Au Nanoparticles on 4-Thiophenol-Electrodeposited Carbon Surfaces for the Simultaneous Detection of 8-Hydroxyguanine and Guanine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Instrumentation
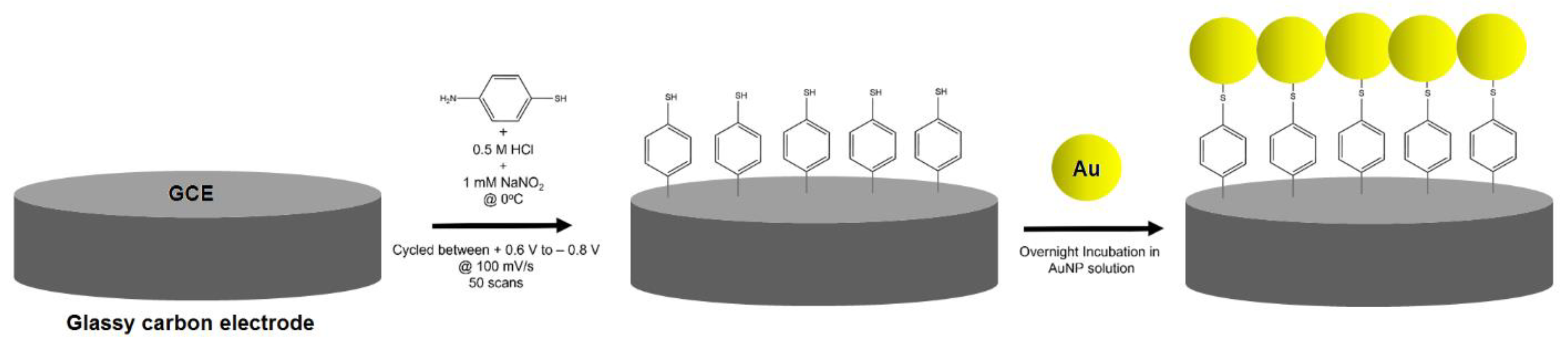
2.3. AuNP Synthesis and Electrode Modification
2.4. Real Sample Preparation
3. Results and Discussion
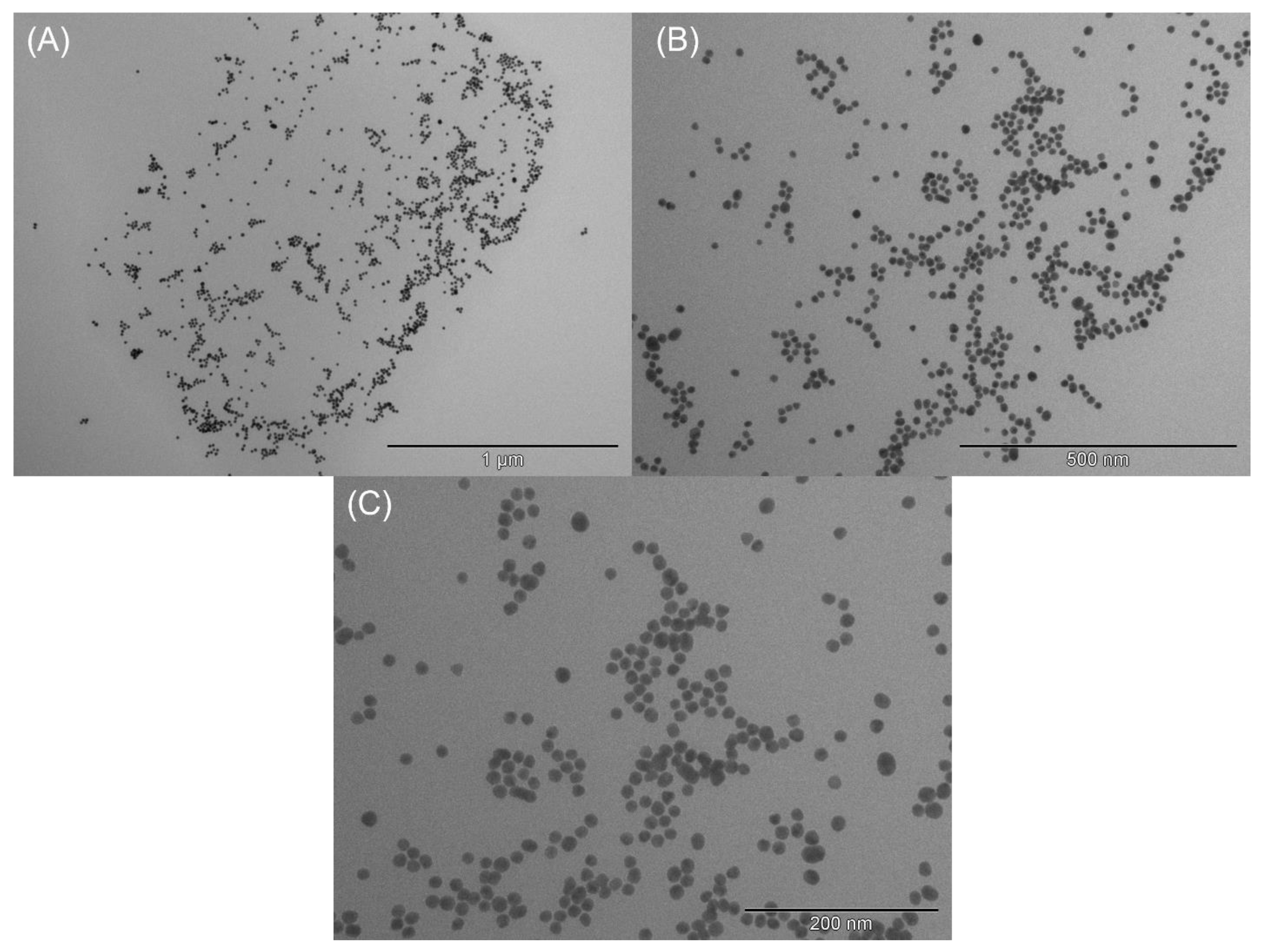
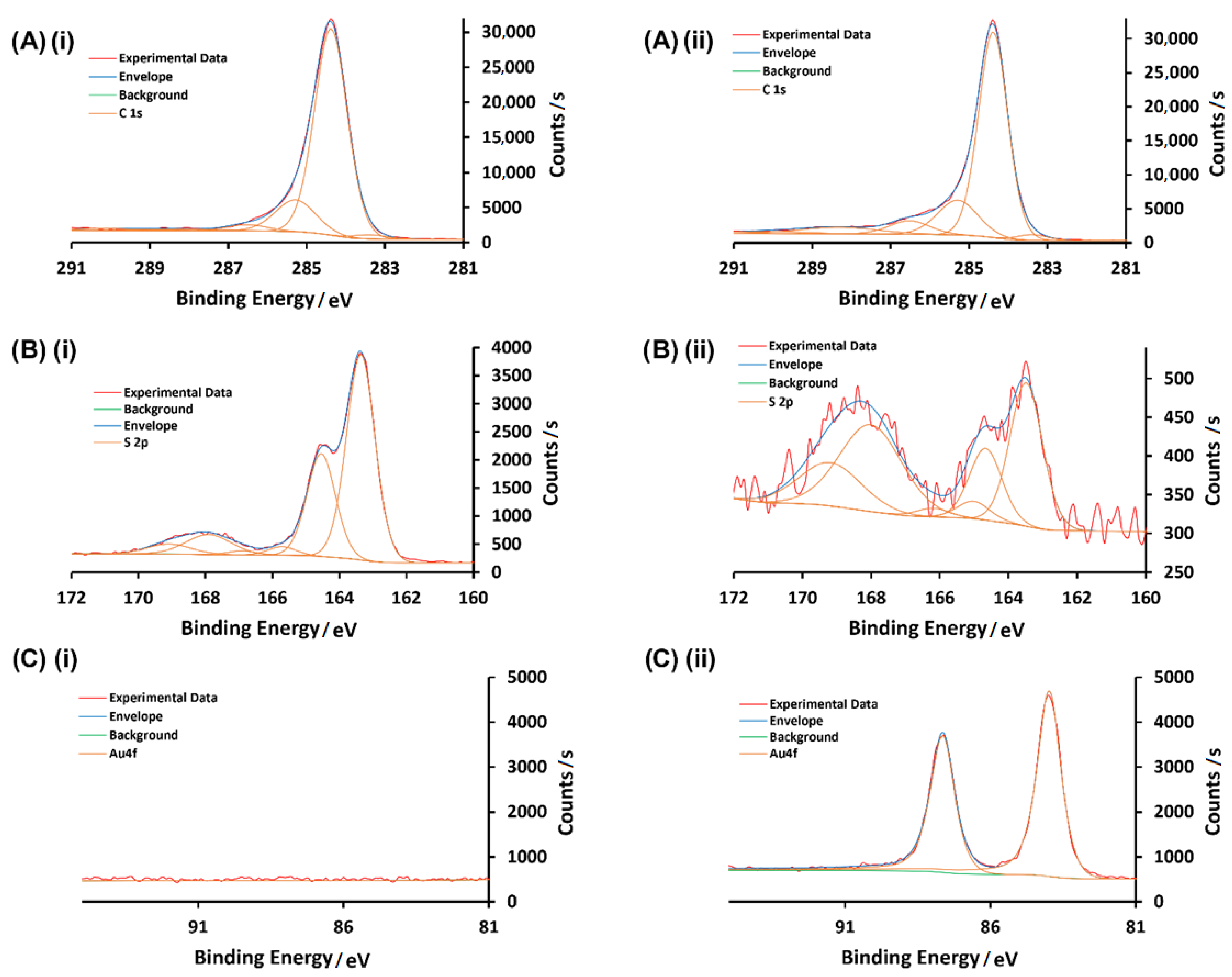
3.1. Characterization of AuNPs and Modified GCEs
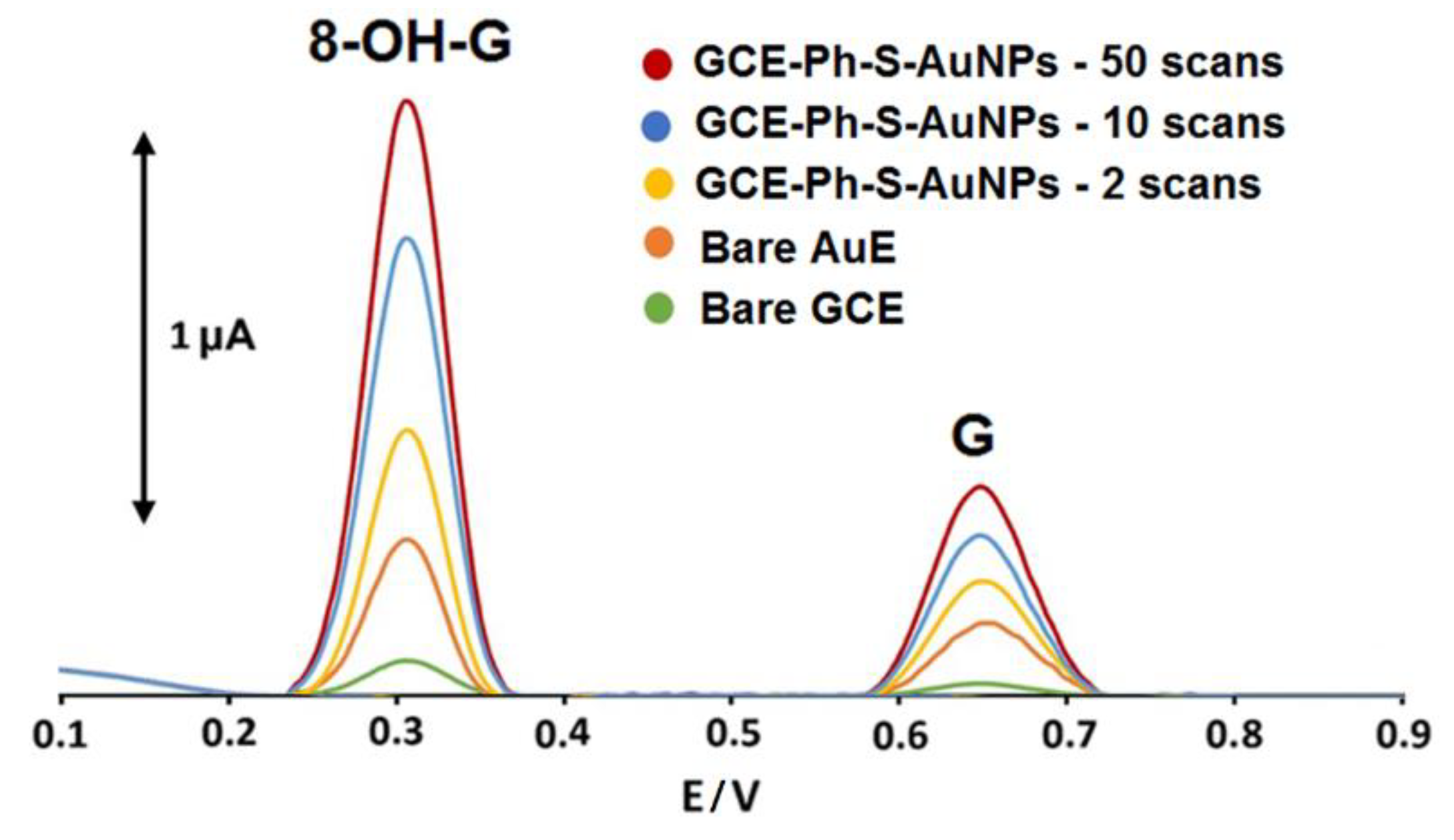
3.2. Comparison between the Modified Surfaces
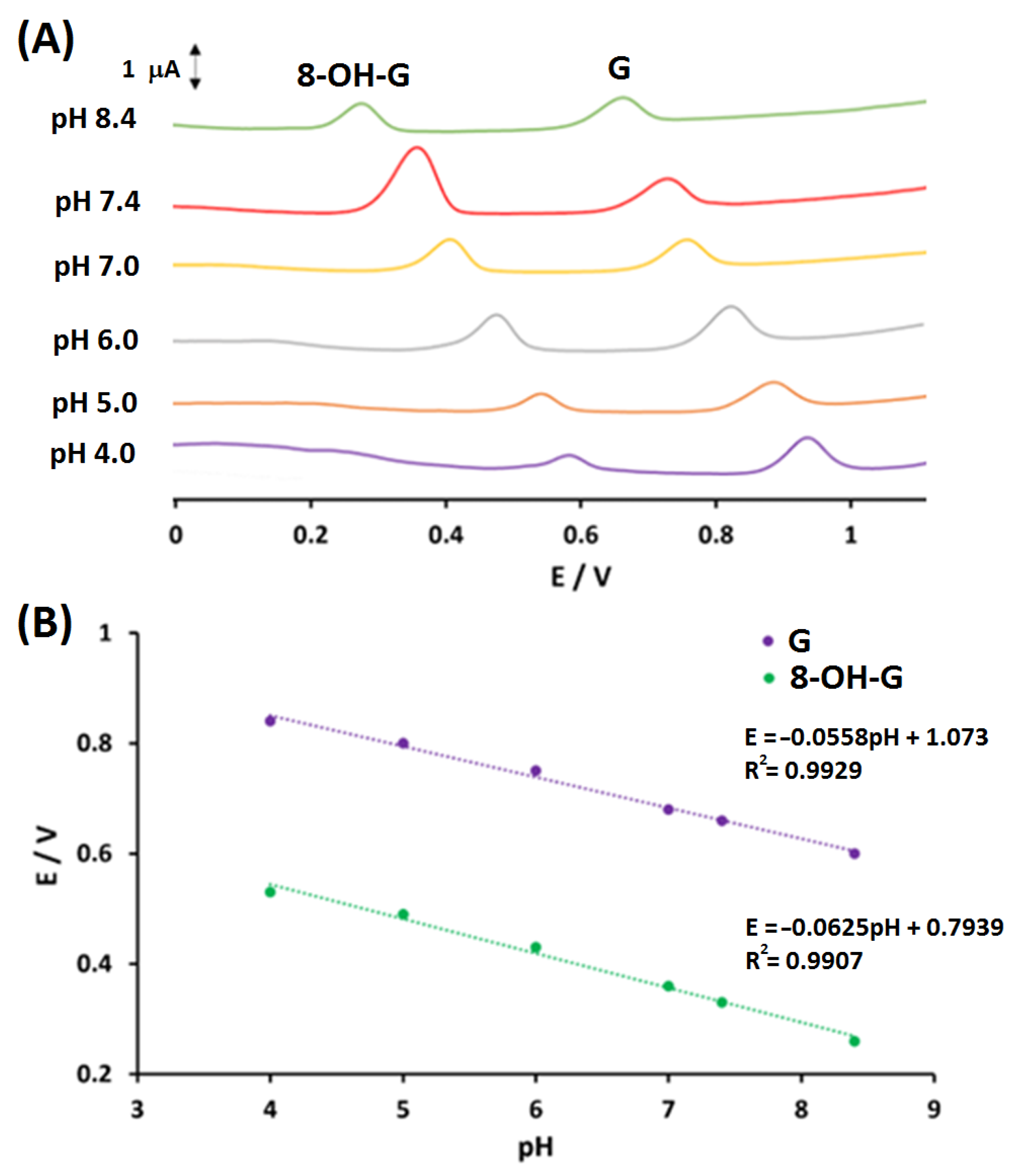
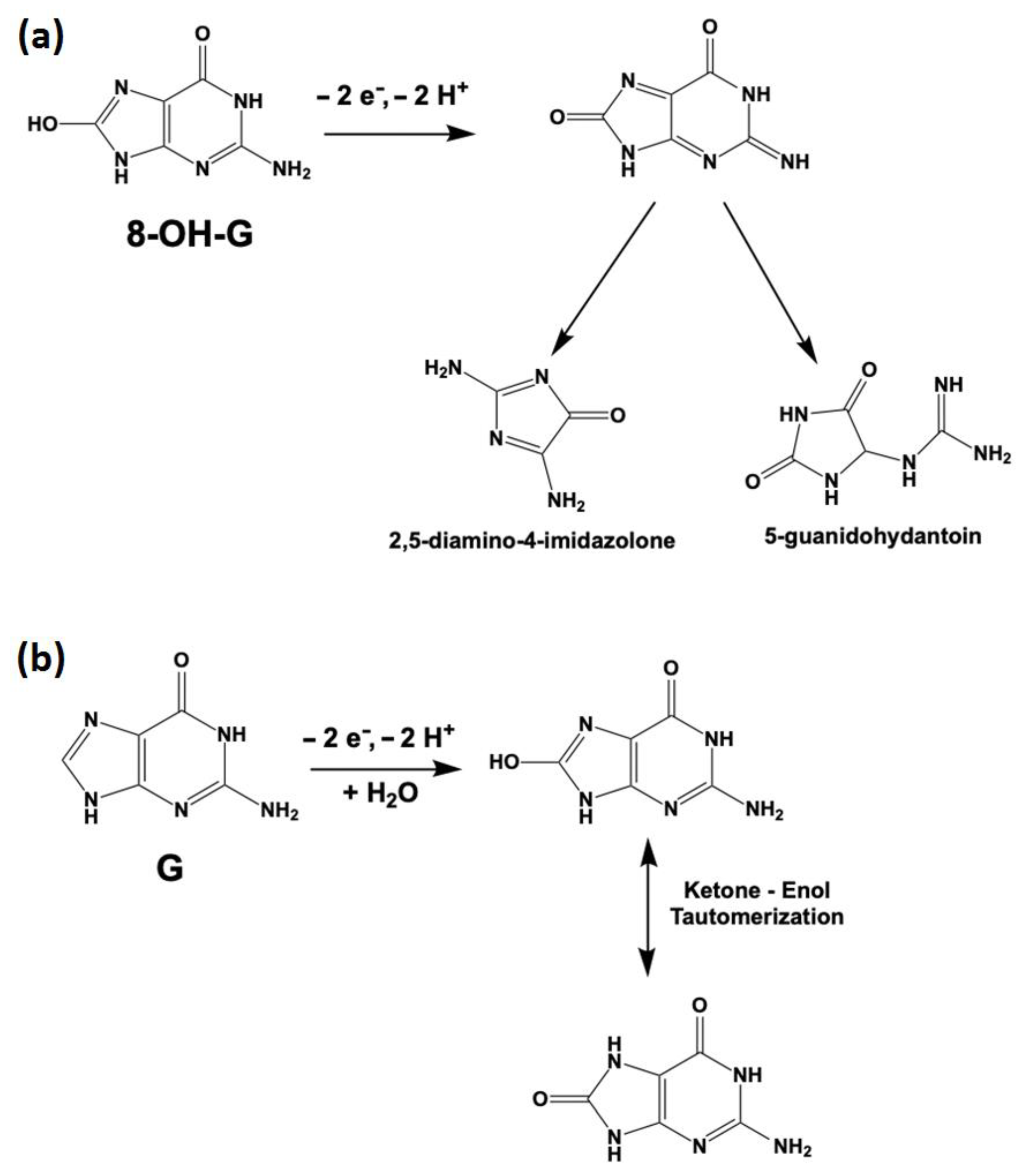
3.3. pH Study
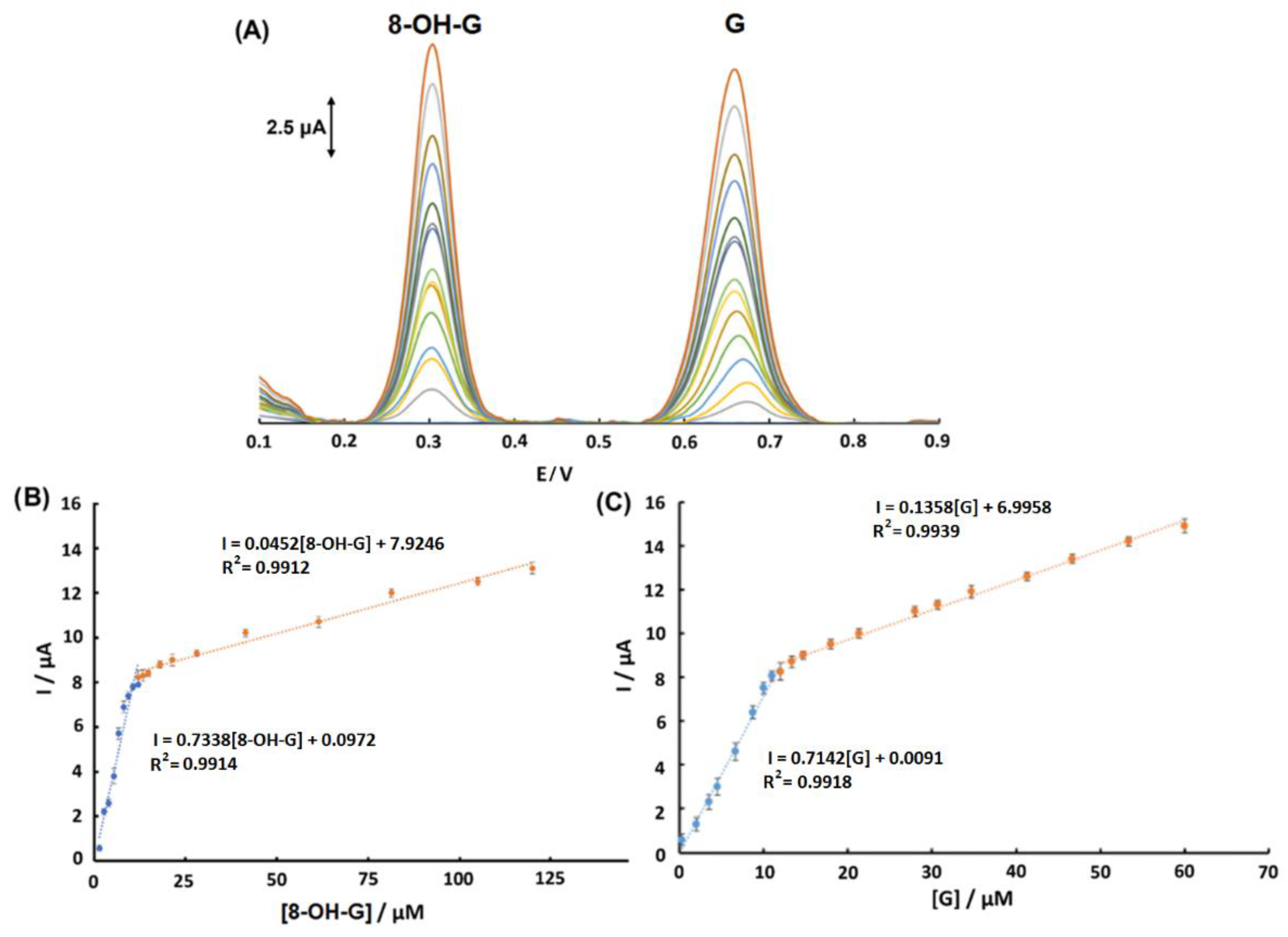
3.4. Calibration Study
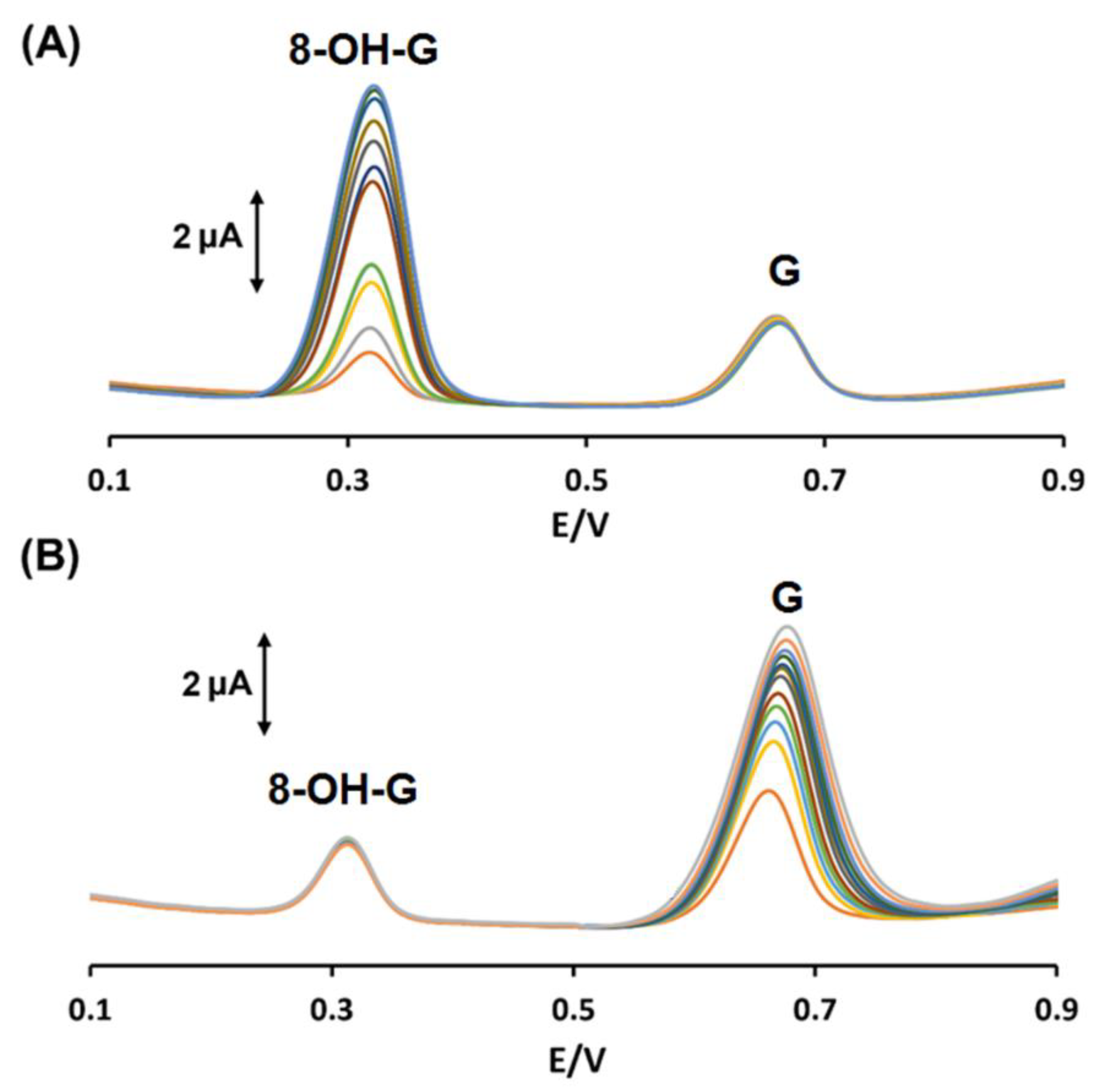
3.5. Interference Study
3.6. Electrochemical Impedance Spectroscopy (EIS) and Scan Rate Study
3.7. Chronoamperometry
3.8. Real Sample Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–562. [Google Scholar] [CrossRef]
- Cobb, M. 60 Years Ago, Francis Crick Changed the Logic of Biology. PLoS Biol. 2017, 15, e2003243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crick, F.; Watson, J. Molecular Structure of Nucleic Acids. Nature 1953, 171, 737–738. [Google Scholar]
- Idzko, M.; Ferrari, D.; Riegel, A.K.; Eltzschig, H.K. Extracellular Nucleobase and Nucleoside Signaling in Vascular and Blood Disease. Blood 2014, 124, 1029–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peplinska-Miaskowska, J.; Wichowicz, H.; Smolenski, R.T.; Jablonska, P.; Kaska, L. Comparison of Plasma Nucleobase Metabolites and Amino Acids Pattern in Patients with Binge Eating Disorder and Obesity. Nucleos. Nucleot. Nucl. 2020, 40, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Kamiya, H. Mutations Induced by 8-Hydroxyguanine (8-Oxo-7,8-Dihydroguanine), a Representative Oxidized Base, in Mammalian Cells. Genes Environ. 2017, 39, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an Abundant Form of Oxidative DNA Damage, Causes G → T and A → C Substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [CrossRef]
- Lovell, M.A.; Markesbery, W.R. Ratio of 8-Hydroxyguanine in Intact DNA to Free 8-Hydroxyguanine Is Increased in Alzheimer Disease Ventricular Cerebrospinal Fluid. Arch. Neurol. 2001, 58, 392–396. [Google Scholar] [CrossRef] [Green Version]
- Lovell, M.A.; Markesbery, W.R. Oxidative DNA Damage in Mild Cognitive Impairment and Late-Stage Alzheimer’s Disease. Nucleic Acids Res. 2007, 35, 7497–7504. [Google Scholar] [CrossRef] [Green Version]
- Alam, Z.I.; Jenner, A.; Daniel, S.E.; Lees, A.J.; Cairns, N.; Marsden, C.D.; Jenner, P.; Halliwell, B. Oxidative DNA Damage in the Parkinsonian Brain: An Apparent Selective Increase in 8-Hydroxyguanine Levels in Substantia Nigra. J. Neurochem. 1997, 69, 1196–1203. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Li, X.; Li, X.; Liu, Q.; Cheng, Y. Oxidative Stress in Parkinson’s Disease: A Systematic Review and Meta-Analysis. Front. Mol. Neurosci. 2018, 11, 236. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Kohno, T.; Kasai, H.; Koda, K.; Sugimura, H.; Yokota, J. Infrequent Mutations of the HOGG1 Gene, That Is Involved in the Excision of 8-Hydroxyguanine in Damaged DNA, in Human Gastric Cancer. Jpn. J. Cancer Res. 1998, 89, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Rozalski, R.; Gackowski, D.; Roszkowski, K.; Foksinski, M.; Olinski, R. The Level of 8-Hydroxyguanine, a Possible Repair Product of Oxidative DNA Damage, Is Higher in Urine of Cancer Patients than in Control Subjects1. Cancer Epidemiol. Biomark. Prev. 2002, 11, 1072–1075. [Google Scholar]
- Arai, T.; Kelly, V.P.; Minowa, O.; Noda, T.; Nishimura, S. High Accumulation of Oxidative DNA Damage, 8-Hydroxyguanine, in Mmh/Ogg1 Deficient Mice by Chronic Oxidative Stress. Carcinogenesis 2002, 23, 2005–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Kawasaki, Y.; Kawai, K. Salivary 8-Hydroxyguanine as a Lifestyle-Related Oxidative Stress Biomarker in Workers. J. Clin. Biochem. Nutr. 2020, 66, 57–61. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, H.; Jurado, J.; Laval, J.; Dizdaroglu, M. Comparison of the Levels of 8-Hydroxyguanine in DNA as Measured by Gas Chromatography Mass Spectrometry Following Hydrolysis of DNA by Escherichia Coli Fpg Protein or Formic Acid. Nucleic Acids Res. 2000, 28, 75. [Google Scholar] [CrossRef] [Green Version]
- England, T.G.; Jenner, A.; Aruoma, O.I.; Halliwell, B. Determination of Oxidative DNA Base Damage by Gas Chromatography-Mass Spectrometry. Effect of Derivatization Conditions on Artifactual Formation of Certain Base Oxidation Products. Free Radic. Res. 1998, 29, 321–330. [Google Scholar] [CrossRef]
- Herbert, K.E.; Evans, M.D.; Finnegan, M.T.V.; Farooq, S.; Mistry, N.; Podmore, I.D.; Farmer, P.; Lunec, J. A Novel HPLC Procedure for the Analysis of 8-Oxoguanine in DNA. Free Radic. Biol. Med. 1996, 20, 467–473. [Google Scholar] [CrossRef]
- Kawai, K.; Kasai, H.; Li, Y.S.; Kawasaki, Y.; Watanabe, S.; Ohta, M.; Honda, T.; Yamato, H. Measurement of 8-Hydroxyguanine as an Oxidative Stress Biomarker in Saliva by HPLC-ECD. Genes Environ. 2018, 40, 2–5. [Google Scholar] [CrossRef]
- Helbock, H.J.; Beckman, K.B.; Ames, B.N. 8-Hydroxydeoxyguanosine and 8-Hydroxyguanine as Biomarkers of Oxidative DNA Damage. Methods Enzym. 1999, 300, 156–166. [Google Scholar] [CrossRef]
- Ripanti, F.; Fasolato, C.; Mazzarda, F.; Palleschi, S.; Ceccarini, M.; Li, C.; Bignami, M.; Bodo, E.; Bell, S.E.J.; Mazzei, F.; et al. Advanced Raman Spectroscopy Detection of Oxidative Damage in Nucleic Acid Bases: Probing Chemical Changes and Intermolecular Interactions in Guanosine at Ultralow Concentration. Anal. Chem. 2021, 93, 10825–10833. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, F.; Cammisuli, F.; Addobbati, R.; Rizzardi, C.; Gessini, A.; Masciovecchio, C.; Rossi, B.; Pascolo, L. Oxidative Damage in DNA Bases Revealed by UV Resonant Raman Spectroscopy. Analyst 2015, 140, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Beaver, K.; Dantanarayana, A.; Minteer, S.D. Materials Approaches for Improving Electrochemical Sensor Performance. J. Phys. Chem. B 2021, 125, 11820–11834. [Google Scholar] [CrossRef] [PubMed]
- Momenbeitollahi, N.; van der Zalm, J.; Chen, A.; Li, H. Entrapping Gold Nanoparticles in Membranes for Simple-to-Use Enhanced Fluorescence Detection of Proteins. Anal. Chim. Acta 2022, 1195, 339443. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.O.; Piedade, J.A.P.; Serrano, S.H.P. Electrochemical Oxidation of 8-Oxoguanine. