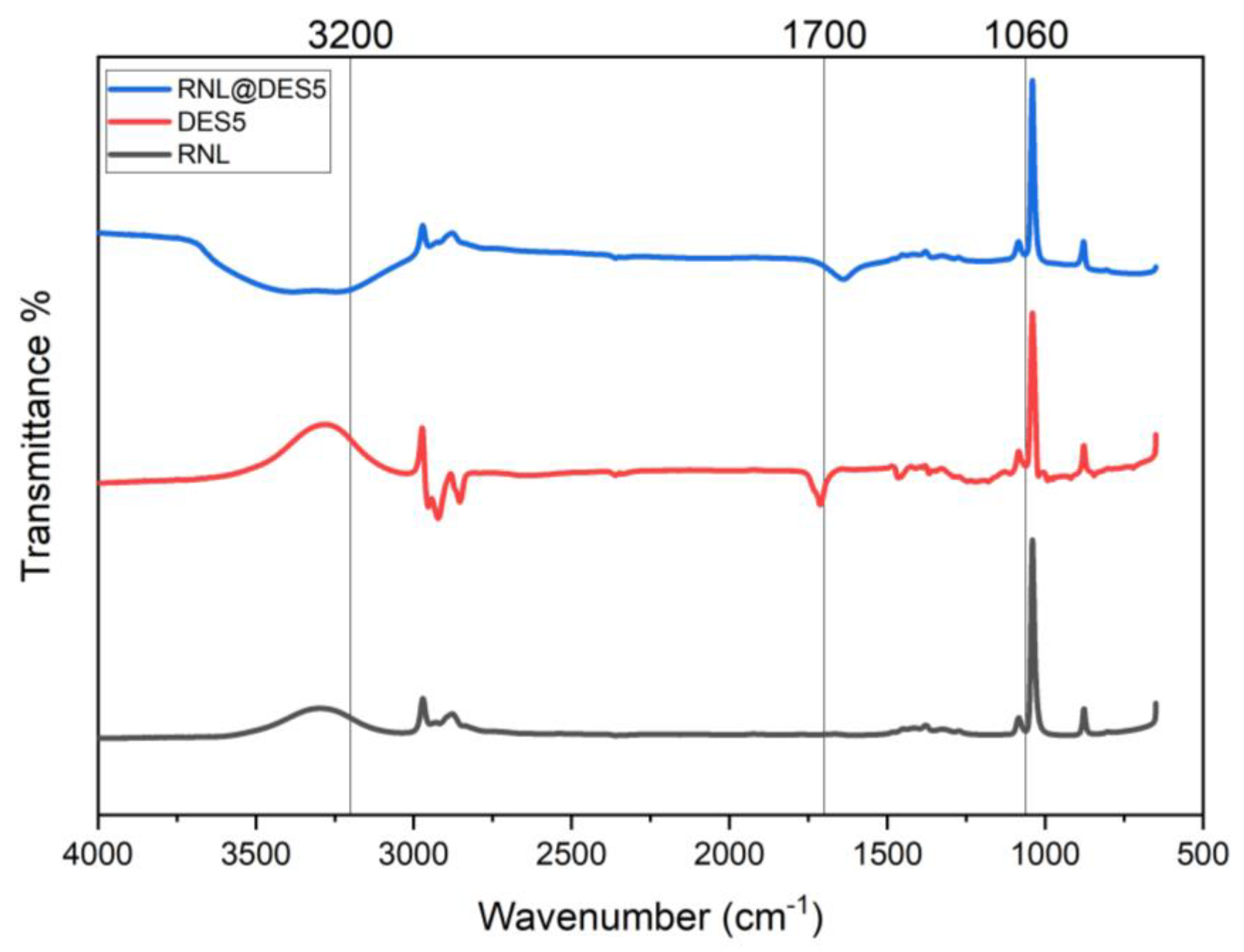
2.8. Activated Lipase and DES Fourier Transformed Infrared (FTIR) Spectroscopy
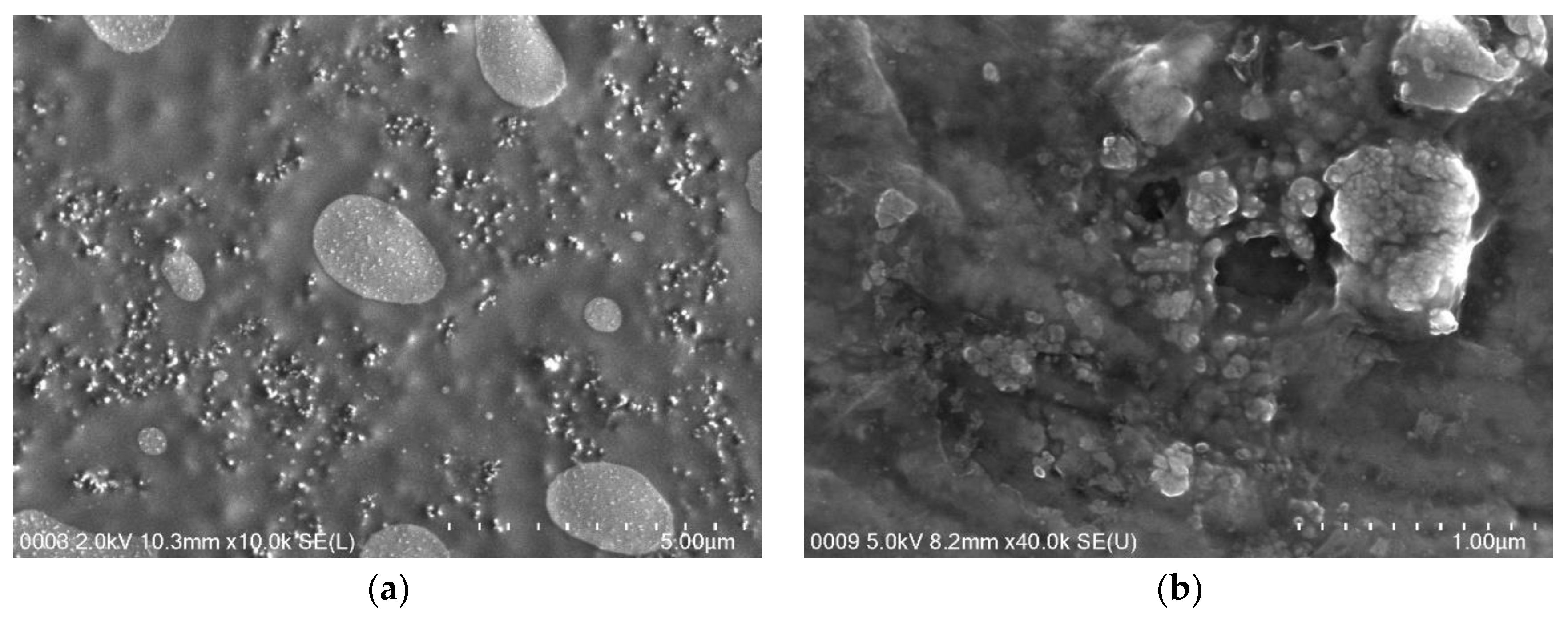
2.9. Morphology of Free and Activated Lipase
2.10. Application in Reverse Reactions: Synthesis
3. Materials and Methods
3.1. Materials
3.2. Menthol-Based DESs Preparation
3.2.1. Solvent Miscibility Test of Hydrophobic DESs
3.2.2. Thermal Analysis of Hydrophobic DESs
3.3. Assay (Enzyme Substrate Solution) Preparation
3.4. Lipase Activity Assay
3.5. Lipases Activities in DES Screening
3.6. Comparison of Lipase Activity in Menthol-based DESs and Other Solvents
3.7. Procedures for Chemical and Analytical Analysis
3.7.1. Protein Content Determination
3.7.2. Measurements of Enzyme Activity and Adsorption
3.7.3. Half-Life (t1/2)
3.8. Total Protein Recyclability and Activated Lipase Recovery in DES
3.9. Lipase Kinetic Parameter in the Presence of DES
3.10. Statistical Analysis
3.11. Characterization
3.11.1. Fourier Transformed Infrared (FTIR) Spectroscopy
3.11.2. Scanning Electron Microscopy (SEM) Analysis
3.12. Application in Reverse Reactions: Synthesis
- Y = fatty acid conversion (%)
- C0 = initial decanoic acid concentration (mM)
- Ct = decanoic acid concentration (mM) after a certain time t (min) of reaction.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, P.A.; Johl, S.K.; Johl, S.K. Does Adoption of ISO 56002-2019 and Green Innovation Reporting Enhance the Firm Sustainable Development Goal Performance? An Emerging Paradigm. Bus. Strategy Environ. 2021, 30, 2922–2936. [Google Scholar] [CrossRef]
- Khan, P.A.; Johl, S.K.; Akhtar, S. Vinculum of Sustainable Development Goal Practices and Firms’ Financial Performance: A Moderation Role of Green Innovation. J. Risk Financ. Manag. 2022, 15, 96. [Google Scholar] [CrossRef]
- Abbott, A.P.; Capper, G.; Davies, D.L.; Rasheed, R.K.; Tambyrajah, V. Novel Solvent Properties of Choline Chloride/Urea Mixtures. Chem. Commun. 2003, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Witkamp, G.J.; Verpoorte, R.; Choi, Y.H. Tailoring Properties of Natural Deep Eutectic Solvents with Water to Facilitate Their Applications. Food Chem. 2015, 187, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Aroso, I.M.; Paiva, A.; Reis, R.L.; Duarte, A.R.C. Natural Deep Eutectic Solvents from Choline Chloride and Betaine—Physicochemical Properties. J. Mol. Liq. 2017, 241, 654–661. [Google Scholar] [CrossRef]
