Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Curd Samples and Lactic Acid Bacteria
2.2. Probiotic Property
2.3. Antagonistic Activity
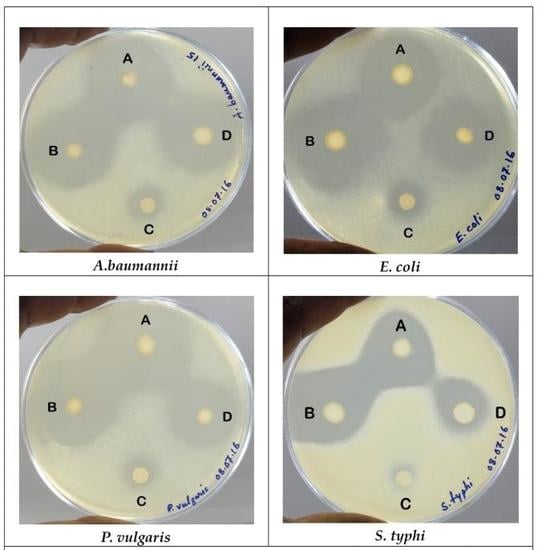
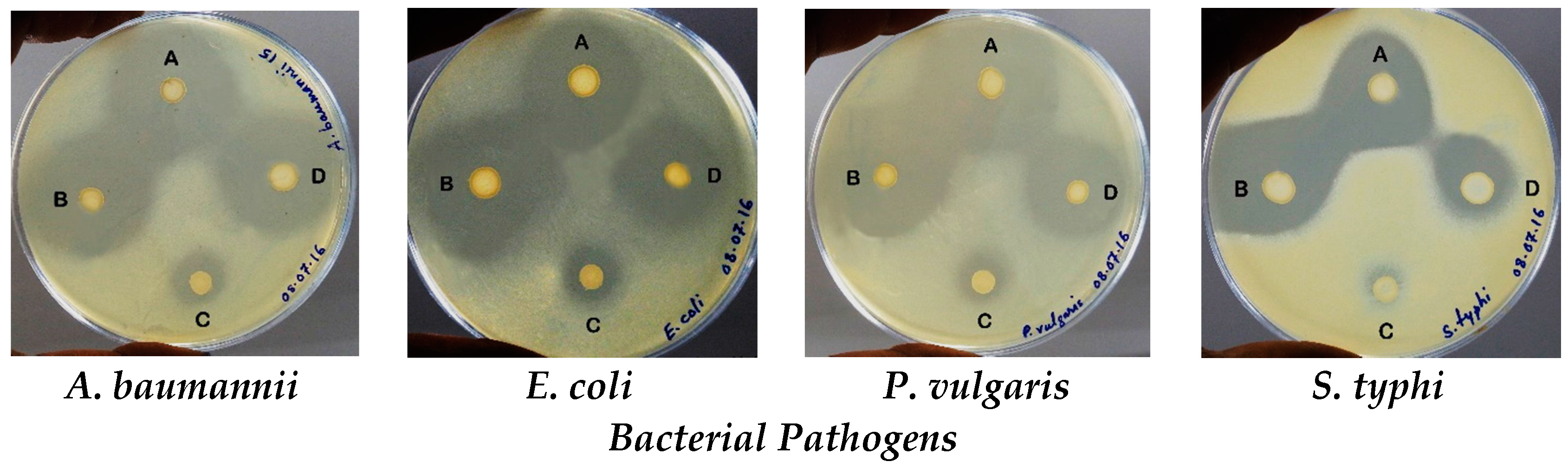
2.3.1. Agar Overlay Method
2.3.2. Agar-Well Diffusion Method
2.4. Safety Profiling
2.4.1. Hemolytic Activity
2.4.2. Antibiotic Susceptibility
2.5. Cumulative Probiotic Potential
3. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fuller, R. Probiotic in man and animals: A review. J. Appl. Bacteriol. 1989, 66, 365–378. [Google Scholar] [PubMed]
- Zheng, J.; Zhao, X.; Lin, X.B.; Michael, G.M. Comparative genomics Lactobacillus reuteri from sourdough reveals adaptation of an intestinal symbiont to food fermentations. Sci. Rep. 2015, 5, 18234. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hou, C.; Zeng, X.; Qiao, S. The use of lactic acid bacteria as a probiotic in swine diets. Pathogens 2015, 4, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Sieladie, D.V.; Zambou, N.F.; Kaktcham, P.M.; Cresci, A.; Fonteh, F. Probiotic properties of lactobacilli strains isolated from raw cow milk in the western highlands of Cameroon. Innov. Romanian Food Biotechnol. 2011, 9, 12–28. [Google Scholar]
- Vuotto, C.; Longo, F.; Donelli, G. Probiotics to counteract biofilm-associated infections: Promising and conflicting data. Int. J. Oral Sci. 2014, 6, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Eid, R.; Jakee, J.E.; Rashidy, A.; Asfour, H.; Omara, S.; Kandil, M.M.; Mahmood, Z.; Hahne, J.; Seida, A.A. Potential antimicrobial activities of probiotic Lactobacillus strains isolated from raw milk. Probiot. Health 2016, 4, 138. [Google Scholar]
- Abdel-Daim, A.; Hassouna, N.; Hafez, M.; Ashor, M.S.A.; Aboulwafa, M.M. Antagonistic activity of Lactobacillus isolates against salmonella typhi in vitro. Biomed. Res. Int. 2013, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Di Cerbo, A.; Palmieri, B.; Aponte, M.; Morales-Medina, J.C.; Iannitti, T. Mechanisms and therapeutic effectiveness of lactobacilli. J. Clin. Pathol. 2015, 69, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Harris, H.M.B.; McCann, A.; Guo, C.; Argimon, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mashak, K. Antimicrobial activity of lactobacillus isolated from kashk-e zard and tarkhineh, two Iranian traditional fermented foods. Int. J. Enteric Pathog. 2016, 4, e34692. [Google Scholar] [CrossRef]
- Goudarzi, L.; Kermanshahi, R.K.; Moosavinezhad, Z. Investigating the effect of probiotic bacteria on urease activity and swarming movement in Proteus species. Curr. Res. Microbiol. Biotechnol. 2016, 4, 827–834. [Google Scholar]
- Halder, D.; Mandal, S. Antibacterial potentiality of commercially available probiotic lactobacilli and curd lactobacilli strains, alone and in combination, against human pathogenic bacteria. Transl. Biomed. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Halder, D.; Mandal, S. Curd lactobacilli with probiotic potentiality. Transl. Biomed. 2015, 6, 1–6. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R. Bergey’s Manual of Systematic Bacteriology; Williams and Wilkins: Baltimore, MD, USA, 1984. [Google Scholar]
