Microfluidic Devices for Label-Free DNA Detection
Abstract
:1. Introduction
2. Flow-Cell Biosensing Approaches
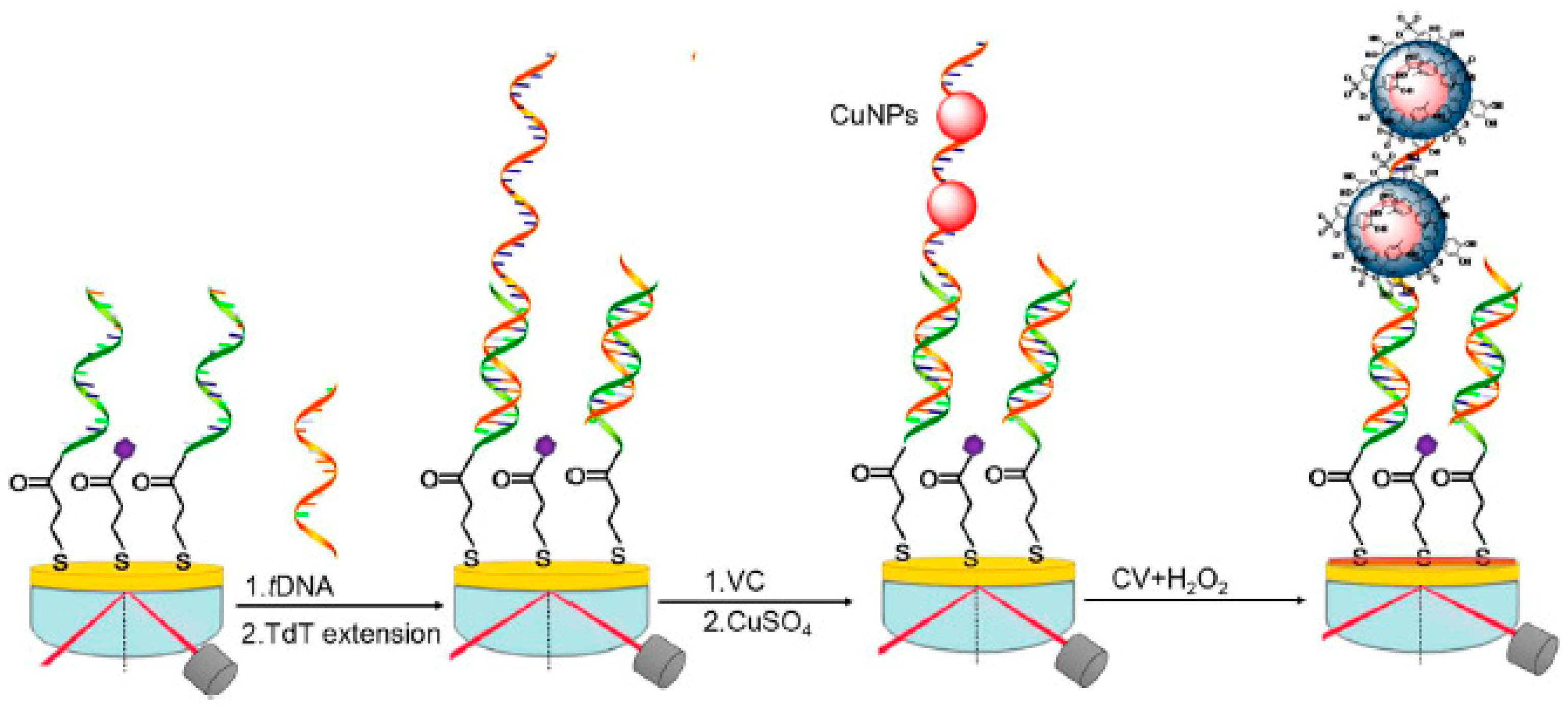
2.1. Optical
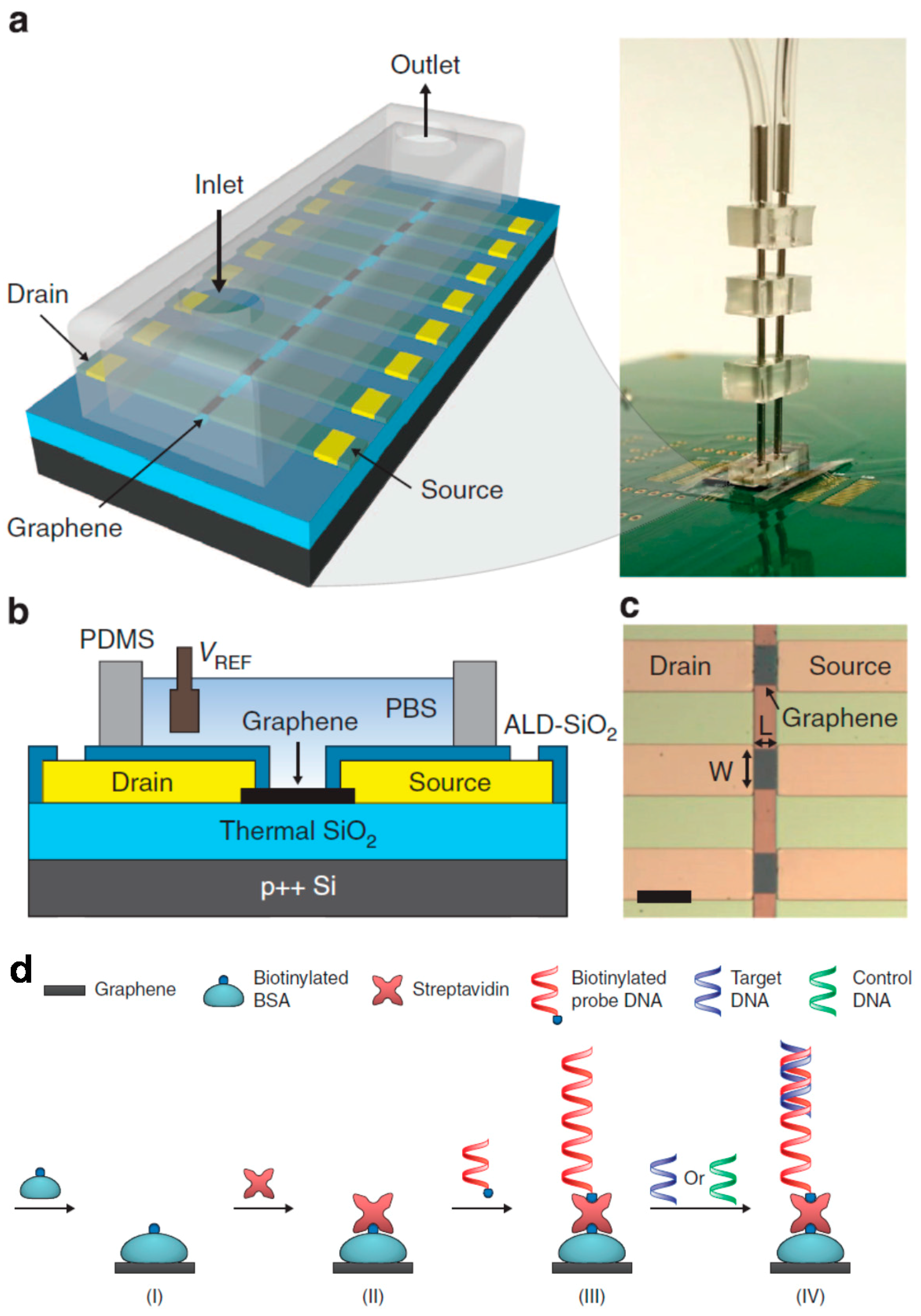
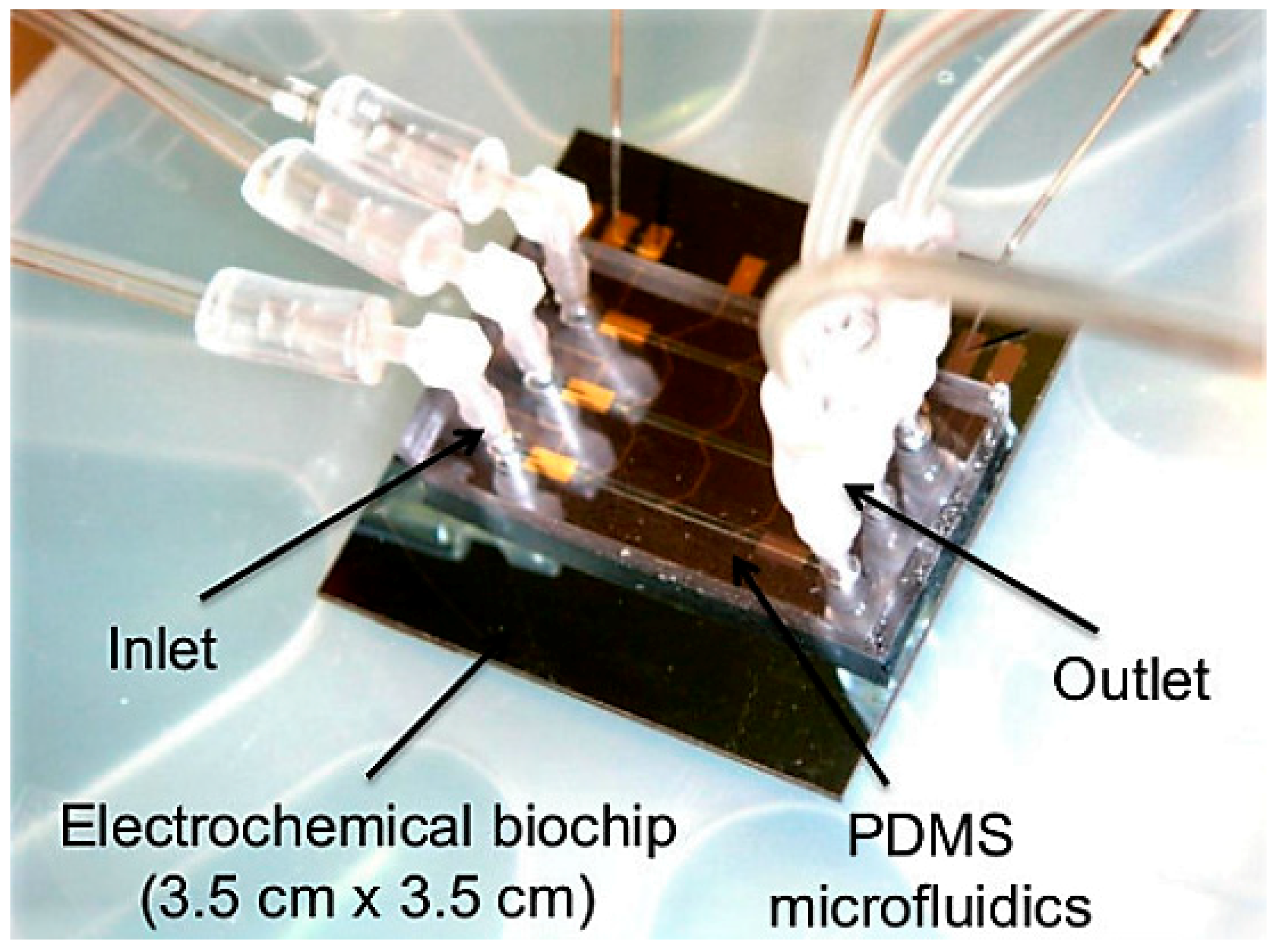
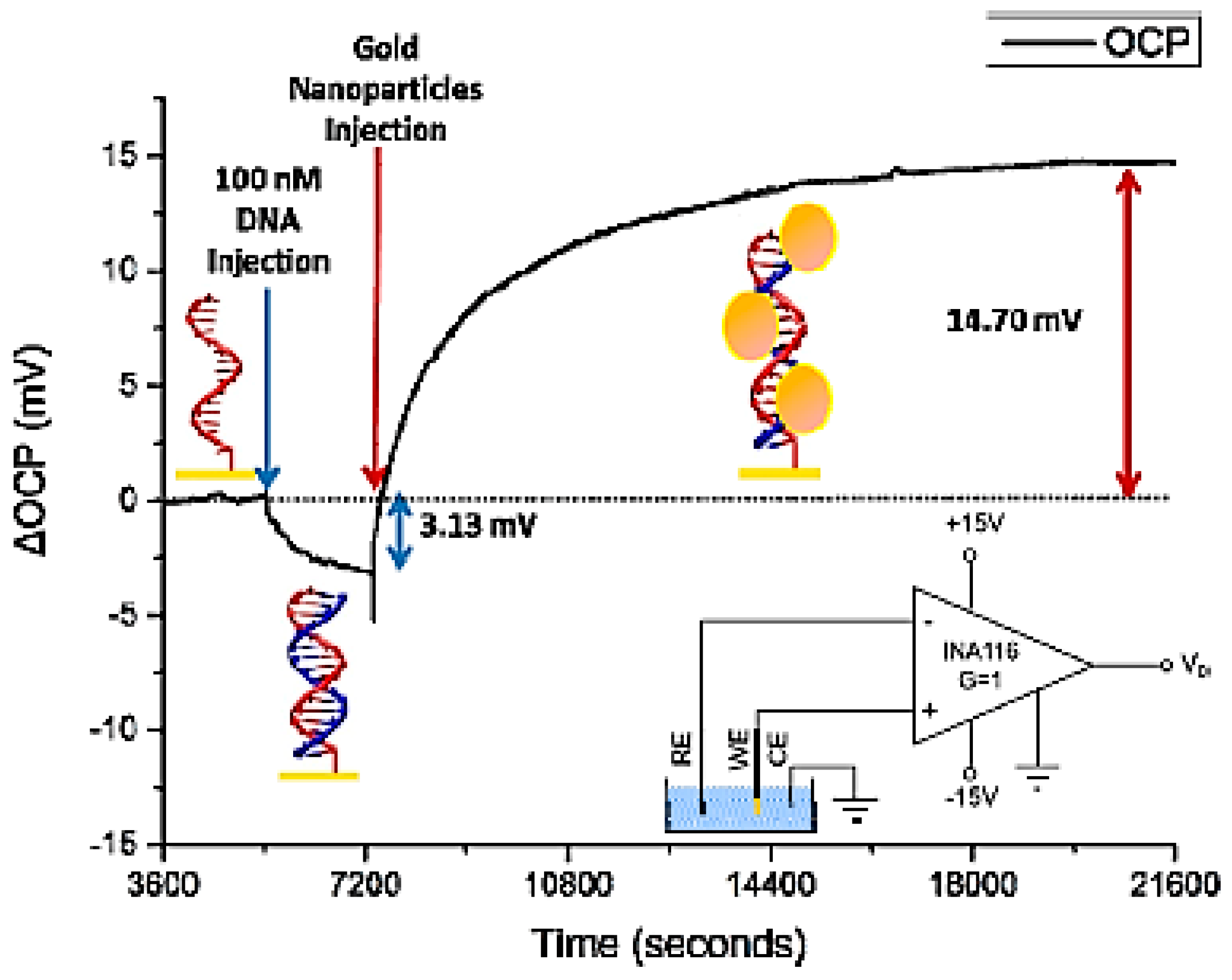
2.2. Electrochemical
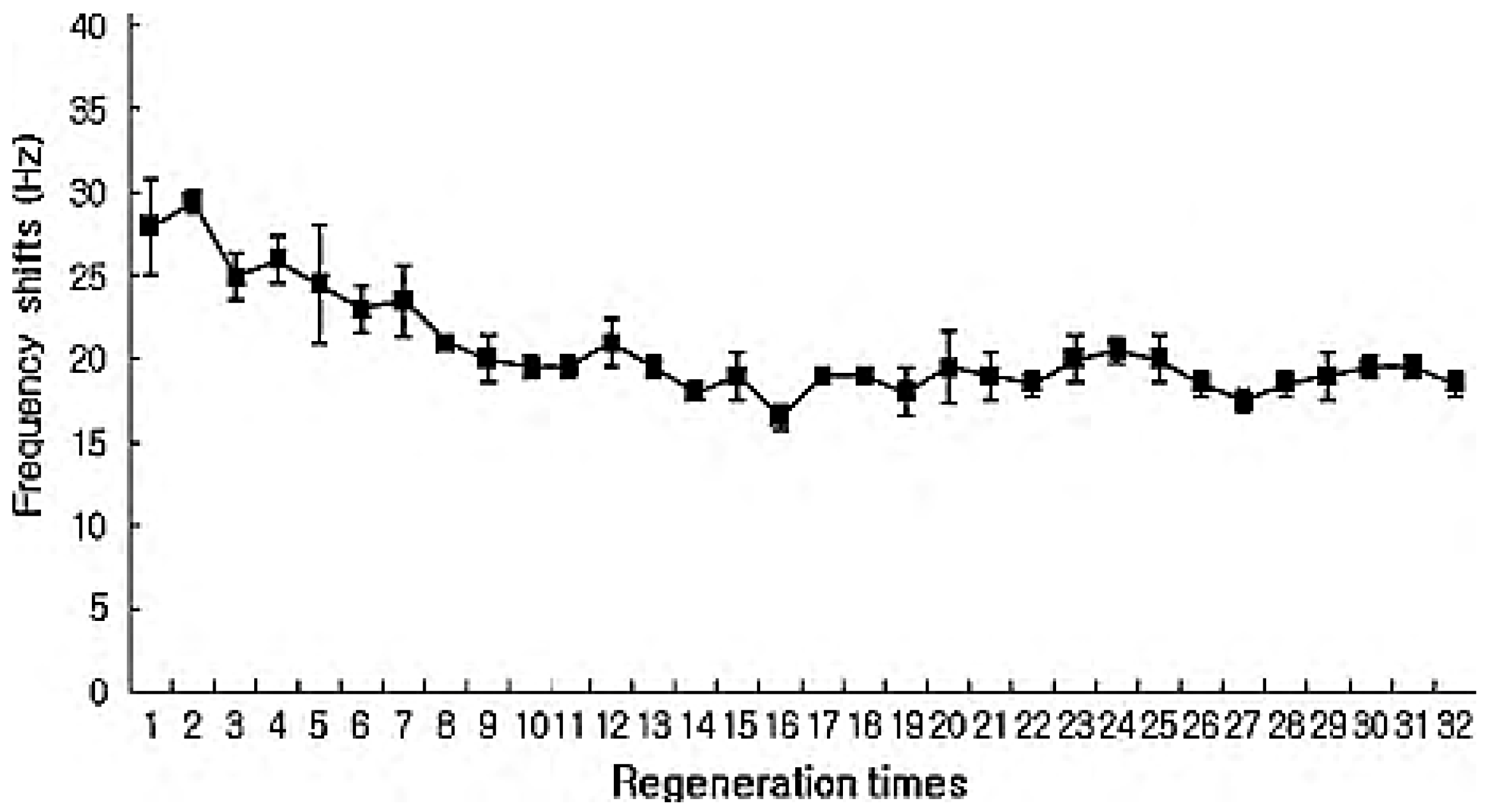
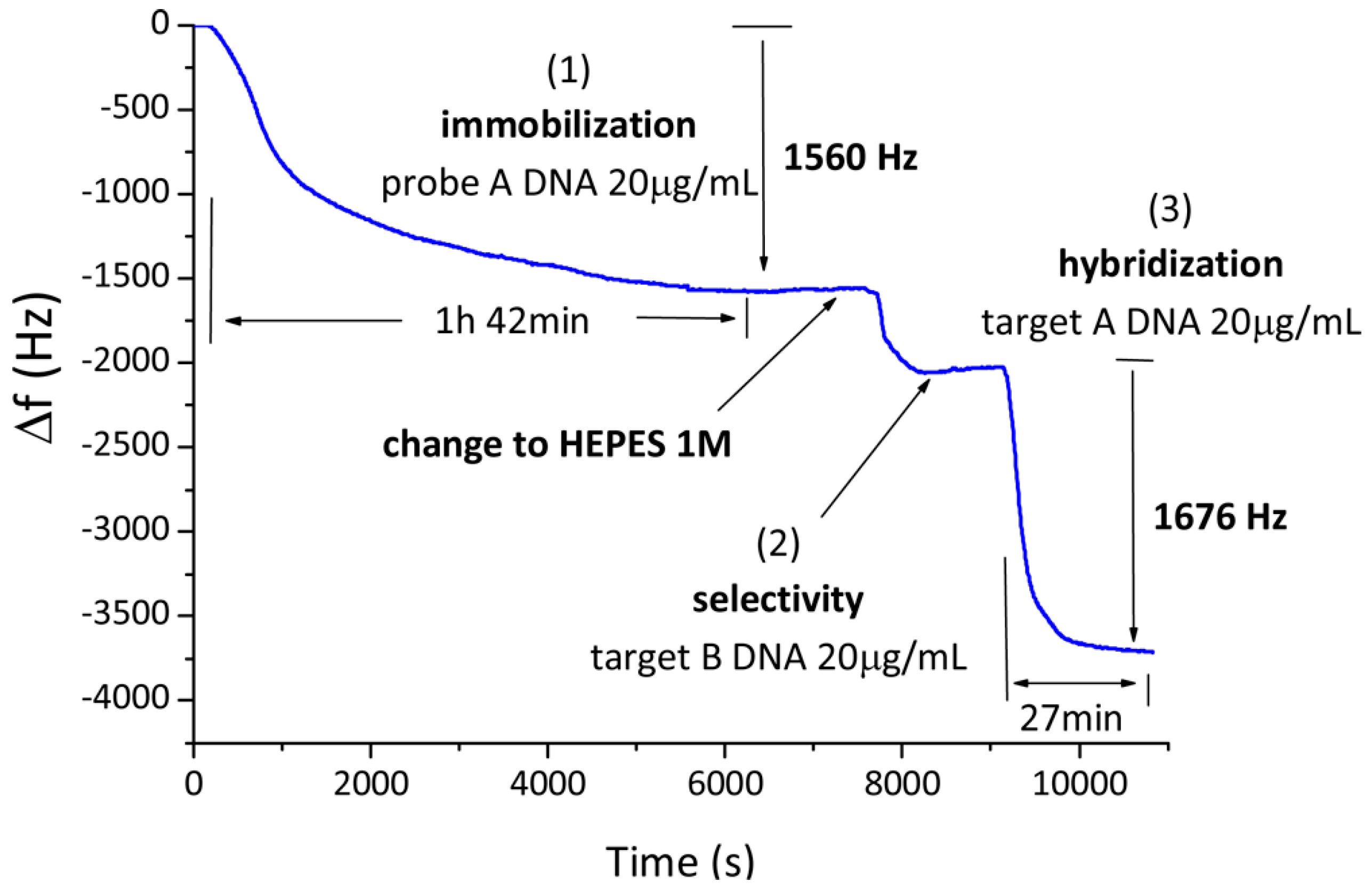
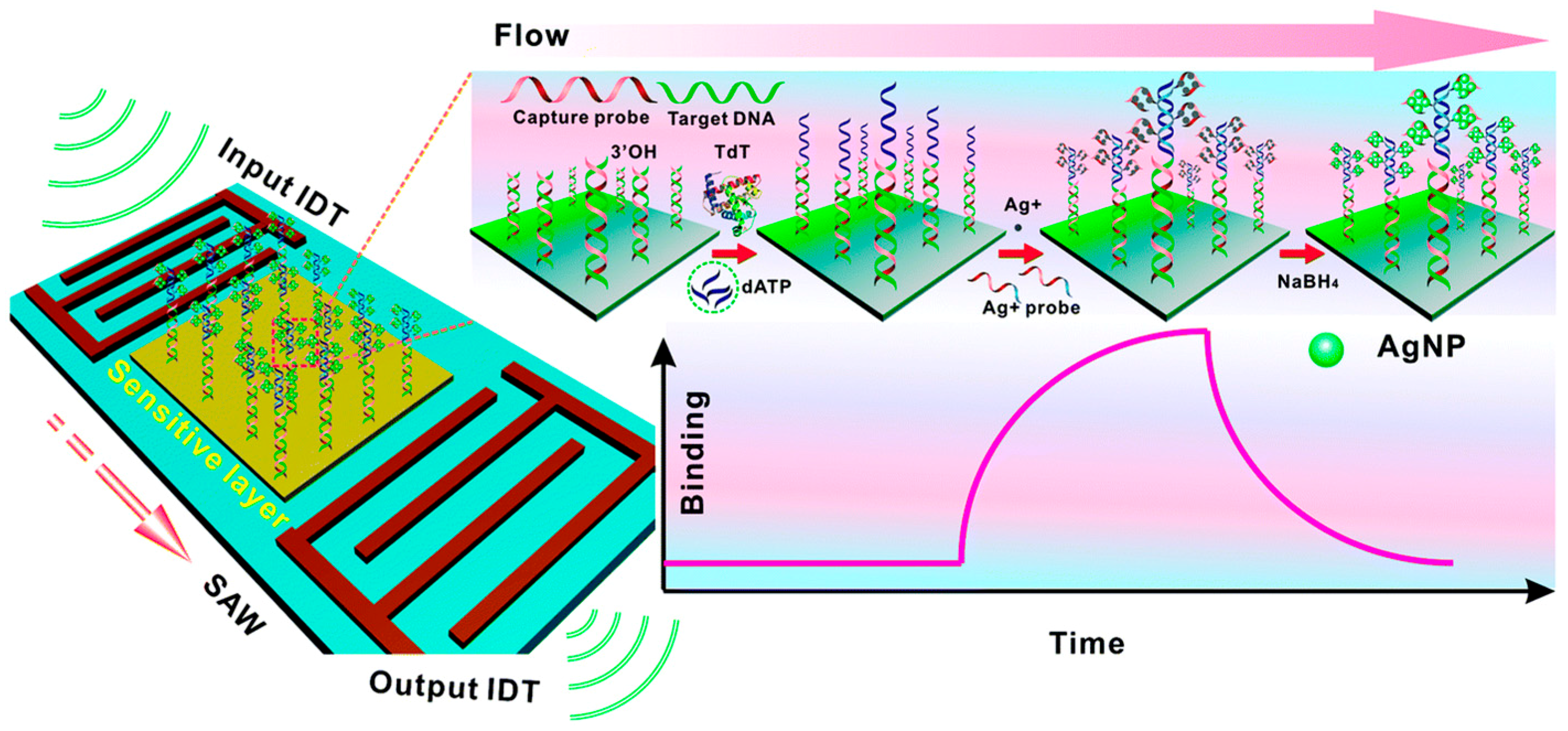
2.3. Mass-Based
3. Microfluidic Modules for Sample Preparation
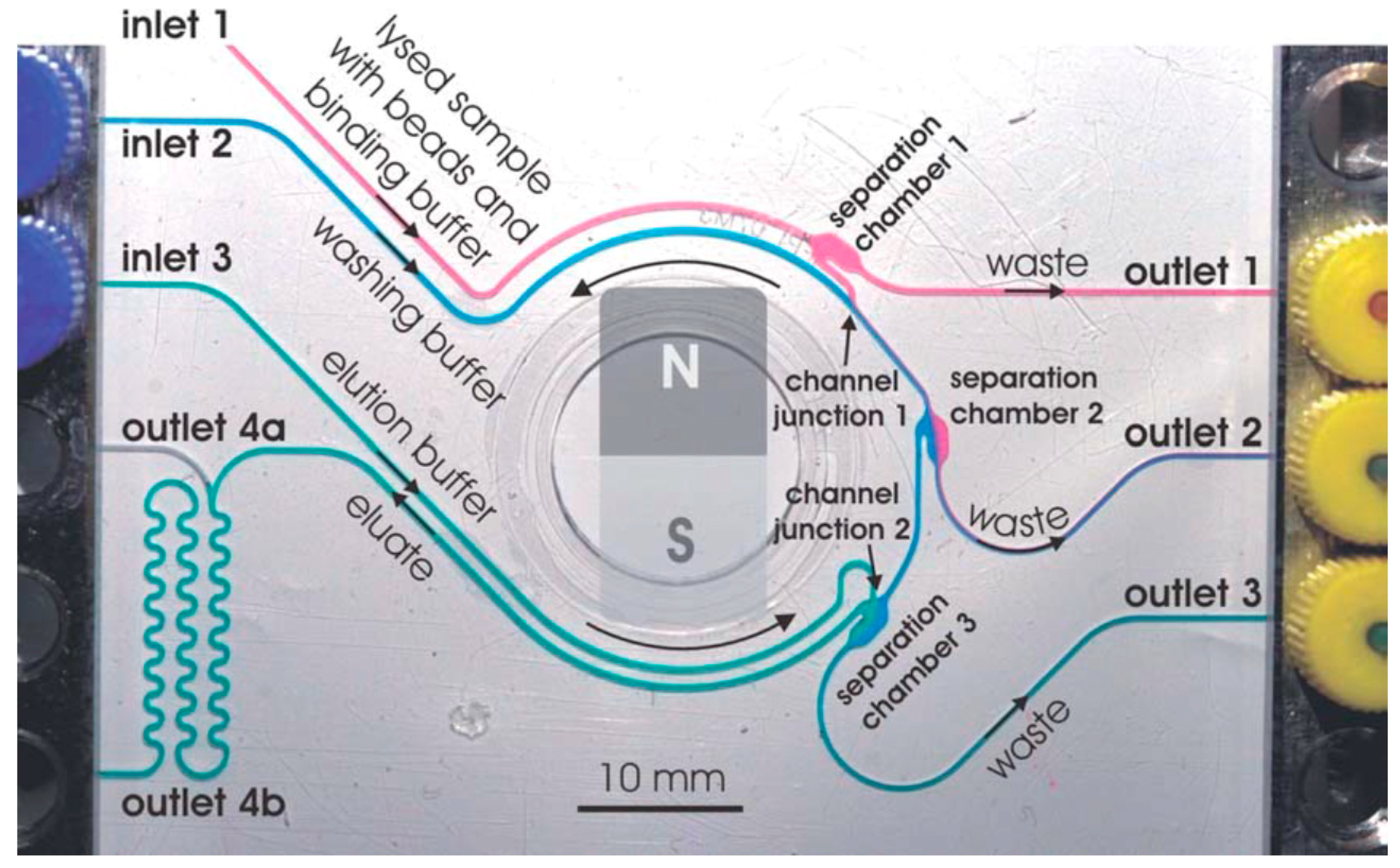
3.1. DNA Extraction
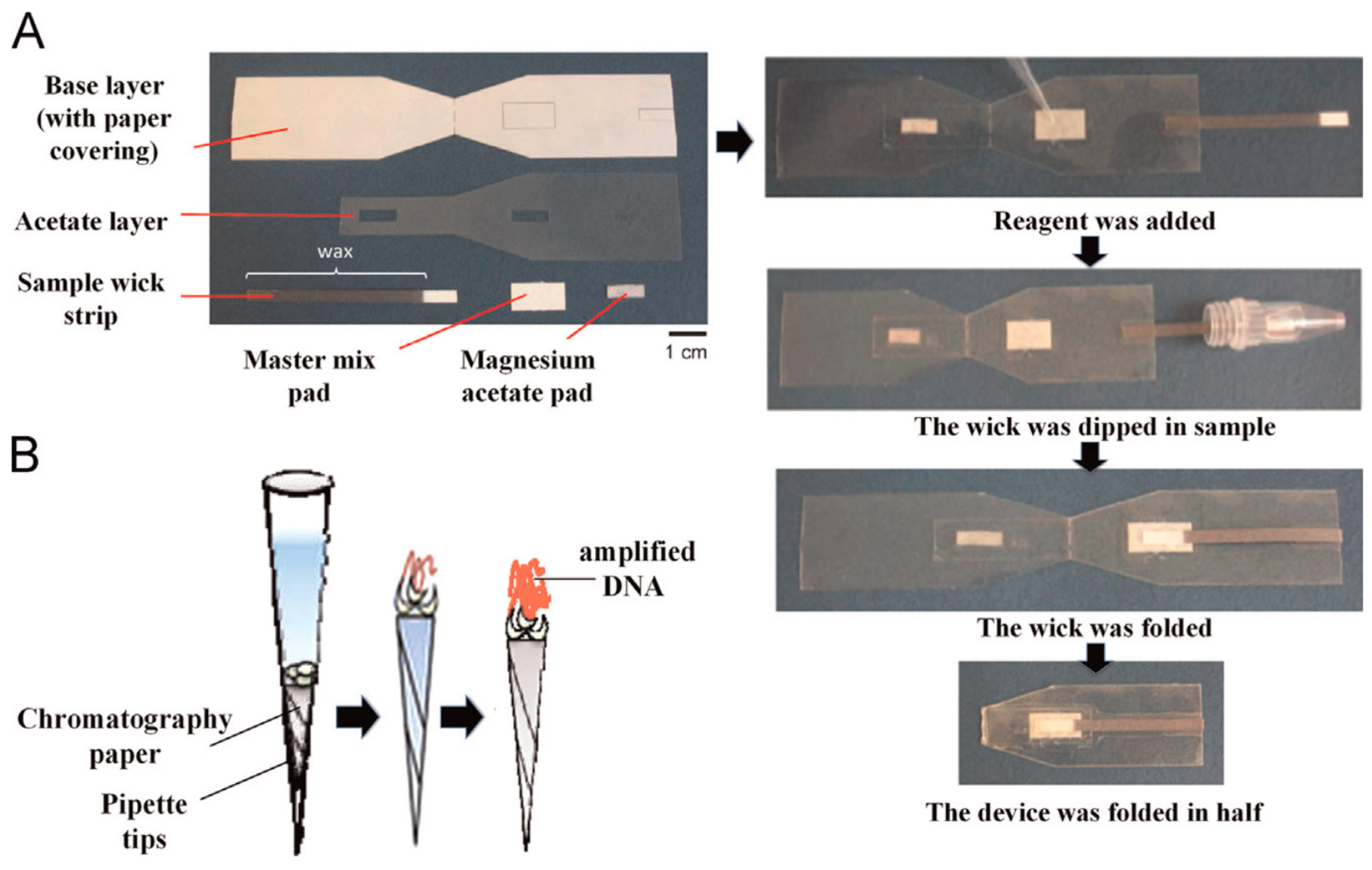
3.2. DNA Amplification
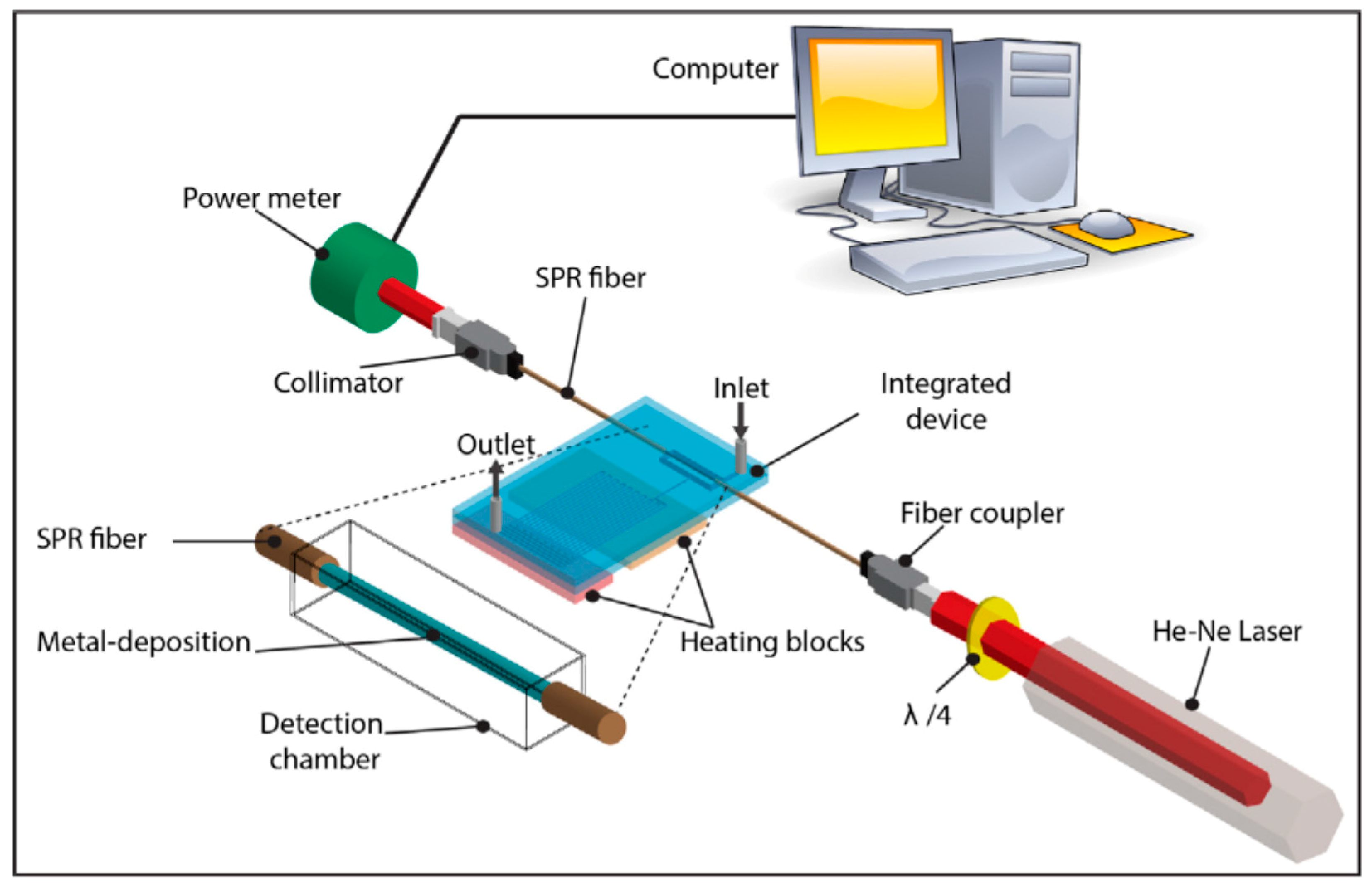
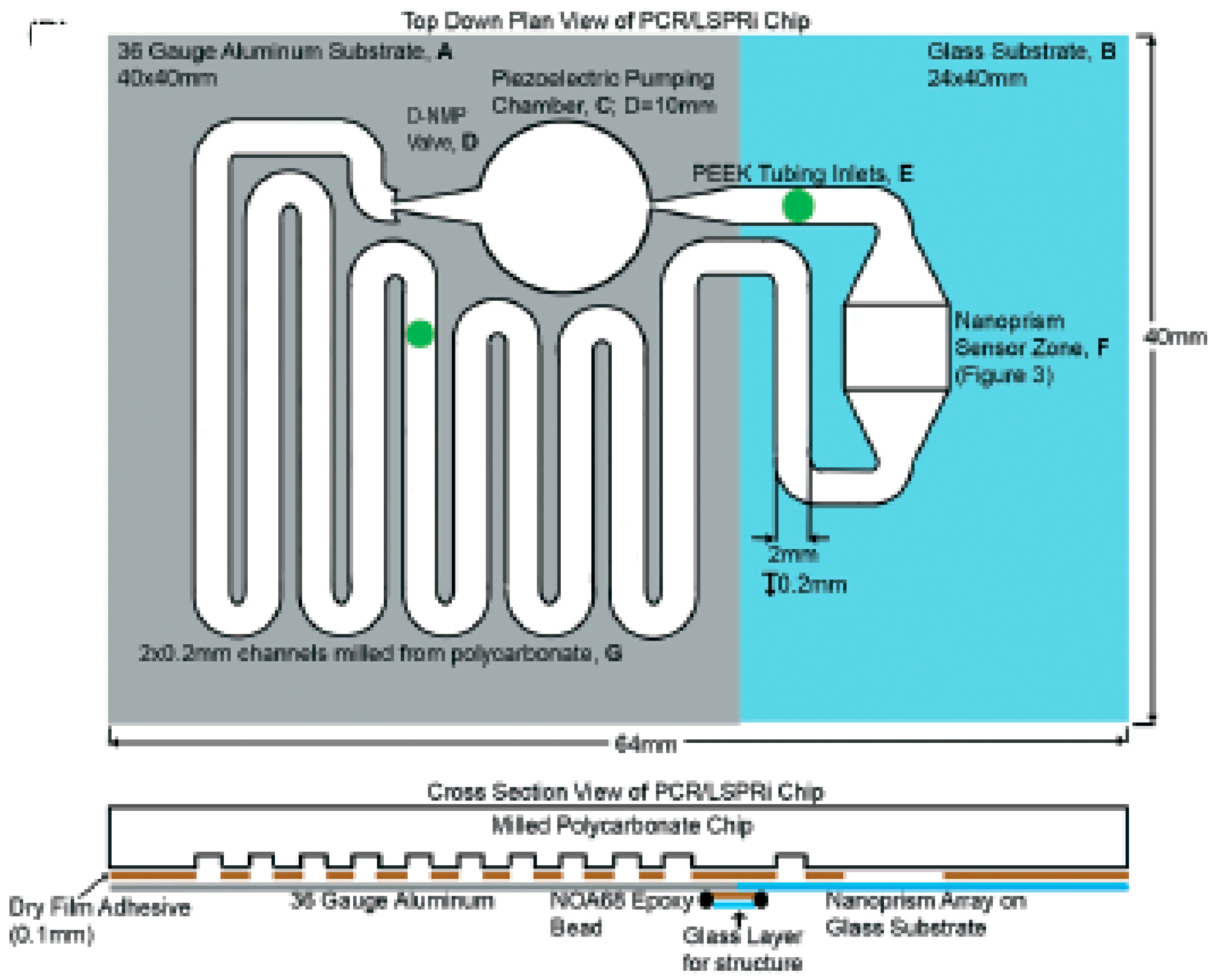
4. Higher Integration Platforms
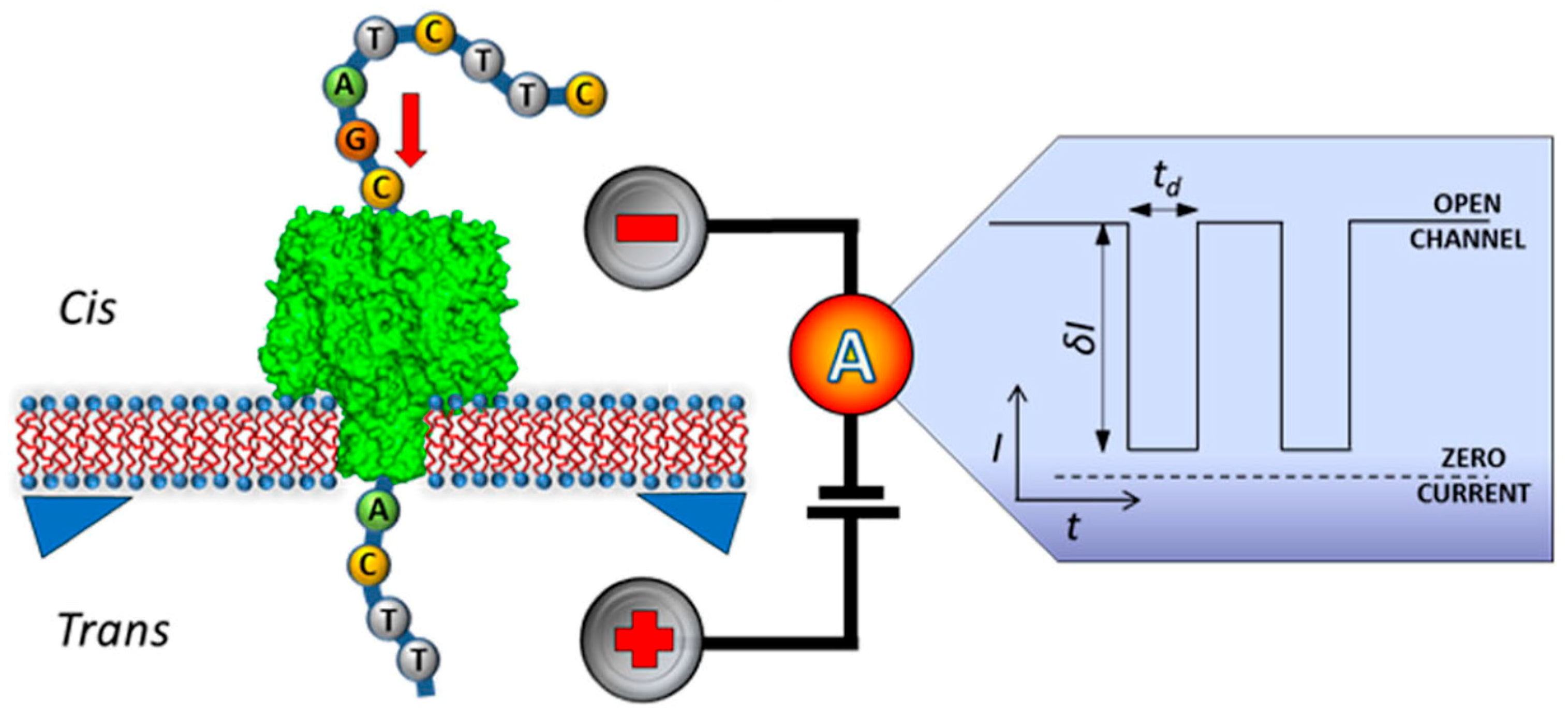
5. Nanopore Technology for Label-Free DNA Sequencing
6. Commercial Systems
7. Conclusions and Future Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Lafleur, J.P.; Jönsson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef] [PubMed]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Huang, R.; He, N.; Li, Z. Recent progresses in DNA nanostructure-based biosensors for detection of tumor markers. Biosens. Bioelectron. 2018, 109, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Bizid, S.; Blili, S.; Mlika, R.; Haj Said, A.; Korri-Youssoufi, H. Direct E-DNA sensor of Mycobacterium tuberculosis mutant strain based on new nanocomposite transducer (Fc-ac-OMPA/MWCNTs). Talanta 2018, 184, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Guijt, R.M.; Manz, A. Miniaturised Total Chemical-Analysis Systems (µTAS) that Periodically Convert Chemical into Electronic Information. Sens. Actuators B Chem. 2018, 273, 1334–1345. [Google Scholar] [CrossRef]
- Sandipan, R.; Gunjan, M.; Sanjeeva, S. Label-free detection techniques for protein microarrays: Prospects, merits and challenges. Proteomics 2010, 10, 731–748. [Google Scholar]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; DeRosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics integrated biosensors: A leading technology towards lab-on-a-chip and sensing applications. Sensors 2015, 15, 30011–30031. [Google Scholar] [CrossRef] [PubMed]
- Dincer, C.; Kling, A.; Chatelle, C.; Armbrecht, L.; Kieninger, J.; Weber, W.; Urban, G.A. Designed miniaturization of microfluidic biosensor platforms using the stop-flow technique. Analyst 2016, 141, 6073–6079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kartanas, T.; Ostanin, V.; Challa, P.K.; Daly, R.; Charmet, J.; Knowles, T.P.J. Enhanced Quality Factor Label-free Biosensing with Micro-Cantilevers Integrated into Microfluidic Systems. Anal. Chem. 2017, 89, 11929–11936. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, S.; Punjabi, N. Label—Free Integrated Optical Biosensors for Multiplexed Analysis. J. Indian Inst. Sci. 2012, 92, 253–294. [Google Scholar]
