Drosophila as a Rapid Screening Model to Evaluate the Hypoglycemic Effects of Dipeptidyl Peptidase 4 (DPP4) Inhibitors: High Evolutionary Conservation of DPP4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequences
2.2. Domain Analysis, Alignments, Phylogenetic Analysis, and Protein Modeling
2.3. Fly Strains and Maintenance
2.4. Treatment of Flies with Drugs
2.5. Capillary Feeding (CAFE) Assay
2.6. Fly Starvation Assay
2.7. Measurement of Fly Hemolymph Glucose
2.8. Measurement of Fly Protein
2.9. Measurement of Fly Triglycerides (TAGs)
2.10. Statistical Analysis
3. Results
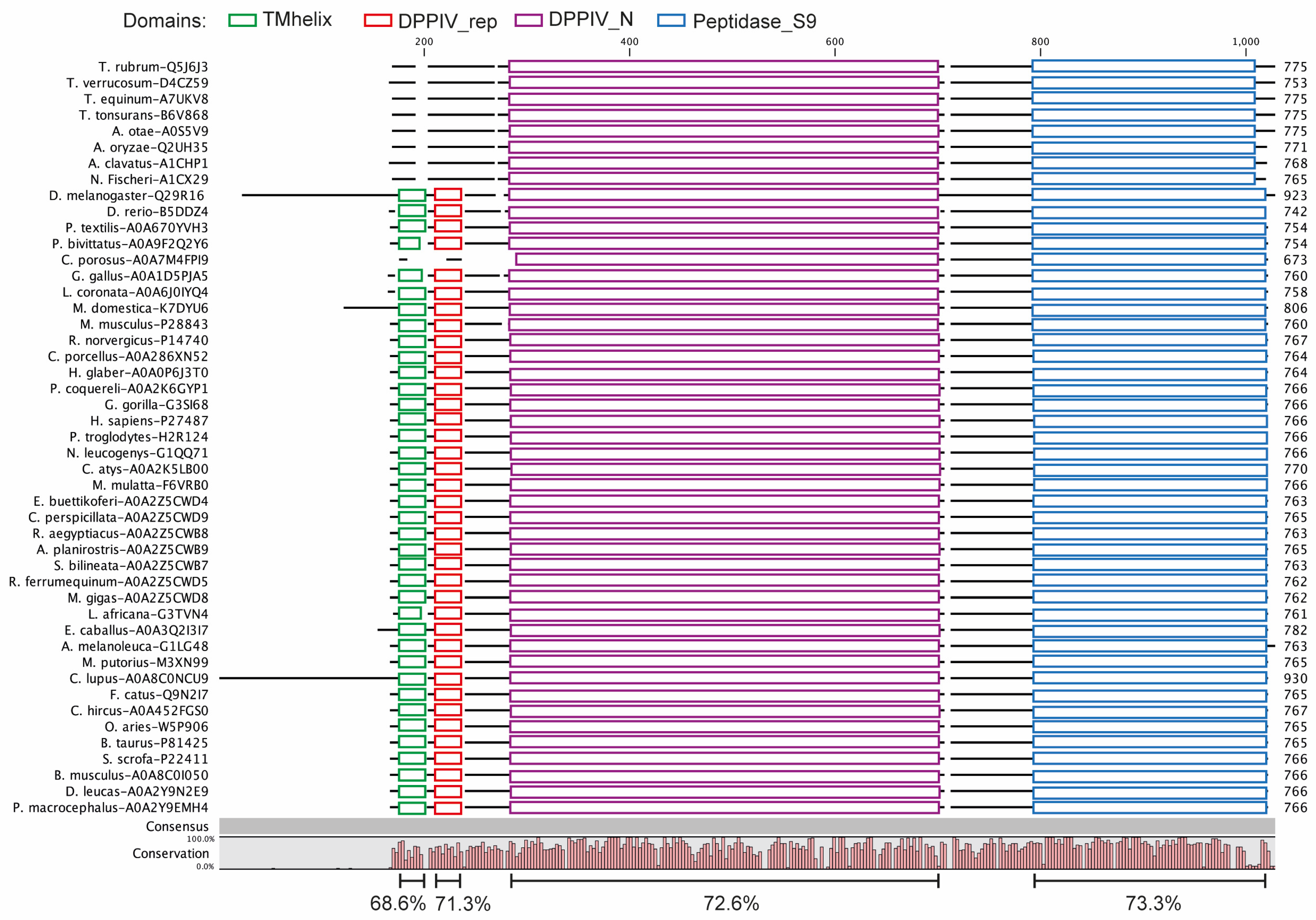
3.1. DPP4 Is an Evolutionarily Conserved Protein
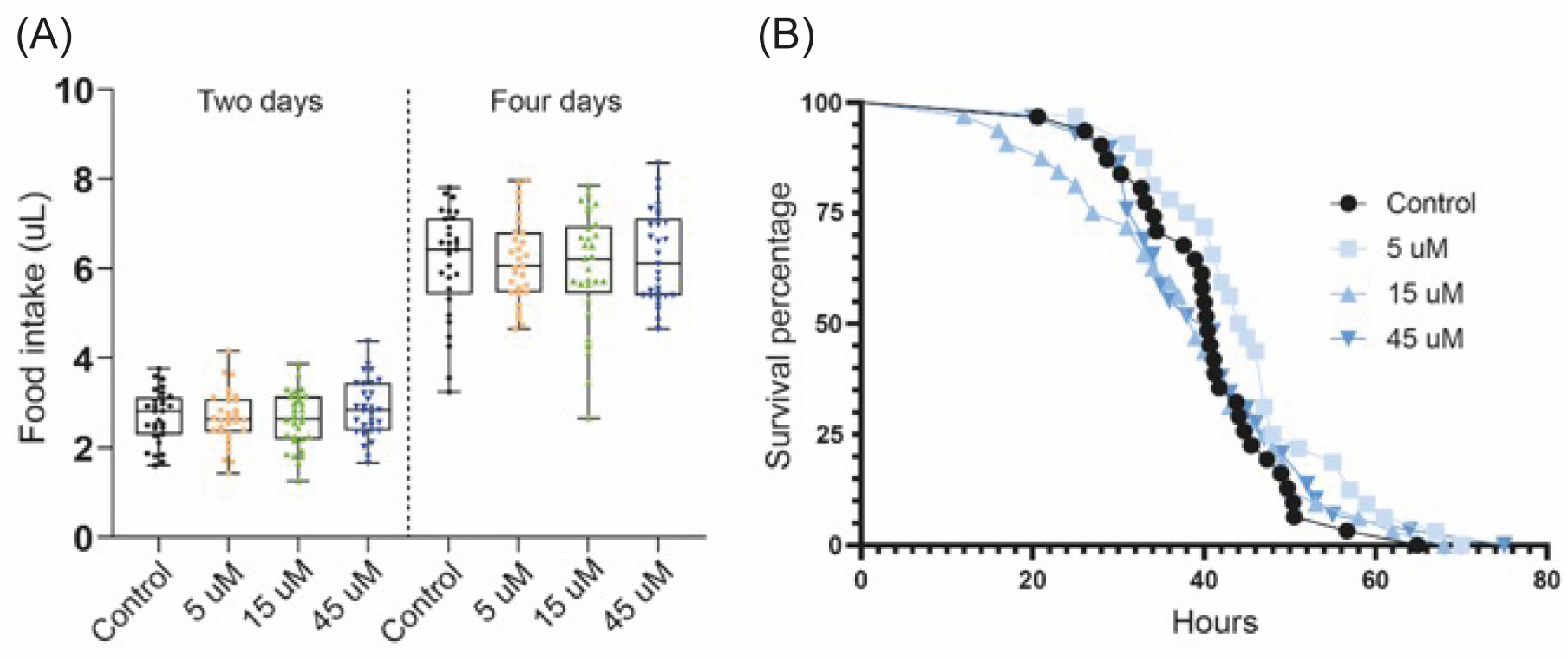
3.2. Diprotin A Does Not Alter Food Intake and Starvation Resistance in Fruit Flies
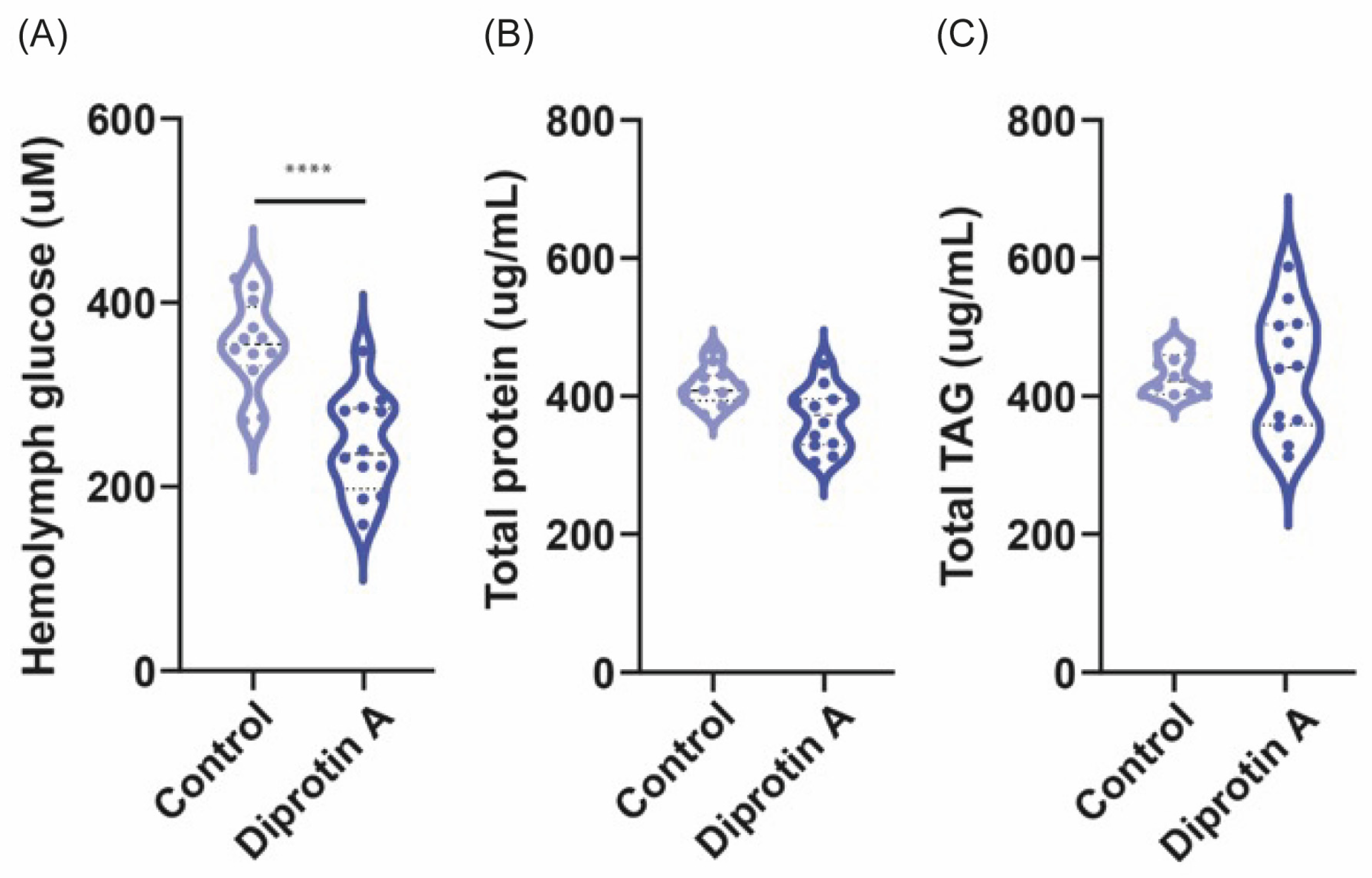
3.3. Diprotin A Reduces Hemolymph Glucose Levels but Not Protein and TAG Content in Fruit Flies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nargis, T.; Chakrabarti, P. Significance of Circulatory DPP4 Activity in Metabolic Diseases. IUBMB Life 2018, 70, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Enz, N.; Vliegen, G.; De Meester, I.; Jungraithmayr, W. CD26/DPP4—A Potential Biomarker and Target for Cancer Therapy. Pharmacol. Ther. 2019, 198, 135–159. [Google Scholar] [CrossRef] [PubMed]
- Röhrborn, D. DPP4 in Diabetes. Front. Immunol. 2015, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Mikhail, N. Safety of Dipeptidyl Peptidase 4 Inhibitors for Treatment of Type 2 Diabetes. Curr. Drug Saf. 2011, 6, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Deacon, C.F. Dipeptidyl Peptidase 4 Inhibitors in the Treatment of Type 2 Diabetes Mellitus. Nat. Rev. Endocrinol. 2020, 16, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Ahrén, B. DPP-4 Inhibition and the Path to Clinical Proof. Front. Endocrinol. 