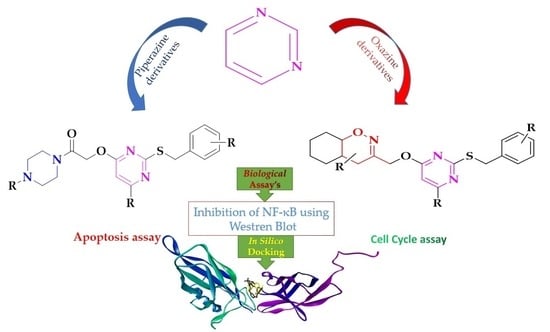
Development of Piperazine- and Oxazine-Linked Pyrimidines as p65 Subunit Binders of NF–κB in Human Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Procedure for the Synthesis of Oxazine and Piperazine Clubbed Pyrimidine Derivatives
2.1.1. Synthesis of Compound 2
2.1.2. Synthesis of Compound 3
2.1.3. Synthesis of Compounds 4/5
2.1.4. Synthesis of Compound 5a
2.1.5. Synthesis of Compound 5b/f/k/o
2.1.6. Synthesis of Compounds 5d/h/m/q
2.1.7. Synthesis of Compounds 5c/g/l/p
2.1.8. Synthesis of Compounds 5e/i/j/n
2.1.9. 6,6–Dimethyl–3–(((2–((3–methylbenzyl)thio)pyrimidin–4–yl)oxy)methyl)–4–phenyl –5,6–dihydro–4H–1,2–oxazine (3a)
2.1.10. 4–(4–Methoxyphenyl)–3–(((2–((3–methylbenzyl)thio)pyrimidin–4–yl)oxy)methyl)-4a,5,–6,7,8,8a–hexahydro–4H–benzo[e][1,2]oxazine (3b)
2.1.11. 4–(4–Chlorophenyl)–6,6–dimethyl–3–(((2–((3–methylbenzyl)thio)pyrimidin–4–yl)oxy)–methyl)–5,6–dihydro–4H–1,2–oxazine (3c)
2.1.12. 3–(((2–((4–Chlorobezyl)thio)pyrimidin–4–yl)oxy)methyl)–6,6–dimethyl–4–phenyl–5,6–dihydro–4H–1,2–oxazine (3d)
2.1.13. 3–(((2–((4–Chlorobezyl)thio)pyrimidin–4–yl)oxy)methyl)–4–(4–methoxyphenyl)–4a,5,6,–7,8,8a–hexahydro–4H–benzo[e][1,2]oxazine (3e)
2.1.14. 3–(((2–((4–Chlorobezyl)thio)pyrimidin–4–yl)oxy)methyl)–4–(4–chlorophenyl)–6,6–di–methyl–5,6–dihydro–4H–1,2–oxazine (3f)
2.1.15. 3–(((2–((4–Fluorobenzyl)thio)pyrimidin–4–yl)oxy)methyl)–6,6–dimethyl–4–phenyl–5,6–dihydro–4H–1,2–oxazine (3g)
2.1.16. 3–(((2–((4–Fluorobenzyl)thio)pyrimidin–4–yl)oxy)methyl)–4–(4–methoxyphenyl)–4a,5,6,–7,8, 8a–hexahydro–4H–benzo[e][1,2]oxazine (3h)
2.1.17. 4–(4–Chlorophenyl)–3–(((2–((4–fluorobenzyl)thio)pyrimidin–4–yl)oxy)methyl)–6,6–dimethyl–5,6–dihydro–4H–1,2–oxazine (3i)
2.1.18. 3–(((2–((4–Fluorobenzyl)thio)–6–methylpyrimidin–4–yl)oxy)methyl)–6,6–dimethyl–4–phenyl–5,6–dihydro–4H–1,2–oxazine (3j)
2.1.19. 3–(((2–((4–Fluorobenzyl)thio)–6–methylpyrimidin–4–yl)oxy)methyl)–4–(4–methoxy–phenyl)–4a,5,6,7,8,8a–hexahydro–4H–benzo[e][1,2]oxazine (3k)
2.1.20. 4–(4–Chlorophenyl)–3–(((2–((4–fluorobenzyl)thio)–6–methylpyrimidin–4–yl)oxy)–methyl)–6,6–dimethyl–5,6–dihydro–4H–1,2–oxazine (3l)
2.1.21. 1,1′–(Piperazine–1,4–diyl)bis(2–((2–((4–methoxybenzyl)thio)pyrimidin–4–yl)oxy)ethan–one) (5a)
2.1.22. 1–(4–Acetylpiperazin–1–yl)–2–((2–((4–methoxybenzyl)thio)pyrimidin–4–yl)oxy)ethan–one (5b)
2.1.23. Tert–butyl 4–(2–((2–((4–Methoxybenzyl)thio)pyrimidin–4–yl)oxy)acetyl)piperazine–1–carboxylate (5c)
2.1.24. 1–(4–(5–Bromopicolinoyl)piperazin–1–yl)–2–((2–((4–methoxybenzyl)thio)pyramidin–4–yl)–oxy)ethanone (5d)
2.1.25. 1–(4–Acetylpiperazin–1–yl)–2–((2–((3–methylbenzyl)thio)pyrimidin–4–yl)oxy)–ethanone (5e)
2.1.26. 2–((2–((3–Methylbenzyl)thio)pyrimidin–4–yl)oxy)–1–(piperazin–1–yl)ethanone (5f)