- Hayyan, A.; Hashim, M.A.; Mjalli, F.S.; Hayyan, M.; AlNashef, I.M. A Novel Phosphonium-Based Deep Eutectic Catalyst for Biodiesel Production from Industrial Low Grade Crude Palm Oil. Chem. Eng. Sci. 2013, 92, 81–88. [Google Scholar] [CrossRef]
- Pena-Pereira, F.; de la Calle, I. Solvents and Eutectic Solvents. In Encyclopedia of Analytical Science, 3rd ed.; Worsfold, P., Poole, C., Townshend, A., Miró, M., Eds.; Academic Press: Oxford, UK, 2019; pp. 184–190. ISBN 978-0-08-101984-9. [Google Scholar]
- He, Y.; Li, K.; Bo, G.; Wang, J.; Xu, L.; Yan, J.; Yang, M.; Yan, Y. Enhancing Biodiesel Production via Liquid Yarrowia Lipolytica Lipase 2 in Deep Eutectic Solvents. Fuel 2022, 316, 123342. [Google Scholar] [CrossRef]
- Lynam, J.G.; Kumar, N.; Wong, M.J. Deep Eutectic Solvents’ Ability to Solubilize Lignin, Cellulose, and Hemicellulose; Thermal Stability; and Density. Bioresour. Technol. 2017, 238, 684–689. [Google Scholar] [CrossRef]
- Piemontese, L.; Perna, F.M.; Logrieco, A.; Capriati, V.; Solfrizzo, M. Deep Eutectic Solvents as Novel and Effective Extraction Media for Quantitative Determination of Ochratoxin A in Wheat and Derived Products. Molecules 2017, 22, 121. [Google Scholar] [CrossRef]
- Ribeiro, B.D.; Florindo, C.; Iff, L.C.; Coelho, M.A.Z.; Marrucho, I.M. Menthol-Based Eutectic Mixtures: Hydrophobic Low Viscosity Solvents. ACS Sustain. Chem. Eng. 2015, 3, 2469–2477. [Google Scholar] [CrossRef]
- Ahmad, I.; Pertiwi, A.S.; Kembaren, Y.H.; Rahman, A.; Mun’im, A. Application of Natural Deep Eutectic Solvent-Based Ultrasonic Assisted Extraction of Total Polyphenolic and Caffeine Content from Coffee Beans (Coffea Beans L.) for Instant Food Products. J. Appl. Pharm. Sci. 2018, 8, 138–143. [Google Scholar] [CrossRef]
- Dai, Y.; Witkamp, G.J.; Verpoorte, R.; Choi, Y.H. Natural Deep Eutectic Solvents as a New Extraction Media for Phenolic Metabolites in Carthamus Tinctorius L. Anal. Chem. 2013, 85, 6272–6278. [Google Scholar] [CrossRef] [PubMed]
- Saravana, P.S.; Ho, T.C.; Chae, S.J.; Cho, Y.J.; Park, J.S.; Lee, H.J.; Chun, B.S. Deep Eutectic Solvent-Based Extraction and Fabrication of Chitin Films from Crustacean Waste. Carbohydr. Polym. 2018, 195, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Baby, J.N.; Sriram, B.; Wang, S.F.; George, M. Effect of Various Deep Eutectic Solvents on the Sustainable Synthesis of MgFe2O4 Nanoparticles for Simultaneous Electrochemical Determination of Nitrofurantoin and 4-Nitrophenol. ACS Sustain. Chem. Eng. 2020, 8, 1479–1486. [Google Scholar] [CrossRef]