2019, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Banerjee, S.; Kumar, S.K.A.; Paira, P. Critical Role of Dipeptidyl Peptidase IV: A Therapeutic Target for Diabetes and Cancer. Mini-Rev. Med. Chem. 2019, 19, 88–97. [Google Scholar] [CrossRef]
- Gupta, V.; Kalra, S. Choosing a Gliptin. Indian J. Endocrinol. Metab. 2011, 15, 298–308. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Deacon, C.F. Dipeptidyl Peptidase-4 Inhibitors in the Treatment of Type 2 Diabetes: A Comparative Review. Diabetes Obes. Metab. 2011, 13, 7–18. [Google Scholar] [CrossRef]
- Marino, A.B.; Cole, S.W. Alogliptin. J. Pharm. Pract. 2015, 28, 99–106. [Google Scholar] [CrossRef]
- Matthews, D.R.; Paldánius, P.M.; Proot, P.; Chiang, Y.; Stumvoll, M.; Del Prato, S. Glycaemic Durability of an Early Combination Therapy with Vildagliptin and Metformin versus Sequential Metformin Monotherapy in Newly Diagnosed Type 2 Diabetes (VERIFY): A 5-Year, Multicentre, Randomised, Double-Blind Trial. Lancet 2019, 394, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Scirica, B.M.; Bhatt, D.L.; Braunwald, E.; Steg, P.G.; Davidson, J.; Hirshberg, B.; Ohman, P.; Frederich, R.; Wiviott, S.D.; Hoffman, E.B.; et al. Saxagliptin and Cardiovascular Outcomes in Patients with Type 2 Diabetes Mellitus. N. Engl. J. Med. 2013, 369, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.; Morgan, J.; Zimmer, Z.; Lam, R.L.H.; O’Neill, E.A.; Kaufman, K.D.; Engel, S.S.; Raji, A. A Randomized Clinical Trial of the Efficacy and Safety of Sitagliptin Compared with Dapagliflozin in Patients with Type 2 Diabetes Mellitus and Mild Renal Insufficiency: The CompoSIT-R Study. Diabetes Obes. Metab. 2018, 20, 2876–2884. [Google Scholar] [CrossRef] [PubMed]