- Liong, M.T.; Shah, N.P. Acid and bile tolerance and cholesterol removal ability of lactobacilli strain. J. Dairy Sci. 2005, 88, 55–66. [Google Scholar] [CrossRef]
- Chowdhury, A.; Hossain, M.N.; Mostazir, N.J.; Fakruddin, M.; Billah, M.; Ahmed, M. Screening of Lactobacillus spp. from buffalo yoghurt for probiotic and antibacterial activity. J. Bacteriol. Parasitol. 2012, 3, 156. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Jahromi, M.F.; Ahmed, M. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed. Res. Int. 2014, 2, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Carasi, P.; Diaz, M.; Racedo, S.M.; Antoni, G.D.; Urdaci, M.C.; Serradell, M.A. Safety characterization and antimicrobial properties ofkefir-isolated Lactobacillus kefiri. Biomed. Res. Int. 2014, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pisano, M.B.; Viale, S.; Conti, S.; Fadda, M.; Deplano, M.; Melis, M.P.; Deiana, M.; Cosentino, S. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. Biomed. Res. Int. 2014, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tagg, J.R.; McGiven, A.R. Assay system for bacteriocins. Appl. Microbiol. 1971, 21, 943–944. [Google Scholar] [PubMed]
- Iyapparaj, P.; Maruthiah, T.; Ramasubburayan, R.; Prakash, S.; Kumar, C.; Immanuel, G.; Palavesam, A. Optimization of bacteriocin production by Lactobacillus sp. MSU3IR against shrimp bacterial pathogens. Aquat. Biosyst. 2013, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.J.; Kirby, W.; Turck, M. Antibiotic susceptibility testing by standardized single disc method. Am. J. Clin. Pathol. 1996, 45, 493–496. [Google Scholar]
- Saha, A.; Mandal, S. In vitro assessment of two commercial honey samples for antibacterial and antioxidant activities. Austin J. Trop. Med. Hyg. 2015, 1, 1002. [Google Scholar]
- Liasi, S.A.; Azmi, T.I.; Hassan, M.D.; Shuhaimi, M.; Rosfarizan, M. Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product Budu. Malays. J. Microbiol. 2009, 5, 33–37. [Google Scholar]
- Vlkova, E.; Rada, V.; Popelarova, P.; Trojanová, I.; Killer, J. Antimicrobial susceptibility of bifidobacteria isolated from gastrointestinal tract of calves. Livest. Sci. 2006, 105, 253–259. [Google Scholar] [CrossRef]
- Tambekar, D.H.; Bhutada, S.A. An evaluation of probiotic potential of Lactobacillus sp. from milk of domestic animals and commercial available probiotic preparations in prevention of enteric bacterial infections. Recent Res. Sci. Technol. 2010, 2, 82–88. [Google Scholar]
- Reardon, S. Microbiome therapy gains market traction. Nature 2014, 509, 269–270. [Google Scholar] [CrossRef] [PubMed]
- Samuel, S.S.; Shukla, S.; Ramteke, P.W.; Sushma. Isolation and identification of antagonistic Lactobacillus spp. isolated from dairy products against selected pathogens. J. Pharm. Innovat. 2016, 5, 8–13. [Google Scholar]
- Sahadeva, R.P.K.; Leong, S.F.; Chua, K.H. Survival of commercial probiotic strains to pH and bile. Int. Food Res. J. 2011, 18, 1515–1522. [Google Scholar]
- Koll, P.; Mandar, R.; Marcotte, H.; Leibur, E.; Mikelsaar, M.; Hammarstrom, L. Characterization of oral lactobacilli as potential probiotics for oral health. Oral Microbiol. Immunol. 2008, 23, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Mourad, K.; Nour-Eddine, K. In vitro preselection criteria for probiotic Lactobacillus plantarum strains of fermented olives origin. Int. J. Probiot. Prebiot. 2006, 1, 27–32. [Google Scholar]
- Ehrmann, M.A.; Kurzak, P.; Bauer, J.; Vogel, R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002, 92, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Jose, N.M.; Bunt, C.R.; Hussain, M.A. Comparison of microbiological and probiotic characteristics of lactobacilli isolates from dairy food products and animal rumen contents. Microorganisms 2015, 3, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, W.; Zhang, Q.; Tian, F.; Wang, G.; Zhang, H.; Chen, W. Screening of lactobacilli with antagonistic activity against enteroinvasive Escherichia coli. Food Control 2013, 30, 563–568. [Google Scholar] [CrossRef]
- Jeronymo-Ceneviva, A.B. Probiotic properties of lactic acid bacteria isolated from water buffalo mozzarella cheese. Probiot. Antimicrob. Protein 2014, 6, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.M.K. Probiotic properties analysis of isolated lactic acid bacteria from buffalo milk. Arch. Clin. Microbiol. 2015, 7, 1. [Google Scholar]
- Sjovall, P. On the concentration of bile acids in the human intestine during absorption, bile acids and sterioids 74. Acta Physiol. Scand. 1959, 46, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopouloua, G.; Miarisa, C.; Kalantzopoulosa, G.; Potb, B. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Hoque, M.Z.; Akter, F.; Hossain, K.M.; Rahman, M.S.M.; Billah, M.M.; Islam, K.M.D. Isolation, identification and analysis of probiotic properties of Lactobacillus spp. from selective regional yoghurts. World J. Dairy Food Sci. 2010, 5, 39–46. [Google Scholar]
- Manzoor, A.; Ul-Haq, I.; Baig, S.; Qazi, J.I.; Seratlic, S. Efficacy of locally isolated lactic acid bacteria against antibiotic-resistant uropathogens. Jundishapur J. Microbiol. 2016, 9, e18952. [Google Scholar] [CrossRef] [PubMed]
- Benavides, A.B.; Ulcuango, M.; Yepez, L.; Tenea, G.N. Assessment of the in vitro bioactive properties of lactic acid bacteria isolated from native ecological niches of Ecuador. Rev. Argent. Microbiol. 2016, 48, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Cadirci, B.H.; Citak, S. A comparison of two methods used for measuring antagonistic activity of lactic acid bacteria. Pak. J. Nutr. 2005, 4, 237–241. [Google Scholar]
- Rahimifard, N.; Naseri, M. Evaluation and comparison of three antimicrobial activity methods using Bifidobacteriabifidum and Bifidobacteria infantis as probiotic bacteria against Salmonella enterica serotype Enteritidis. J. Bacteriol. Mycol. 2016, 2, 00024. [Google Scholar] [CrossRef]
- Shehata, M.G.; Sohaimy, S.A.E.; El-Sahn, M.A.; Youssef, M.M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016, 61, 65–75. [Google Scholar] [CrossRef]
- Kaktcham, N.F.; Zambou, F.M.; El-Soda, T.M.; Choudhary, M.I. Antimicrobial and safety properties of lactobacilli isolated from two Cameroonian traditional fermented foods. Sci. Pharm. 2012, 80, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, R.; Yocheva, L.; Tserovska, L.; Zhelezova, G.; Stefanova, N.; Atanasova, A. Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol. Biotechnol. Equip. 2015, 29, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Salminen, S.; von Wright, A.; Morelli, L.; Marteau, P.; Brassart, D.; de Vos, W.M. Demonstration of safety of probiotics—A review. Int. J. Food Microbiol. 1998, 44, 93–106. [Google Scholar] [CrossRef]
- Tynkkynen, S.; Singh, K.V.; Varmanen, P. Vancomycin resistance factor of Lactobacillus rhamnosus GG in relation to enterococcal vancomycin resistance (van) genes. Int. J. Food Microbiol. 1998, 41, 195–204. [Google Scholar] [CrossRef]
- Gautam, N.; Sharma, S. Characterization of bacteriocin producer Lactobacillus brevis as potential probiotic strain. J. Microbiol. Biotechnol. Food Sci. 2015, 5, 216–220. [Google Scholar] [CrossRef]
- Handa, S.; Sharma, N. Evaluation of health benefits of lassi (Buttermilk): A traditional non alcoholic beverage of northern India. J. Innov. Biol. 2016, 3, 297–301. [Google Scholar]
- Sharma, N.; Gupta, A.; Handa, S. An exploration of rich microbial diversity of rare traditional functional foods of Trans Himalayan state of India with proven additional probiotic effect. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 999–1014. [Google Scholar]
- FAO and WHO. Guidelines for the Evaluation of Probiotics in Food: Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food; Food and Agriculture Organization; World Health Organization: London, ON, Canada, 2002. [Google Scholar]
| Lactobacilli Strains | NaCl (%) | pH | Bile Salt (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 6.5 | 2 | 3 | 4 | 0.125 | 0.25 | 0.5 | |
| L. animalis LMEM6 | + | + | + | + | + | + | + | + | + |
| L. plantarum LMEM7 | + | + | w | − | w | + | + | + | − |
| L. acidophilus LMEM8 | + | + | w | − | w | + | + | + | W |
| L. rhamnosus LMEM9 | + | + | + | w | w | + | + | + | + |
| Lactobacillus Isolates | ZDI (mm) ± SD for Indicator Bacteria | |||
|---|---|---|---|---|
| A. baumannii | E. coli | P. vulgaris | S. typhi | |
| L. animalis LMEM6 | 29.67 ± 0.58 | 28.25 ± 0.82 | 34.50 ± 0.71 | 30.50 ± 0.71 |
| L. plantarum LMEM7 | 32.33 ± 0.58 | 30.00 ± 1.71 | 35.67 ± 2.52 | 25.25 ± 1.71 |
| L. acidophilus LMEM8 | 15.25 ± 0.96 | 12.50 ± 1.29 | 12.50 ± 0.71 | 11.33 ± 0.58 |
| L. rhamnosus LMEM9 | 31.33 ± 1.53 | 22.67 ± 1.53 | 26.00 ± 2.16 | 20.67 ± 1.53 |
| Lactobacillus Isolates | R Value (mm), Mean ± SD, for the Pathogenic Indicator Bacteria | |||
|---|---|---|---|---|
| A. baumannii | E. coli | P. vulgaris | S. typhi | |
| L. animalis LMEM6 | 12.3 ± 0.29 | 11.50 ± 0.41 | 14.75 ± 0.35 | 12.75 ± 0.35 |