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef] [PubMed]
- Liedberg, B.; Nylander, C.; Lunström, I. Surface plasmon resonance for gas detection and biosensing. Sens. Actuators 1983, 4, 299–304. [Google Scholar] [CrossRef]
- Yuan, P.-X.; Deng, S.Y.; Zheng, C.Y.; Cosnier, S.; Shan, D. In situ formed copper nanoparticles templated by TdT-mediated DNA for enhanced SPR sensor-based DNA assay. Biosens. Bioelectron. 2017, 97, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kurita, R.; Yanagisawa, H.; Yoshioka, K.; Niwa, O. On-chip sequence-specific immunochemical epigenomic analysis utilizing outward-turned cytosine in a DNA bulge with handheld surface plasmon resonance equipment. Anal. Chem. 2015, 87, 11581–11586. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.L.; Bhalla, N.; Rafiee, S.D.; Estrela, P. Localized surface plasmon resonance as a biosensing platform for developing countries. Biosensors 2014, 4, 172–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavela, A.F.; Grajales García, D.; Ramirez, J.C.; Lechuga, L.M. Last Advances in Silicon-Based Optical Biosensors. Sensors 2016, 16, 285. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Zhang, H.; Liu, B.; Lin, W.; Wu, J. Label-free in-situ real-time DNA hybridization kinetics detection employing microfiber-assisted Mach-Zehnder interferometer. Biosens. Bioelectron. 2016, 81, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Kussrow, A.; Enders, C.S.; Bornhop, D.J. Interferometric Methods for Label-Free Molecular Interaction Studies. Anal. Chem. 2011, 84, 779–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahn, I.J.; Žukovskaja, O.; Zheng, X.S.; Weber, K.; Bocklitz, T.W.; Cialla-May, D.; Popp, J. Surface-enhanced Raman spectroscopy and microfluidic platforms: Challenges, solutions and potential applications. Analyst 2017, 142, 1022–1047. [Google Scholar] [CrossRef] [PubMed]
- Strelau, K.K.; Kretschmer, R.; Möller, R.; Fritzsche, W.; Popp, J. SERS as tool for the analysis of DNA-chips in a microfluidic platform. Anal. Bioanal. Chem. 2010, 396, 1381–1384. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Zeng, J.; Zhao, F.; Lin, S.H.; Raja, B.; Strych, U.; Willson, R.C.; Shih, W.C. Label-free, in situ SERS monitoring of individual DNA hybridization in microfluidics. Nanoscale 2014, 6, 8521–8526. [Google Scholar] [CrossRef] [PubMed]
- Novara, C.; Chiadò, A.; Paccotti, N.; Catuogno, S.; Esposito, C.L.; Condorelli, G.; De Franciscis, V.; Geobaldo, F.; Rivolo, P.; Giorgis, F. SERS-active metal-dielectric nanostructures integrated in microfluidic devices for label-free quantitative detection of miRNA. Faraday Discuss. 2017, 205, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.L.; Formisano, N.; Estrela, P.; Carrara, S.; Tkac, J. Electrochemical biosensors and nanobiosensors. Essays Biochem. 2016, 60, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rackus, D.G.; Shamsi, M.H.; Wheeler, A.R. Electrochemistry, biosensors and microfluidics: A convergence of fields. Chem. Soc. Rev. 2015, 44, 5320–5340. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, J.Y.; Ahn, J.H.; Choi, J.M.; Im, M.; Choi, Y.K. Integration of field effect transistor-based biosensors with a digital microfluidic device for a lab-on-a-chip application. Lab Chip 2012, 12, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Abbott, J.; Qin, L.; Yeung, K.Y.; Song, Y.; Yoon, H.; Kong, J.; Ham, D. Electrophoretic and field-effect graphene for all-electrical DNA array technology. Nat. Commun. 2014, 5, 4866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Yoav, H.; Dykstra, P.H.; Gordonov, T.; Bentley, W.E.; Ghodssi, R. A microfluidic-based electrochemical biochip for label-free DNA hybridization analysis. Biosens. Bioelectron. 2012, 38, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Batistuti, M.R.; Miodek, A.; Zhurauski, P.; Mulato, M.; Lindsay, M.A.; Estrela, P. A simple and highly sensitive electrochemical platform for detection of MicroRNAs. In Proceedings of the IEEE SENSORS, Busan, Korea, 1–4 November 2015; pp. 803–806. [Google Scholar]
- Wu, C.-C.; Huang, W.-C.; Hu, C.-C. An ultrasensitive label-free electrochemical impedimetric DNA biosensing chip integrated with a DC-biased AC electroosmotic vortex. Sens. Actuators B Chem. 2015, 209, 61–68. [Google Scholar] [CrossRef]
- Hong, S.-R.; Jeong, H.-D.; Hong, S. QCM DNA biosensor for the diagnosis of a fish pathogenic virus VHSV. Talanta 2010, 82, 899–903. [Google Scholar] [CrossRef] [PubMed]
- García-Martinez, G.; Bustabad, E.A.; Perrot, H.; Gabrielli, C.; Bucur, B.; Lazerges, M.; Rose, D.; Rodriguez-Pardo, L.; Fariña, J.; Compère, C.; Vives, A.A. Development of a mass sensitive quartz crystal microbalance (QCM)-based DNA biosensor using a 50 MHz electronic oscillator circuit. Sensors 2011, 11, 7656–7664. [Google Scholar] [CrossRef] [PubMed]
- Gronewold, T.M.; Baumgartner, A.; Quandt, E.; Famulok, M. Discrimination of single mutations in cancer-related gene fragments with a surface acoustic wave sensor. Anal. Chem. 2006, 78, 4865–4871. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.-Y.; Mao, X.-B.; Ning, Y.; Zhang, G.-J. Single-shot analytical assay based on graphene-oxide-modified surface acoustic wave biosensor for detection of single-nucleotide polymorphisms. Anal. Chem. 2015, 87, 9352–9359. [Google Scholar] [CrossRef] [PubMed]
- Go, D.B.; Atashbar, M.Z.; Ramshani, Z.; Chang, H.C. Surface acoustic wave devices for chemical sensing and microfluidics: A review and perspective. Anal. Methods 2017, 9, 4112–4134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, F.; Sun, Z.; Li, Y.T.; Zhang, G.J. A surface acoustic wave biosensor synergizing DNA-mediated in situ silver nanoparticle growth for a highly specific and signal-amplified nucleic acid assay. Analyst 2017, 142, 3468–3476. [Google Scholar] [CrossRef] [PubMed]
- Ayoib, A.; Hashim, U.; Gopinath, S.C.B.; Md Arshad, M.K. DNA extraction on bio-chip: History and preeminence over conventional and solid-phase extraction methods. Appl. Microbiol. Biotechnol. 2017, 101, 8077–8088. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: The past and the present. BioMed Res. Int. 2009, 2009, 574398. [Google Scholar]
- Ali, N.; Rampazzo, R.C.P.; Costa, A.D.T.; Krieger, M.A. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. BioMed Res. Int. 2017, 2017, 9306564. [Google Scholar] [CrossRef] [PubMed]
- Breadmore, M.C.; Wolfe, K.A.; Arcibal, I.G.; Leung, W.K.; Dickson, D.; Giordano, B.C.; Power, M.E.; Ferrance, J.P.; Feldman, S.H.; Norris, P.M.; et al. Microchip-based purification of DNA from biological samples. Anal. Chem. 2003, 75, 1880–1886. [Google Scholar] [CrossRef] [PubMed]
- Cady, N.C.; Stelick, S.; Batt, C.A. Nucleic acid purification using microfabricated silicon structures. Biosens. Bioelectron. 2003, 19, 59–66. [Google Scholar] [CrossRef]
- Chen, X.; Cui, D.F.; Sun, J.H.; Zhang, L.L.; Li, H. Microdevice-based DNA extraction method using green reagent. Key Eng. Mater. 2013, 562–565, 1111–1115. [Google Scholar] [CrossRef]
- Chen, X.; Cui, D.-F.; Liu, C.-C. High purity DNA extraction with a SPE microfluidic chip using KI as the binding salt. Chin. Chem. Lett. 2006, 17, 1101–1104. [Google Scholar]
- Chung, Y.-C.; Jan, M.S.; Lin, Y.C.; Lin, J.H.; Cheng, W.C.; Fan, C.Y. Microfluidic chip for high efficiency DNA extraction. Lab Chip 2004, 4, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.A.; Breadmore, M.C.; Ferrance, J.P.; Power, M.E.; Conroy, J.F.; Norris, P.M.; Landers, J.P. Toward a microchip-based solid-phase extraction method for isolation of nucleic acids. Electrophoresis 2002, 23, 727–733. [Google Scholar] [CrossRef]
- Wu, J.; Kodzius, R.; Cao, W. Extraction, amplification and detection of DNA in microfluidic chip-based assays. Microchim. Acta 2014, 181, 1611–1631. [Google Scholar] [CrossRef]
- Karle, M.; Miwa, J.; Czilwik, G.; Auwärter, V.; Roth, G.; Zengerle, R.; von Stetten, F. Continuous microfluidic DNA extraction using phase-transfer magnetophoresis. Lab Chip 2010, 10, 3284–3290. [Google Scholar] [CrossRef] [PubMed]