- McMurray, J.J.V.; Ponikowski, P.; Bolli, G.B.; Lukashevich, V.; Kozlovski, P.; Kothny, W.; Lewsey, J.D.; Krum, H. Effects of Vildagliptin on Ventricular Function in Patients With Type 2 Diabetes Mellitus and Heart Failure. JACC Heart Fail. 2018, 6, 8–17. [Google Scholar] [CrossRef]
- DeVries, J.H.; Rosenstock, J. DPP-4 Inhibitor–Related Pancreatitis: Rare but Real! Diabetes Care 2017, 40, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Dicembrini, I.; Montereggi, C.; Nreu, B.; Mannucci, E.; Monami, M. Pancreatitis and Pancreatic Cancer in Patientes Treated with Dipeptidyl Peptidase-4 Inhibitors: An Extensive and Updated Meta-Analysis of Randomized Controlled Trials. Diabetes Res. Clin. Pract. 2020, 159, 107981. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, E.; Li, W.; Wang, J. Association between Dipeptidyl Peptidase-4 Inhibitor Drugs and Risk of Acute Pancreatitis. Medicine 2017, 96, e8952. [Google Scholar] [CrossRef] [PubMed]
- Men, P.; He, N.; Song, C.; Zhai, S. Dipeptidyl Peptidase-4 Inhibitors and Risk of Arthralgia: A Systematic Review and Meta-Analysis. Diabetes Metab. 2017, 43, 493–500. [Google Scholar] [CrossRef]
- Sayiner, Z. DPP-4 Inhibitors Increase the Incidence of Arthritis/Arthralgia but Do Not Affect Autoimmunity. Acta Endocrinol. 2018, 14, 473–476. [Google Scholar] [CrossRef]
- Wang, C.Y.; Fu, S.H.; Yang, R.S.; Hsiao, F.Y. Use of Dipeptidyl Peptidase-4 Inhibitors and the Risk of Arthralgia: Population-Based Cohort and Nested Case–Control Studies. Pharmacoepidemiol. Drug Saf. 2019, 28, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Freires, I.A.; Sardi, J.d.C.O.; de Castro, R.D.; Rosalen, P.L. Alternative Animal and Non-Animal Models for Drug Discovery and Development: Bonus or Burden? Pharm. Res. 2017, 34, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Su, T.T. Drug Screening in Drosophila; Why, When, and When Not? WIREs Dev. Biol. 2019, 8, e346. [Google Scholar] [CrossRef]
- Tickoo, S. Drosophila Melanogaster as a Model System for Drug Discovery and Pathway Screening. Curr. Opin. Pharmacol. 2002, 2, 555–560. [Google Scholar] [CrossRef]
- Hoyer-Fender, S. Transgenerational Effect of Drug-Mediated Inhibition of LSD1 on Eye Pigment Expression in Drosophila. BMC Ecol. 2020, 20, 62. [Google Scholar] [CrossRef] [PubMed]
- Brogiolo, W.; Stocker, H.; Ikeya, T.; Rintelen, F.; Fernandez, R.; Hafen, E. An Evolutionarily Conserved Function of the Drosophila Insulin Receptor and Insulin-like Peptides in Growth Control. Curr. Biol. 2001, 11, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Laiolo, P.; Pato, J.; Jiménez-Alfaro, B.; Obeso, J.R. Evolutionary Conservation of Within-Family Biodiversity Patterns. Nat. Commun. 2020, 11, 882. [Google Scholar] [CrossRef]
- Chihara, C.J.; Song, C.; LaMonte, G.; Fetalvero, K.; Hinchman, K.; Phan, H.; Pineda, M.; Robinson, K.; Schneider, G.P. Identification and Partial Characterization of the Enzyme of Omega: One of Five Putative DPP IV Genes in Drosophila Melanogaster. J. Insect Sci. 2005, 5, 26. [Google Scholar] [CrossRef]
- Peters, I.D.; Rancourt, D.E.; Davies, P.L.; Walker, V.K. Isolation and Characterization of an Antifreeze Protein Precursor from Transgenic Drosophila: Evidence for Partial Processing. Biochim. Biophys. Acta-Gene Struct. Expr. 1993, 1171, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Ja, W.W.; Carvalho, G.B.; Mak, E.M.; De La Rosa, N.N.; Fang, A.Y.; Liong, J.C.; Brummel, T.; Benzer, S. Prandiology of Drosophila and the CAFE Assay. Proc. Natl. Acad. Sci. USA 2007, 104, 8253–8256. [Google Scholar] [CrossRef] [PubMed]
- Mattila, J.; Hietakangas, V. Regulation of Carbohydrate Energy Metabolism in Drosophila Melanogaster. Genetics 2017, 207, 1231–1253. [Google Scholar] [CrossRef]
- Kawai, T.; Choi, U.; Liu, P.-C.; Whiting-Theobald, N.L.; Linton, G.F.; Malech, H.L. Diprotin A Infusion into Nonobese Diabetic/Severe Combined Immunodeficiency Mice Markedly Enhances Engraftment of Human Mobilized CD34 + Peripheral Blood Cells. Stem Cells Dev. 2007, 16, 361–370. [Google Scholar] [CrossRef]
- Shiina, Y.; Muto, T.; Zhang, Z.; Baihaqie, A.; Yoshizawa, T.; Lee, H.J.; Park, E.; Tsukiji, S.; Takimoto, K. Fly DPP10 Acts as a Channel Ancillary Subunit and Possesses Peptidase Activity. Sci. Rep. 2016, 6, 26290. [Google Scholar] [CrossRef]
- Sun, C.; Xiao, Y.; Li, J.; Ge, B.; Chen, X.; Liu, H.; Zheng, T. Nonenzymatic Function of DPP4 in Diabetes-associated Mitochondrial Dysfunction and Cognitive Impairment. Alzheimer’s Dement. 2021, 18, 966–987. [Google Scholar] [CrossRef]
- Ricklefs, R.E.; Renner, S.S. Global Correlations in Tropical Tree Species Richness and Abundance Reject Neutrality. Science 2012, 335, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Xu, Q.; Yu, X.; Pan, R.; Chen, Y. Dipeptidyl Peptidase 4 Inhibitors and Their Potential Immune Modulatory Functions. Pharmacol. Ther. 2020, 209, 107503. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Arora, T.; Joshi, V.; Rathor, N.; Mehta, A.; Mehta, K.; Mediratta, P. Substitute of Animals in Drug Research: An Approach towards Fulfillment of 4R′s. Indian J. Pharm. Sci. 2011, 73, 1. [Google Scholar] [CrossRef] [PubMed]
- MacArthur Clark, J. The 3Rs in Research: A Contemporary Approach to Replacement, Reduction and Refinement. Br. J. Nutr. 2018, 120, S1–S7. [Google Scholar] [CrossRef] [PubMed]
- Jessen, L.; Aulinger, B.A.; Hassel, J.L.; Roy, K.J.; Smith, E.P.; Greer, T.M.; Woods, S.C.; Seeley, R.J.; D’Alessio, D.A. Suppression of Food Intake by Glucagon-Like Peptide-1 Receptor Agonists: Relative Potencies and Role of Dipeptidyl Peptidase-4. Endocrinology 2012, 153, 5735–5745. [Google Scholar] [CrossRef] [PubMed]