- Krishnan, S.K.; Chipatecua Godoy, Y. Deep Eutectic Solvent-Assisted Synthesis of Au Nanostars Supported on Graphene Oxide as an Efficient Substrate for SERS-Based Molecular Sensing. ACS Omega 2020, 5, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Putra, S.S.S.; Basirun, W.J.; Elgharbawy, A.A.M.; Hayyan, A.; Hayyan, M.; Mohammed, M.A. Nanocellulose and Natural Deep Eutectic Solvent as Potential Biocatalyst System toward Enzyme Immobilization. Mol. Catal. 2022, 528, 112422. [Google Scholar] [CrossRef]
- Shehata, M.; Unlu, A.; Sezerman, U.; Timucin, E. Lipase and Water in a Deep Eutectic Solvent: Molecular Dynamics and Experimental Studies of the Effects of Water-in-Deep Eutectic Solvents on Lipase Stability. J. Phys. Chem. B 2020, 124, 8801–8810. [Google Scholar] [CrossRef]
- Cao, J.; Wu, R.; Zhu, F.; Dong, Q.; Su, E. How to Improve the Efficiency of Biocatalysis in Non-Aqueous Pure Deep Eutectic Solvents: A Case Study on the Lipase-Catalyzed Transesterification Reaction. Biochem. Eng. J. 2022, 179, 108336. [Google Scholar] [CrossRef]
- Hayyan, A.; Hadj-Kali, M.K.; Salleh, M.Z.M.; Hashim, M.A.; Rubaidi, S.R.; Hayyan, M.; Zulkifli, M.Y.; Rashid, S.N.; Mirghani, M.E.S.; Ali, E.; et al. Characterization of Tetraethylene Glycol-Based Deep Eutectic Solvents and Their Potential Application for Dissolving Unsaturated Fatty Acids. J. Mol. Liq. 2020, 312, 113284. [Google Scholar] [CrossRef]
- Florindo, C.; Romero, L.; Rintoul, I.; Branco, L.C.; Marrucho, I.M. From Phase Change Materials to Green Solvents: Hydrophobic Low Viscous Fatty Acid-Based Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2018, 6, 3888–3895. [Google Scholar] [CrossRef]
- Rashid, S.N.; Hayyan, A.; Hayyan, M.; Hashim, M.A.; Elgharbawy, A.A.M.; Sani, F.S.; Basirun, W.J.; Lee, V.S.; Alias, Y.; Mohammed, A.K.; et al. Ternary Glycerol-Based Deep Eutectic Solvents: Physicochemical Properties and Enzymatic Activity. Chem. Eng. Res. Des. 2021, 169, 77–85. [Google Scholar] [CrossRef]
- Chin Ting, H.; Warsi Khan, H.; Vijaya Bhaskar Reddy, A.; Goto, M.; Moniruzzaman, M. Extraction of Salicylic Acid from Wastewater Using Ionic Liquid-Based Green Emulsion Liquid Membrane: COSMO-RS Prediction and Experimental Verification. J. Mol. Liq. 2022, 347, 118280. [Google Scholar] [CrossRef]