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 2010, 363, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ozdemir, P. Microfluidic DNA amplification—A review. Anal. Chim. Acta 2009, 638, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capretto, L.; Cheng, W.; Hill, M.; Zhang, X. Microfluidics Technologies and Applications. Top. Curr. Chem. 2011, 304, 27–68. [Google Scholar] [PubMed]
- Giuffrida, M.C.; Spoto, G. Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosens. Bioelectron. 2017, 90, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Tsaloglou, M.-N.; Nemiroski, A.; Camci-Unal, G.; Christodouleas, D.C.; Murray, L.P.; Connelly, J.T.; Whitesides, G.M. Handheld isothermal amplification and electrochemical detection of DNA in resource-limited settings. Anal. Biochem. 2018, 543, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Tang, R.; Wang, S.; Wan Abas, W.A.; Pingguan-Murphy, B.; Xu, F. Paper-based sample-to-answer molecular diagnostic platform for point-of-care diagnostics. Biosens. Bioelectron. 2015, 74, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.H.; Ramalingam, N.; Xian-Dui, D.; Ngin, T.S.; Xianting, Z.; Lai Kuan, A.T.; Peng Huat, E.Y.; Hai-Qing, G. Real-time PCR microfluidic devices with concurrent electrochemical detection. Biosens. Bioelectron. 2009, 24, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Woolley, A.T.; Hadley, D.; Landre, P.; de Mello, A.J.; Mathies, R.A.; Northrup, M.A. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal. Chem. 1996, 68, 4081–4086. [Google Scholar] [CrossRef] [PubMed]
- Wilding, P.; Kricka, L.J.; Cheng, J.; Hvichia, G.; Shoffner, M.A.; Fortina, P. Integrated cell isolation and polymerase chain reaction analysis using silicon microfilter chambers. Anal. Biochem. 1998, 257, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, G.; Murasova, P.; Hamiot, A.; Tsougeni, K.; Kaprou, G.; Eck, M.; Rabus, D.; Bilkova, Z.; Dupuy, B.; Jobst, G.; et al. Micro-nano-bio acoustic system for the detection of foodborne pathogens in real samples. Biosens. Bioelectron. 2018, 111, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rauch, C.B.; Stevens, R.L.; Lenigk, R.; Yang, J.; Rhine, D.B.; Grodzinski, P. DNA amplification and hybridization assays in integrated plastic monolithic devices. Anal. Chem. 2002, 74, 3063–3070. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Trinh, K.L.; Yoon, W.J.; Lee, N.Y.; Ju, H. Integration of a microfluidic polymerase chain reaction device and surface plasmon resonance fiber sensor into an inline all-in-one platform for pathogenic bacteria detection. Sens. Actuators B Chem. 2017, 242, 1–8. [Google Scholar] [CrossRef]
- Kao, L.T.-H.; Shankar, L.; Kang, T.G.; Zhang, G.; Tay, G.K.; Rafei, S.R.; Lee, C.W. Multiplexed detection and differentiation of the DNA strains for influenza A (H1N1 2009) using a silicon-based microfluidic system. Biosens. Bioelectron. 2011, 26, 2006–2011. [Google Scholar] [CrossRef] [PubMed]
- Medina-Sánchez, M.; Ibarlucea, B.; Pérez, N.; Karnaushenko, D.D.; Weiz, S.M.; Baraban, L.; Cuniberti, G.; Schmidt, O.G. High-performance three-dimensional tubular nanomembrane sensor for DNA detection. Nano Lett. 2016, 16, 4288–4296. [Google Scholar] [CrossRef] [PubMed]
- Haber, J.; Gascoyne, P.; Sokolov, K. Rapid real-time recirculating PCR using localized surface plasmon resonance (LSPR) and piezo-electric pumping. Lab Chip 2017, 17, 2821–2830. [Google Scholar] [CrossRef] [PubMed]
- Dak, P.; Ebrahimi, A.; Swaminathan, V.; Duarte-Guevara, C.; Bashir, R.; Alam, M.A. Droplet-based biosensing for lab-on-a-chip, open microfluidics platforms. Biosensors 2016, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Malic, L.; Veres, T.; Tabrizian, M. Nanostructured digital microfluidics for enhanced surface plasmon resonance imaging. Biosens. Bioelectron. 2011, 26, 2053–2059. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, A.T.-H.; Pan, P.J.-H.; Lee, A.P. Rapid label-free DNA analysis in picoliter microfluidic droplets using FRET probes. Microfluid. Nanofluid. 2009, 6, 391. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Dak, P.; Salm, E.; Dash, S.; Garimella, S.V.; Bashir, R.; Alam, M.A. Nanotextured superhydrophobic electrodes enable detection of attomolar-scale DNA concentration within a droplet by non-faradaic impedance spectroscopy. Lab Chip 2013, 13, 4248–4256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudr, J.; Skalickova, S.; Nejdl, L.; Moulick, A.; Ruttkay-Nedecky, B.; Adam, V.; Kizek, R. Fabrication of solid-state nanopores and its perspectives. Electrophoresis 2015, 36, 2367–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Reisner, W. Fabrication and characterization of nanopore-interfaced nanochannel devices. Nanotechnology 2015, 26, 455301. [Google Scholar] [CrossRef] [PubMed]
- Freedman, K.J.; Otto, L.M.; Ivanov, A.P.; Barik, A.; Oh, S.H.; Edel, J.B. Nanopore sensing at ultra-low concentrations using single-molecule dielectrophoretic trapping. Nat. Commun. 2016, 7, 10217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sze, J.Y.Y.; Kumar, S.; Ivanov, A.P.; Oh, S.H.; Edel, J.B. Fine tuning of nanopipettes using atomic layer deposition for single molecule sensing. Analyst 2015, 140, 4828–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Li, C.; Zhou, Y.; Jin, Y. Controllable Shrinking of Glass Capillary Nanopores Down to sub-10 nm by Wet-Chemical Silanization for Signal-Enhanced DNA Translocation. ACS Sens. 2017, 2, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- Briggs, K.; Madejski, G.; Magill, M.; Kastritis, K.; de Haan, H.W.; McGrath, J.L.; Tabard-Cossa, V. DNA translocations through nanopores under nanoscale preconfinement. Nano Lett. 2017, 18, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Tahvildari, R.; Beamish, E.; Tabard-Cossa, V.; Godin, M. Integrating nanopore sensors within microfluidic channel arrays using controlled breakdown. Lab Chip 2015, 15, 1407–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenstein, M. Oxford Nanopore announcement sets sequencing sector abuzz. Nat. Biotechnol. 2012, 30, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Laver, T.; Harrison, J.; O’Neill, P.A.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Studholme, D.J. Assessing the performance of the oxford nanopore technologies minion. Biomol. Detect. Quantif. 2015, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lemon, J.K.; Khil, P.P.; Frank, K.M.; Dekker, J.P. Rapid nanopore sequencing of plasmids and resistance gene detection in clinical isolates. J. Clin. Microbiol. 2017, 55, 3530–3543. [Google Scholar] [CrossRef] [PubMed]
- Mikheyev, A.S.; Tin, M.M. A first look at the Oxford Nanopore MinION sequencer. Mol. Ecol. Resour. 2014, 14, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; Hoon, J.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ion Torrent™ Next-Gen Sequencing Technology. Available online: https://www.youtube.com/watch?v=WYBzbxIfuKs (accessed on 31 July 2018).
- Ion Torrent. Available online: https://www.thermofisher.com/uk/en/home/brands/ion-torrent.html.html (accessed on 2 November 2017).
- Oxford Nanopore Technologies, Company History. Available online: https://nanoporetech.com/about-us/history (accessed on 2 November 2017).
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A.; et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016, 530, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Vu, C.L.; Chan, J.; Todaro, M.; Skafidas, S.; Kwan, P. Point-of-care molecular diagnostic devices: An overview. Pharmacogenomics 2015, 16, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- QuantuMDx, Q-POCTM. Available online: http://quantumdx.com/systems-o/q-poc-2 (accessed on 2 November 2017).
- QuantuMDx, Applications. Available online: http://quantumdx.com/applications (accessed on 2 November 2017).
- Burn, J. Company profile: QuantuMDx group limited. Pharmacogenomics 2013, 14, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Anscombe, N. Unravelling DNA. Electro Optics. 2016. Available online: https://www.electrooptics.com/feature/unravelling-dna (accessed on 2 November 2017).
- Atlas Genetics, io® System. Available online: http://www.atlasgenetics.com/solutions/systems.html (accessed on 30 July 2018).
- Edwards, T.R.K.; Mullarkey, T.C.E.; NealeJay, K.D.; Taylor, J.K.; Arlett, B. Fluidic Cartridge for Nucleic Acid Amplification and Detection. U.S. Patent 9,816,135, 14 November 2017. [Google Scholar]
- Hillier, S.C.; Flower, S.E.; Frost, C.G.; Jenkins, A.T.A.; Keay, R.; Braven, H. An electrochemical gene detection assay utilising T7 exonuclease activity on complementary probe–target oligonucleotide sequences. Electrochem. Commun. 2004, 6, 1227–1232. [Google Scholar] [CrossRef]
- Marsh, B.J.; Hampton, L.; Goggins, S.; Frost, C.G. Fine-tuning of ferrocene redox potentials towards multiplex DNA detection. New J. Chem. 2014, 38, 5260–5263. [Google Scholar] [CrossRef] [Green Version]
- Muir, A.; Jenkins, A.T.; Forrest, G.; Clarkson, J.; Wheals, A. Rapid electrochemical identification of pathogenic Candida species. J. Med. Microbiol. 2009, 58, 1182–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muir, A.; Forrest, G.; Clarkson, J.; Wheals, A. Detection of Candida albicans DNA from blood samples using a novel electrochemical assay. J. Med. Microbiol. 2011, 60, 467–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearce, D.M.; Shenton, D.P.; Holden, J.; Gaydos, C.A. Evaluation of a novel electrochemical detection method for Chlamydia trachomatis: Application for point-of-care diagnostics. IEEE Trans. Biomed. Eng. 2011, 58, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.M.; Styles, D.N.; Hardick, J.P.; Gaydos, C.A. A new rapid molecular point-of-care assay for Trichomonas vaginalis: Preliminary performance data. Sex. Transm. Infect. 2013, 89, 495–497. [Google Scholar] [CrossRef] [PubMed]
- DNAe. Available online: https://www.dnae.com/ (accessed on 2 November 2017).
- Toumazou, C.; Shepherd, L.M.; Reed, S.C.; Chen, G.I.; Patel, A.; Garner, D.M.; Wang, C.J.; Ou, C.P.; Amin-Desai, K.; Athanasiou, P.; et al. Simultaneous DNA amplification and detection using a pH-sensing semiconductor system. Nat. Methods 2013, 10, 641–646. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dutta, G.; Rainbow, J.; Zupancic, U.; Papamatthaiou, S.; Estrela, P.; Moschou, D. Microfluidic Devices for Label-Free DNA Detection. Chemosensors 2018, 6, 43. https://doi.org/10.3390/chemosensors6040043
Dutta G, Rainbow J, Zupancic U, Papamatthaiou S, Estrela P, Moschou D. Microfluidic Devices for Label-Free DNA Detection. Chemosensors. 2018; 6(4):43. https://doi.org/10.3390/chemosensors6040043
Chicago/Turabian StyleDutta, Gorachand, Joshua Rainbow, Uros Zupancic, Sotirios Papamatthaiou, Pedro Estrela, and Despina Moschou. 2018. "Microfluidic Devices for Label-Free DNA Detection" Chemosensors 6, no. 4: 43. https://doi.org/10.3390/chemosensors6040043