2.1.27. Tert–butyl–4–(2–((2–((3–methylbenzyl)thio)pyrimidin–4–yl)oxy)acetyl)piperazine–1–car–boxylate (5g)
2.1.28. 1–(4–(5–Bromopicolinoyl)piperazin–1–yl)–2–((2–((3–methylbenzyl)thio)pyrimidin–4–yl)–oxy)–ethanone (5h)
2.1.29. 1–(4–(5–Bromopicolinoyl)piperazin–1–yl)–2–((2–((4–fluorobenzyl)thio)pyrimidin–4–yl)–oxy)–ethanone (5i)
2.1.30. 1–(4–Acetylpiperazin–1–yl)–2–((2–((4–fluorobenzyl)thio)pyrimidin–4–yl)oxy)–ethanone (5j)
2.1.31. 2–((2–((4–Fluorobenzyl)thio)pyrimidin–4–yl)oxy)–1–(piperazin–1–yl)ethanone (5k)
2.1.32. Tert–butyl–4–(2–((2–((4–fluorobenzyl)thio)pyrimidin–4–yl)oxy)acetyl)piperazine–1–car–boxylate (5l)
2.1.33. 1–(4–(5–Bromopicolinoyl)piperazin–1–yl)–2–((2–((4–chlorobenzyl)thio)pyrimidin–4–yl)–oxy)–ethanone (5m)
2.1.34. 1–(4–Acetylpiperazin–1–yl)–2–((2–((4–chlorobenzyl)thio)pyrimidin–4–yl)oxy)–ethanone (5n)
2.1.35. 2–((2–((4–Chlorobenzyl)thio)pyrimidin–4–yl)oxy)–1–(piperazin–1–yl)ethanone (5o)
2.1.36. Tert–butyl–4–(2–((2–((4–chlorobenzyl)thio)pyrimidin–4–yl)oxy)acetyl)piperazine–1–car–boxylate (5p)
2.2. Cell Viability Assay
2.3. Annexin V Apoptosis and Cell Cycle Analysis Assay
2.4. Western Blot Analysis
2.5. Data Analysis and Statistics
2.6. Molecular Docking
3. Results
3.1. Synthesis of Piperazine- and Oxazine-Linked Pyrimidine Derivatives
3.2. Efficacy of Pyrimidine Derivatives in Breast Cancer Cells
3.3. Title Compounds Induce Apoptosis in MCF–7 Cells
3.4. Lead Compounds Arrest MCF–7 Cell Cycle at the Sub-G1 Phase
3.5. Lead Compounds Inhibited the Phosphorylation of Human p65 Protein (Serine–536 Amino Acid) of NF–κB Subunit in MCF–7 Cells
3.6. In Silico Analysis of Novel Compounds 3a and 5b Targeting the NF–Kappa B p65 Subunit
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2019. CA A Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA A Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Baltimore, D. Multiple Nuclear Factors Interact with the Immunoglobulin Enhancer Sequences. Cell 1986, 46, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, F.; Sarabi, P.Z.; Athari, S.S.; Esmaeilzadeh, A. Therapeutics Strategies against Cancer Stem Cell in Breast Cancer. Int. J. Biochem. Cell Biol. 2019, 109, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, J.; Chen, H.; Li, X.; Ye, C.; Zhang, F.; Zhang, Z.; Yao, Q.; Guo, Y. Curcumin Targeting NF–κB/Ubiquitin–Proteasome–System Axis Ameliorates Muscle Atrophy in Triple–Negative Breast Cancer Cachexia Mice. Mediat. Inflamm. 2022, 2022, 2567150. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Ghosh, S. The NF–κB Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF–κB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef]
- Miller, S.C.; Huang, R.; Sakamuru, S.; Shukla, S.J.; Attene–Ramos, M.S.; Shinn, P.; Van Leer, D.; Leister, W.; Austin, C.P.; Xia, M. Identification of Known Drugs That Act as Inhibitors of NF–κB Signaling and Their Mechanism of Action. Biochem. Pharmacol. 2010, 79, 1272–1280. [Google Scholar] [CrossRef]
- Sun, X.-M.; Bratton, S.B.; Butterworth, M.; MacFarlane, M.; Cohen, G.M. Bcl–2 and Bcl–xL Inhibit CD95–Mediated Apoptosis by Preventing Mitochondrial Release of Smac/DIABLO and Subsequent Inactivation of X–Linked Inhibitor–of–Apoptosis Protein. J. Biol. Chem. 2002, 277, 11345–11351. [Google Scholar] [CrossRef]
- Perkins, N.D.; Felzien, L.K.; Betts, J.C.; Leung, K.; Beach, D.H.; Nabel, G.J. Regulation of NF–κB by Cyclin–Dependent Kinases Associated with the P300 Coactivator. Science 1997, 275, 523–527. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin–Dependent Kinases in Cell–Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [PubMed]
- Baud, V.; Karin, M. Is NF–κB a Good Target for Cancer Therapy? Hopes and Pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Frantz, B.; O’Neill, E.A. The Effect of Sodium Salicylate and Aspirin on NF–κB. Science 1995, 270, 2017–2018. [Google Scholar] [CrossRef] [PubMed]
- Kopp, E.; Ghosh, S. Inhibition of NF–κB by Sodium Salicylate and Aspirin. Science 1994, 265, 956–959. [Google Scholar] [CrossRef] [PubMed]
- Mercogliano, M.F.; Bruni, S.; Elizalde, P.V.; Schillaci, R. Tumor Necrosis Factor α Blockade: An Opportunity to Tackle Breast Cancer. Front. Oncol. 2020, 10, 584. [Google Scholar] [CrossRef] [PubMed]
- Chylińska, J.B.; Janowiec, M.; Urbański, T. Antibacterial Activity of Dihydro–1,3–Oxazine Derivatives Condensed with Aromatic Rings in Positions 5, 6. Br. J. Pharmacol. 1971, 43, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-H.; Guo, H.-Y.; Deng, H.; Li, J.; Quan, Z.-S. Piperazine Skeleton in the Structural Modification of Natural Products: A Review. J. Enzym. Inhib. Med. Chem. 2021, 36, 1165–1197. [Google Scholar] [CrossRef] [PubMed]
- Jalageri, M.D.; Nagaraja, A.; Puttaiahgowda, Y.M. Piperazine Based Antimicrobial Polymers: A Review. RSC Adv. 2021, 11, 15213–15230. [Google Scholar] [CrossRef]
- Akl, L.; Abd El-Hafeez, A.A.; Ibrahim, T.M.; Salem, R.; Marzouk, H.M.M.; El-Domany, R.A.; Ghosh, P.; Eldehna, W.M.; Abou-Seri, S.M. Identification of Novel Piperazine-Tethered Phthalazines as Selective CDK1 Inhibitors Endowed with in Vitro Anticancer Activity toward the Pancreatic Cancer. Eur. J. Med. Chem. 2022, 243, 114704. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, W.; Ding, X.; Zhang, L.; Xia, Q.; Wang, Q.; Chen, Q.; Gao, Q.; Yan, J.; Lesyk, R.; et al. Design, Synthesis, and Anticancer Evaluation of Nitrobenzoxadiazole-Piperazine Hybrids as Potent pro-Apoptotic Agents. Tetrahedron 2023, 138, 133393. [Google Scholar] [CrossRef]