- Khan, H.W.; Reddy, A.V.B.; Bustam, M.A.; Goto, M.; Moniruzzaman, M. Development and Optimization of Ionic Liquid-Based Emulsion Liquid Membrane Process for Efficient Recovery of Lactic Acid from Aqueous Streams. Biochem. Eng. J. 2021, 176, 108216. [Google Scholar] [CrossRef]
- Silva, J.M.; Pereira, C.V.; Mano, F.; Silva, E.; Castro, V.I.B.; Sá-Nogueira, I.; Reis, R.L.; Paiva, A.; Matias, A.A.; Duarte, A.R.C. Therapeutic Role of Deep Eutectic Solvents Based on Menthol and Saturated Fatty Acids on Wound Healing. ACS Appl. Bio. Mater. 2019, 2, 4346–4355. [Google Scholar] [CrossRef]
- Simakova, I.L.; Vajglová, Z.; Mäki-Arvela, P.; Eränen, K.; Hupa, L.; Peurla, M.; Mäkilä, E.M.; Wärnå, J.; Murzin, D.Y. Citral-to-Menthol Transformations in a Continuous Reactor over Ni/Mesoporous Aluminosilicate Extrudates Containing a Sepiolite Clay Binder. Org. Process. Res. Dev. 2022, 26, 387–403. [Google Scholar] [CrossRef]
- Sharma, A.; Tyagi, V.V.; Chen, C.R.; Buddhi, D. Review on Thermal Energy Storage with Phase Change Materials and Applications. Renew. Sustain. Energy Rev. 2009, 13, 318–345. [Google Scholar] [CrossRef]
- Hayyan, A.; Mjalli, F.S.; Alnashef, I.M.; Al-Wahaibi, Y.M.; Al-Wahaibi, T.; Hashim, M.A. Glucose-Based Deep Eutectic Solvents: Physical Properties. J. Mol. Liq. 2013, 178, 137–141. [Google Scholar] [CrossRef]
- McGaughy, K.; Reza, M.T. Liquid–Liquid Extraction of Furfural from Water by Hydrophobic Deep Eutectic Solvents: Improvement of Density Function Theory Modeling with Experimental Validations. ACS Omega 2020, 5, 22305–22313. [Google Scholar] [CrossRef]
- Longeras, O.; Gautier, A.; Ballerat-Busserolles, K.; Andanson, J.-M. Deep Eutectic Solvent with Thermo-Switchable Hydrophobicity. ACS Sustain. Chem. Eng. 2020, 8, 12516–12520. [Google Scholar] [CrossRef]
- Schmitz, D.; Shubert, V.A.; Betz, T.; Schnell, M. Exploring the Conformational Landscape of Menthol, Menthone, and Isomenthone: A Microwave Study. Front. Chem. 2015, 3, 15. [Google Scholar] [CrossRef]
- Hümmer, M.; Kara, S.; Liese, A.; Huth, I.; Schrader, J.; Holtmann, D. Synthesis of (-)-Menthol Fatty Acid Esters in and from (-)-Menthol and Fatty Acids—Novel Concept for Lipase Catalyzed Esterification Based on Eutectic Solvents. Mol. Catal. 2018, 458, 67–72. [Google Scholar] [CrossRef]
- Hollenbach, R.; Ochsenreither, K.; Syldatk, C. Enzymatic Synthesis of Glucose Monodecanoate in a Hydrophobic Deep Eutectic Solvent. Int. J. Mol. Sci. 2020, 21, 4342. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wu, R.; Dong, Q.; Zhao, L.; Cao, F.; Su, E. Effective Release of Intracellular Enzymes by Permeating the Cell Membrane with Hydrophobic Deep Eutectic Solvents. ChemBioChem 2020, 21, 672–680. [Google Scholar] [CrossRef] [PubMed]
- López-Fernández, J.; Benaiges, M.D.; Valero, F. Rhizopus Oryzae Lipase, a Promising Industrial Enzyme: Biochemical Characteristics, Production and Biocatalytic Applications. Catalysts 2020, 10, 1277. [Google Scholar] [CrossRef]
- Elgharbawy, A.A.; Hayyan, A.; Hayyan, M.; Rashid, S.N.; Nor, M.R.M.; Zulkifli, M.Y.; Alias, Y.; Mirghani, M.E.S. Shedding Light on Lipase Stability in Natural Deep Eutectic Solvents. Chem. Biochem. Eng. Q. 2018, 32, 359–370. [Google Scholar] [CrossRef]
- Khan, H.W.; Elgharbawy, A.A.M.; Bustam, M.A.; Goto, M.; Moniruzzaman, M. Vegetable Oil–Ionic Liquid-Based Emulsion Liquid Membrane for the Removal of Lactic Acid from Aqueous Streams: Emulsion Size, Membrane Breakage, and Stability Study. ACS Omega 2022, 7, 32176–32183. [Google Scholar] [CrossRef]
- Zhao, H. What Do We Learn from Enzyme Behaviors in Organic Solvents?—Structural Functionalization of Ionic Liquids for Enzyme Activation and Stabilization. Biotechnol. Adv. 2020, 45, 107638. [Google Scholar] [CrossRef]
- Pätzold, M.; Weimer, A.; Liese, A.; Holtmann, D. Optimization of Solvent-Free Enzymatic Esterification in Eutectic Substrate Reaction Mixture. Biotechnol. Rep. 2019, 22, e00333. [Google Scholar] [CrossRef]
- Toledo, M.L.; Pereira, M.M.; Freire, M.G.; Silva, J.P.A.; Coutinho, J.A.P.; Tavares, A.P.M. Laccase Activation in Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2019, 7, 11806–11814. [Google Scholar] [CrossRef]
- Lorsch, J.R. Chapter One—Practical Steady-State Enzyme Kinetics. In Laboratory Methods in Enzymology: Protein Part A; Lorsch, J.B.T.-M., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 536, pp. 3–15. ISBN 0076-6879. [Google Scholar]
- Fabrizio, A.; Corminboeuf, C. How Do London Dispersion Interactions Impact the Photochemical Processes of Molecular Switches? J. Phys. Chem. Lett. 2018, 9, 464–470. [Google Scholar] [CrossRef]
- Strauss, M.A.; Wegner, H.A. Exploring London Dispersion and Solvent Interactions at Alkyl–Alkyl Interfaces Using Azobenzene Switches. Angew. Chem. Int. Ed. 2019, 58, 18552–18556. [Google Scholar] [CrossRef] [PubMed]
- Sonesson, A.W.; Elofsson, U.M.; Brismar, H.; Callisen, T.H. Adsorption and Mobility of a Lipase at a Hydrophobic Surface in the Presence of Surfactants. Langmuir 2006, 22, 5810–5817. [Google Scholar] [CrossRef] [PubMed]
- Mozhaev, V. Engineering Stability of Enzymes in Systems with Organic Solvents. In Stability and Stabilization of Biocatalysts; Ballesteros, A., Plou, F.J., Iborra, J.L., Halling, P.J., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; Volume 15, pp. 355–363. ISBN 0921-0423. [Google Scholar]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef] [PubMed]
- Di, X.; Zhang, Y.; Fu, J.; Yu, Q.; Wang, Z.; Yuan, Z. Ionic Liquid-Strengthened Immobilized Rhizomucor Miehei Lipase for Catalytic Esterification of Itaconic Acid in Aqueous Media. ACS Sustain. Chem. Eng. 2020, 8, 1805–1812. [Google Scholar] [CrossRef]
- Bingham, R.J.; Ballone, P. Computational Study of Room-Temperature Ionic Liquids Interacting with a POPC Phospholipid Bilayer. J. Phys. Chem. B 2012, 116, 11205–11216. [Google Scholar] [CrossRef]
- Makoś, P.; Słupek, E.; Gębicki, J. Hydrophobic Deep Eutectic Solvents in Microextraction Techniques—A Review. Microchem. J. 2020, 152, 104384. [Google Scholar] [CrossRef]
- Ge, D.; Wang, Y.; Jiang, Q.; Dai, E. A Deep Eutectic Solvent as an Extraction Solvent to Separate and Preconcentrate Parabens in Water Samples Using in Situ Liquid-Liquid Microextraction. J. Braz. Chem. Soc. 2019, 30, 1203–1210. [Google Scholar] [CrossRef]
- Florindo, C.; Lima, F.; Branco, L.C.; Marrucho, I.M. Hydrophobic Deep Eutectic Solvents: A Circular Approach to Purify Water Contaminated with Ciprofloxacin. ACS Sustain. Chem. Eng. 2019, 7, 14739–14746. [Google Scholar] [CrossRef]
- Cao, J.; Wu, R.; Zhu, F.; Dong, Q.; Su, E. Enzymes in Nearly Anhydrous Deep Eutectic Solvents: Insight into the Biocompatibility and Thermal Stability. Enzym. Microb. Technol. 2022, 157, 110022. [Google Scholar] [CrossRef]
- Guajardo, N.; Ahumada, K.; Domínguez de María, P. Immobilized Lipase-CLEA Aggregates Encapsulated in Lentikats® as Robust Biocatalysts for Continuous Processes in Deep Eutectic Solvents. J. Biotechnol. 2020, 310, 97–102. [Google Scholar] [CrossRef]
- Tang, W.; Dai, Y.; Row, K.H. Evaluation of Fatty Acid/Alcohol-Based Hydrophobic Deep Eutectic Solvents as Media for Extracting Antibiotics from Environmental Water. Anal. Bioanal. Chem. 2018, 410, 7325–7336. [Google Scholar] [CrossRef] [PubMed]
- Elgharbawy, A.A.; Riyadi, F.A.; Alam, M.Z.; Moniruzzaman, M. Ionic Liquids as a Potential Solvent for Lipase-Catalysed Reactions: A Review. J. Mol. Liq. 2018, 251, 150–166. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Z.; Ge, X.; Zhang, J. Effects of Lignin and Surfactant on Adsorption and Hydrolysis of Cellulases on Cellulose. Biotechnol. Biofuels 2016, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Noori, M.S.; Karimi, K. Detailed Study of Efficient Ethanol Production from Elmwood by Alkali Pretreatment. Biochem. Eng. J. 2016, 105, 197–204. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, S.; Yu, H.; Kim, J.H.; Kim, H.J.; Yang, Y.H.; Kim, Y.H.; Kim, K.J.; Kan, E.; Lee, S.H. Effect of Deep Eutectic Solvent Mixtures on Lipase Activity and Stability. J. Mol. Catal. B Enzym. 2016, 128, 65–72. [Google Scholar] [CrossRef]
- Hou, C.; Zhu, H.; Wu, D.; Li, Y.; Hou, K.; Jiang, Y.; Li, Y. Immobilized Lipase on Macroporous Polystyrene Modified by PAMAM-Dendrimer and Their Enzymatic Hydrolysis. Process Biochem. 2014, 49, 244–249. [Google Scholar] [CrossRef]
- Elgharbawy, A.A.M.; Syed Putra, S.S.; Hayyan, M.; Hayyan, A.; Basirun, W.J.; Mirghani, M.E.S.; Majrashi, A.A.; Nawawi, W.M.F.W.; Alias, Y.; Mohd Salleh, H.; et al. Polyamidoamine Dendrimers: Favorable Polymeric Nanomaterials for Lipase Activation. Mater. Today Commun. 2020, 25, 101492. [Google Scholar] [CrossRef]
- Bassi, J.J.; Todero, L.M.; Lage, F.A.P.; Khedy, G.I.; Ducas, J.D.; Custódio, A.P.; Pinto, M.A.; Mendes, A.A. Interfacial Activation of Lipases on Hydrophobic Support and Application in the Synthesis of a Lubricant Ester. Int. J. Biol. Macromol. 2016, 92, 900–909. [Google Scholar] [CrossRef]





| HBD, HBA/DES | Tm (°C) |
|---|---|
| Menthol | 42.96 |
| Propanoic acid | −21.00 |
| Butanoic acid | −7.90 |
| Hexanoic acid | −3.40 |
| Octanoic acid | 16.70 |
| Decanoic acid | 31.60 |
| DES 1 | 1.37 |
| DES 2 | 133.90 |
| DES 3 | 152.44 |
| DES 4 | −14.61 |
| DES 5 | −5.72 |
| Sample | Specific Activity U. mg−1 Protein | Activity Yield (%) | Absorption mg mL−1 | Desorption mg mL−1 | Desorption (%) |
|---|---|---|---|---|---|
| * Free RNL (Control) | 6.17 ± 0.05 | 93.47 ± 1.2 | 50.22 ± 3.5 | 47.16 ± 3.5 | 6.09 ± 1.0 |
| RNL@DES-5 | 7.30 ± 0.04 | 167.58 ± 2.5 | 125.75 ± 5.6 | 87.99 ± 5.5 | 30.03 ± 2.5 |
| Free RNL | RNL@DES-5 | |
|---|---|---|
| Best-fit values | ||
| * Y0 | 98.50 | 108.30 |
| * Plateau | −710.30 | −39,281 |
| k | 0.03952 | 1.765 × 10−5 |
| Half-life (days) | 9 | 29 |
| Tau | 25.30 | 56,671 |
| Span | 142.6 | 202,937 |
| Goodness of Fit | ||
| R squared | 0.9780 | 0.8909 |
| Sum of squares | 287.4 | 1366 |
| Lipase | Reaction Medium | KM (mM) | Vmax (mM min−1) | kcat min–1 | kcat/KM (min mM–1) |
|---|---|---|---|---|---|
| RNL | Control | 1.27 | 3.39 | 12.69 | 9.99 |
| DES 4 | 0.27 | 59.56 | 21.71 | 80.41 | |
| DES 5 | 0.078 | 22.61 | 25.98 | 333.08 |
| Wavenumber (cm−1) | Bond/Stretching |
|---|---|
| 3600–3200 | O-H stretching vibration |
| 3022–2850 | C-H stretching vibration |
| 1712–1700 | C=O stretching vibration |
| 1206–1171 | C-O stretching vibration |
| 1065–1061 | S=O stretching vibration |
| 991–842 | C=C stretching vibration |
| Lipases | Description | Abbreviation |
|---|---|---|
| Lipase from Rhizopus Niveus (Sigma-Aldrich) | ≥1.5 U mg−1 | RNL |
| Lipase from porcine pancreas (Sigma-Aldrich) | 100–500 U mg−1 protein | PPL |
| Lipase from Candida Rugosa (Sigma-Aldrich) | ≥700 U mg−1 solid | CRL |
| Amano lipase PS, from Burkholderia Cepacia (Sigma-Aldrich) | ≥30,000 U g−1 | AML |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elgharbawy, A.A.M.; Syed Putra, S.S.; Khan, H.W.; Azmi, N.A.N.; Sani, M.S.A.; Ab llah, N.; Hayyan, A.; Jewaratnam, J.; Basirun, W.J. Menthol and Fatty Acid-Based Hydrophobic Deep Eutectic Solvents as Media for Enzyme Activation. Processes 2023, 11, 547. https://doi.org/10.3390/pr11020547
Elgharbawy AAM, Syed Putra SS, Khan HW, Azmi NAN, Sani MSA, Ab llah N, Hayyan A, Jewaratnam J, Basirun WJ. Menthol and Fatty Acid-Based Hydrophobic Deep Eutectic Solvents as Media for Enzyme Activation. Processes. 2023; 11(2):547. https://doi.org/10.3390/pr11020547
Chicago/Turabian StyleElgharbawy, Amal A. M., Sharifah Shahira Syed Putra, Huma Warsi Khan, Nor Azrini Nadiha Azmi, Muhamad Shirwan Abdullah Sani, Nazurah Ab llah, Adeeb Hayyan, Jegalakshimi Jewaratnam, and Wan Jefrey Basirun. 2023. "Menthol and Fatty Acid-Based Hydrophobic Deep Eutectic Solvents as Media for Enzyme Activation" Processes 11, no. 2: 547. https://doi.org/10.3390/pr11020547