| L. plantarum LMEM7 | 13.67 ± 0.29 | 12.63 ± 0.85 | 15.33 ± 1.26 | 10.13 ± 0.85 |
| L. acidophilus LMEM8 | 5.13 ± 0.48 | 3.75 ± 0.65 | 3.75 ± 0.35 | 3.17 ± 0.29 |
| L. rhamnosus LMEM9 | 13.17 ± 0.76 | 8.83 ± 0.76 | 10.50 ± 1.08 | 7.83 ± 0.76 |
| Lactobacillus Isolates | ZDI (mm) ± SD for Indicator Bacteria | |||
|---|---|---|---|---|
| A. baumannii | E. coli | P. vulgaris | S. typhi | |
| L. animalis LMEM6 | 14.67 ± 0.58 | 21.67 ± 1.53 | 23.67 ± 1.53 | 18.33 ± 1.53 |
| L. plantarum LMEM7 | 16.00 ± 2.16 | 19.75 ± 1.89 | 14.67 ± 1.53 | 19.50 ± 1.30 |
| L. acidophilus LMEM8 | 13.67 ± 0.58 | 21.00 ± 1.00 | 15.67 ± 1.53 | 19.67 ± 0.58 |
| L. rhamnosus LMEM9 | 20.33 ± 1.53 | 29.50 ± 2.10 | 14.33 ± 1.53 | 20.00 ± 1.00 |
| Lactobacillus Isolates | Antagonistic Activity (AU/mL) | Average (AU/mL) | |||
|---|---|---|---|---|---|
| ST | PV | EC | AB | ||
| L. animalis LMEM6 | 244.45 ± 20.37 | 315.55 ± 20.37 | 288.89 ± 20.37 | 195.56 ± 7.70 | 261.11 ± 52.63 |
| L. plantarum LMEM7 | 260.00 ± 17.21 | 204.45 ± 20.37 | 263.34 ± 25.24 | 213.33 ± 28.81 | 235.28 ± 30.72 |
| L. acidophilus LMEM8 | 262.22 ± 7.70 | 208.90 ± 20.37 | 280.00 ± 13.33 | 182.22 ± 7.70 | 233.34 ± 45.54 |
| L. rhamnosus LMEM9 | 266.67 ± 13.34 | 191.11 ± 20.37 | 393.34 ± 27.76 | 271.11 ± 20.37 | 280.56 ± 83.67 |
| Strain | R (ZDI: ≤15 mm) | IS (ZDI: 16–20 mm) | S (ZDI: ≥21 mm) |
|---|---|---|---|
| L. animalis LMEM6 | Vm: 6; Ax: 15 | – | Am: 22; Tc 19; Gm: 19 |
| L. plantarum LMEM7 | Vm: 6 | Gm: 18 | Ax: 37; Am: 43; Tc: 29 |
| L. acidophilus LMEM8 | Vm: 6 | Gm: 20 | Ax: 27; Am: 31; Tc: 31 |
| L. rhamnosus LMEM9 | Vm: 6 | Am: 20; Gm: 18 | Ax: 23; Tc: 21 |
| Probiotic Characters | Indicator Score | Individual Probiotic Isolate Score | |||
|---|---|---|---|---|---|
| LMEM6 | LMEM7 | LMEM8 | LMEM9 | ||
| Acidic pH tolerance | Resistant = 1 Sensitive = 0 | 1 | 1 | 1 | 1 |
| Bile salt tolerance | Resistant = 1 Sensitive = 0 | 1 | 1 | 1 | 1 |
| Antagonistic activity(Average) | AU/mL 150–< 200 = 0.5; AU/mL ≥ 200 = 1 | 1 | 1 | 1 | 1 |
| ntibiotic sensitivity | Intrinsic resistance/Sensitive = 1 Other resistance = 0 | 0 | 1 | 1 | 1 |
| Hemolytic activity | β-hemolytic = 0 α-hemolytic = 0 γ-hemolytic = 1 | 1 | 1 | 1 | 1 |
| Total score | 5 | 4 | 5 | 5 | 5 |
| CPP for the lactobacilli | 80% | 100% | 100% | 100% | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halder, D.; Mandal, M.; Chatterjee, S.S.; Pal, N.K.; Mandal, S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines 2017, 5, 31. https://doi.org/10.3390/biomedicines5020031
Halder D, Mandal M, Chatterjee SS, Pal NK, Mandal S. Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria. Biomedicines. 2017; 5(2):31. https://doi.org/10.3390/biomedicines5020031
Chicago/Turabian StyleHalder, Debashis, Manisha Mandal, Shiv Sekhar Chatterjee, Nishith Kumar Pal, and Shyamapada Mandal. 2017. "Indigenous Probiotic Lactobacillus Isolates Presenting Antibiotic like Activity against Human Pathogenic Bacteria" Biomedicines 5, no. 2: 31. https://doi.org/10.3390/biomedicines5020031