- Walayat, K.; Mohsin, N.-A.; Aslam, S.; Ahmad, M. An Insight into the Therapeutic Potential of Piperazine-Based Anticancer Agents. Turk. J. Chem. 2019, 43, 1–23. [Google Scholar] [CrossRef]
- Samie, N.; Muniandy, S.; Kanthimathi, M.S.; Haerian, B.S.; Raja Azudin, R.E. Novel Piperazine Core Compound Induces Death in Human Liver Cancer Cells: Possible Pharmacological Properties. Sci. Rep. 2016, 6, 24172. [Google Scholar] [CrossRef] [PubMed]
- Rinne, M.; Mätlik, K.; Ahonen, T.; Vedovi, F.; Zappia, G.; Moreira, V.M.; Yli-Kauhaluoma, J.; Leino, S.; Salminen, O.; Kalso, E.; et al. Mitoxantrone, Pixantrone and Mitoxantrone (2-Hydroxyethyl)Piperazine Are Toll-like Receptor 4 Antagonists, Inhibit NF-κB Activation, and Decrease TNF-Alpha Secretion in Primary Microglia. Eur. J. Pharm. Sci. 2020, 154, 105493. [Google Scholar] [CrossRef] [PubMed]
- Deveshegowda, S.N.; Metri, P.K.; Shivakumar, R.; Yang, J.-R.; Rangappa, S.; Swamynayaka, A.; Shanmugam, M.K.; Nagaraja, O.; Madegowda, M.; Babu Shubha, P.; et al. Development of 1-(4-(Substituted)Piperazin-1-Yl)-2-((2-((4-Methoxybenzyl)Thio)Pyrimidin-4-Yl)Oxy)Ethanones That Target Poly (ADP-Ribose) Polymerase in Human Breast Cancer Cells. Molecules 2022, 27, 2848. [Google Scholar] [CrossRef] [PubMed]
- Ramadass, V.; Vaiyapuri, T.; Tergaonkar, V. Small Molecule NF–κB Pathway Inhibitors in Clinic. Int. J. Mol. Sci. 2020, 21, 5164. [Google Scholar] [CrossRef]
- Mansouri, S.G.; Zali-Boeini, H.; Zomorodian, K.; Khalvati, B.; Pargali, R.H.; Dehshahri, A.; Rudbari, H.A.; Sahihi, M.; Chavoshpour, Z. Synthesis of Novel Naphtho [1,2-e][1,3]Oxazines Bearing an Arylsulfonamide Moiety and Their Anticancer and Antifungal Activity Evaluations. Arab. J. Chem. 2020, 13, 1271–1282. [Google Scholar] [CrossRef]
- Yousif, M.N.M.; Fathy, U.; Yousif, N.M. Synthesis and Anticancer Activity of Novel Chromene Derivatives, Chromeno[2,3-d][1,3]Oxazines, and Chromeno[2,3-d]Pyrimidines. Med. Chem. 2023, 19, 578–585. [Google Scholar] [CrossRef]
- Olivera, A.; Moore, T.W.; Hu, F.; Brown, A.P.; Sun, A.; Liotta, D.C.; Snyder, J.P.; Yoon, Y.; Shim, H.; Marcus, A.I.; et al. Inhibition of the NF-κB Signaling Pathway by the Curcumin Analog, 3,5-Bis(2-Pyridinylmethylidene)-4-Piperidone (EF31): Anti-Inflammatory and Anti-Cancer Properties. Int. Immunopharmacol. 2012, 12, 368–377. [Google Scholar] [CrossRef]
- Ananthula, S.; Parajuli, P.; Behery, F.; Ayoubi, A.; El Sayed, K.; Nazzal, S.; Sylvester, P. Abstract P3-03-11: Oxazine Derivatives of g- and D- Tocotrienols Display Potent Anticancer Effects in Vivo. Cancer Res. 2013, 73, P3-03. [Google Scholar] [CrossRef]
- Ansari, N.; Khodagholi, F.; Amini, M.; Shaerzadeh, F. Attenuation of LPS-Induced Apoptosis in NGF-Differentiated PC12 Cells via NF-κB Pathway and Regulation of Cellular Redox Status by an Oxazine Derivative. Biochimie 2011, 93, 899–908. [Google Scholar] [CrossRef]