- Kohno, D.; Furusawa, K.; Kitamura, T. Anagliptin Suppresses Diet-Induced Obesity through Enhancing Leptin Sensitivity and Ameliorating Hyperphagia in High-Fat High-Sucrose Diet Fed Mice. Endocr. J. 2020, 67, 523–529. [Google Scholar] [CrossRef]
- Liao, X.; Song, L.; Zeng, B.; Liu, B.; Qiu, Y.; Qu, H.; Zheng, Y.; Long, M.; Zhou, H.; Wang, Y.; et al. Alteration of Gut Microbiota Induced by DPP-4i Treatment Improves Glucose Homeostasis. EBioMedicine 2019, 44, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Neyrinck, A.M.; Pötgens, S.A.; Beaumont, M.; Salazar, N.; Cani, P.D.; Bindels, L.B.; Delzenne, N.M. The DPP-4 Inhibitor Vildagliptin Impacts the Gut Microbiota and Prevents Disruption of Intestinal Homeostasis Induced by a Western Diet in Mice. Diabetologia 2018, 61, 1838–1848. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, X.; Li, M.; Yu, M.; Ping, F.; Zheng, J.; Wang, T.; Wang, X. Vildagliptin Increases Butyrate-Producing Bacteria in the Gut of Diabetic Rats. PLoS ONE 2017, 12, e0184735. [Google Scholar] [CrossRef]
- Chaston, J.M.; Dobson, A.J.; Newell, P.D.; Douglas, A.E. Host Genetic Control of the Microbiota Mediates the Drosophila Nutritional Phenotype. Appl. Environ. Microbiol. 2016, 82, 671–679. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.-H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.-A.; Yoon, J.-H.; Ryu, J.-H.; Lee, W.-J. Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Chaston, J.M.; Newell, P.D.; Douglas, A.E. Metagenome-Wide Association of Microbial Determinants of Host Phenotype in Drosophila Melanogaster. mBio 2014, 5, e01631-14. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Yoshida, H. Drosophila as a Model Organism. In Drosophila Models for Human Diseases; Springer Nature: Singapore, 2018; pp. 1–10. [Google Scholar]
- Graham, P.; Pick, L. Drosophila as a Model for Diabetes and Diseases of Insulin Resistance. In Current Topics in Developmental Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 121, pp. 397–419. [Google Scholar]
- Inoue, Y.H.; Katsube, H.; Hinami, Y. Drosophila Models to Investigate Insulin Action and Mechanisms Underlying Human Diabetes Mellitus. In Drosophila Models for Human Diseases; Springer Nature: Singapore, 2018; pp. 235–256. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagunas-Rangel, F.A.; Liao, S.; Williams, M.J.; Trukhan, V.; Fredriksson, R.; Schiöth, H.B. Drosophila as a Rapid Screening Model to Evaluate the Hypoglycemic Effects of Dipeptidyl Peptidase 4 (DPP4) Inhibitors: High Evolutionary Conservation of DPP4. Biomedicines 2023, 11, 3032. https://doi.org/10.3390/biomedicines11113032
Lagunas-Rangel FA, Liao S, Williams MJ, Trukhan V, Fredriksson R, Schiöth HB. Drosophila as a Rapid Screening Model to Evaluate the Hypoglycemic Effects of Dipeptidyl Peptidase 4 (DPP4) Inhibitors: High Evolutionary Conservation of DPP4. Biomedicines. 2023; 11(11):3032. https://doi.org/10.3390/biomedicines11113032
Chicago/Turabian StyleLagunas-Rangel, Francisco Alejandro, Sifang Liao, Michael J. Williams, Vladimir Trukhan, Robert Fredriksson, and Helgi B. Schiöth. 2023. "Drosophila as a Rapid Screening Model to Evaluate the Hypoglycemic Effects of Dipeptidyl Peptidase 4 (DPP4) Inhibitors: High Evolutionary Conservation of DPP4" Biomedicines 11, no. 11: 3032. https://doi.org/10.3390/biomedicines11113032
APA StyleLagunas-Rangel, F. A., Liao, S., Williams, M. J., Trukhan, V., Fredriksson, R., & Schiöth, H. B. (2023). Drosophila as a Rapid Screening Model to Evaluate the Hypoglycemic Effects of Dipeptidyl Peptidase 4 (DPP4) Inhibitors: High Evolutionary Conservation of DPP4. Biomedicines, 11(11), 3032. https://doi.org/10.3390/biomedicines11113032