- Nirvanappa, A.C.; Mohan, C.D.; Rangappa, S.; Ananda, H.; Sukhorukov, A.Y.; Shanmugam, M.K.; Sundaram, M.S.; Nayaka, S.C.; Girish, K.S.; Chinnathambi, A.; et al. Novel Synthetic Oxazines Target NF–κB in Colon Cancer In Vitro and Inflammatory Bowel Disease In Vivo. PLoS ONE 2016, 11, e0163209. [Google Scholar] [CrossRef] [PubMed]
- Somu, C.; Mohan, C.D.; Ambekar, S.; Dukanya; Rangappa, S.; Baburajeev, C.; Sukhorukov, A.; Mishra, S.; Shanmugam, M.K.; Chinnathambi, A.; et al. Identification of a Novel 1,2 Oxazine That Can Induce Apoptosis by Targeting NF–κB in Hepatocellular Carcinoma Cells. Biotechnol. Rep. 2020, 25, e00438. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Bharathkumar, H.; Dukanya; Rangappa, S.; Shanmugam, M.K.; Chinnathambi, A.; Alharbi, S.A.; Alahmadi, T.A.; Bhattacharjee, A.; Lobie, P.E.; et al. N–Substituted Pyrido–1,4–Oxazin–3–Ones Induce Apoptosis of Hepatocellular Carcinoma Cells by Targeting NF–κB Signaling Pathway. Front. Pharmacol. 2018, 9, 1125. [Google Scholar] [CrossRef] [PubMed]
- Al-Khawaldeh, I.; Al Yasiri, M.J.; Aldred, G.G.; Basmadjian, C.; Bordoni, C.; Harnor, S.J.; Heptinstall, A.B.; Hobson, S.J.; Jennings, C.E.; Khalifa, S.; et al. An Alkynylpyrimidine-Based Covalent Inhibitor That Targets a Unique Cysteine in NF-κB-Inducing Kinase. J. Med. Chem. 2021, 64, 10001–10018. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Gao, Z.-F.; Yan, W.-B.; Yao, B.-R.; Xin, W.-Y.; Wang, C.-H.; Meng, Q.-G.; Hou, G.-G. Discovery of Novel NF–κB Inhibitor Based on Scaffold Hopping: 1,4,5,6,7,8–Hexahydropyrido[4,3–d]Pyrimidine. Eur. J. Med. Chem. 2020, 198, 112366. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.-H.; Kim, J.S.; Kim, B.M. Novel Heterocycle-Substituted Pyrimidines as Inhibitors of NF-κB Transcription Regulation Related to TNF-α Cytokine Release. Bioorganic Med. Chem. Lett. 2008, 18, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Basappa, B.; Chumadathil Pookunoth, B.; Shinduvalli Kempasiddegowda, M.; Knchugarakoppal Subbegowda, R.; Lobie, P.E.; Pandey, V. Novel Biphenyl Amines Inhibit Oestrogen Receptor (ER)–α in ER–Positive Mammary Carcinoma Cells. Molecules 2021, 26, 783. [Google Scholar] [CrossRef] [PubMed]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A Semiempirical Free Energy Force Field with Charge-Based Desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA Dassault Systèmes. Discovery Studio Visualizer, 21.1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2020. [Google Scholar]
- Schrödinger, L.L.C.; DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 15 February 2022).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Parajuli, B.; Lee, H.-G.; Kwon, S.-H.; Cha, S.-D.; Shin, S.-J.; Lee, G.-H.; Bae, I.; Cho, C.-H. Salinomycin Inhibits Akt/NF–κB and Induces Apoptosis in Cisplatin Resistant Ovarian Cancer Cells. Cancer Epidemiol. 2013, 37, 512–517. [Google Scholar] [CrossRef]








| Entry | R1 | R2 | R3 | Yield in % | IC50 in μM |
|---|---|---|---|---|---|
| 3a | H | 3Me | I | 96 | 9.17 |
| 3b | H | 3Me | II | 96 | >100 |
| 3c | H | 3Me | III | 95 | 22.68 |
| 3d | H | 4Cl | I | 94 | 23.53 |
| 3e | H | 4Cl | II | 91 | >100 |
| 3f | H | 4Cl | III | 96 | 48.42 |
| 3g | H | 4F | I | 95 | 13.87 |
| 3h | H | 4F | II | 96 | 50.74 |
| 3i | H | 4F | III | 94 | ND |
| 3j | Me | 4F | I | 94 | 86.46 |
| 3k | Me | 4F | II | 96 | ND |
| 3l | Me | 4F | III | 97 | ND |
| Doxorubicin | 2.96 | ||||
| Tamoxifen | 1.84 | ||||
| Entry | R1 | R2 | R4 | Yield in % | IC50 in μM |
|---|---|---|---|---|---|
| 5a | H | 4OMe | IV | 95 | 16.38 |
| 5b | H | 4OMe | V | 90 | 6.29 |
| 5c | H | 4OMe | VI | 90 | 17.26 |
| 5d | H | 4OMe | VIII | 94 | 29.38 |
| 5e | H | 3Me | V | 90 | >100 |
| 5f | H | 3Me | VII | 80 | 79.00 |
| 5g | H | 3Me | VI | 89 | >100 |
| 5h | H | 3Me | VIII | 90 | 30.09 |
| 5i | H | 4F | VIII | 89 | 83.30 |
| 5j | H | 4F | V | 95 | >100 |
| 5k | H | 4F | VII | 85 | >100 |
| 5l | H | 4F | VI | 80 | >100 |
| 5m | H | 4Cl | VIII | 90 | 14.58 |
| 5n | H | 4Cl | V | 88 | >100 |
| 5o | H | 4Cl | VII | 92 | 42.00 |
| 5p | H | 4Cl | VI | 80 | >100 |
| Doxorubicin | 2.96 | ||||
| Tamoxifen | 1.84 | ||||
| Entry | MDA–MB–231 | BT–549 | SUM159PT |
|---|---|---|---|
| 3a | 7.34 | 5.98 | 14.81 |
| 5b | 57.42 | 37.54 | 47.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravish, A.; Narasimhachar, B.C.; Xi, Z.; Vishwanath, D.; Mohan, A.; Gaonkar, S.L.; Chandrashekara, P.G.; Ahn, K.S.; Pandey, V.; Lobie, P.E.; et al. Development of Piperazine- and Oxazine-Linked Pyrimidines as p65 Subunit Binders of NF–κB in Human Breast Cancer Cells. Biomedicines 2023, 11, 2716. https://doi.org/10.3390/biomedicines11102716
Ravish A, Narasimhachar BC, Xi Z, Vishwanath D, Mohan A, Gaonkar SL, Chandrashekara PG, Ahn KS, Pandey V, Lobie PE, et al. Development of Piperazine- and Oxazine-Linked Pyrimidines as p65 Subunit Binders of NF–κB in Human Breast Cancer Cells. Biomedicines. 2023; 11(10):2716. https://doi.org/10.3390/biomedicines11102716
Chicago/Turabian StyleRavish, Akshay, Bhanuprakash C. Narasimhachar, Zhang Xi, Divakar Vishwanath, Arunkumar Mohan, Santosh L. Gaonkar, Paduvalahippe Gowdegowda Chandrashekara, Kwang Seok Ahn, Vijay Pandey, Peter E. Lobie, and et al. 2023. "Development of Piperazine- and Oxazine-Linked Pyrimidines as p65 Subunit Binders of NF–κB in Human Breast Cancer Cells" Biomedicines 11, no. 10: 2716. https://doi.org/10.3390/biomedicines11102716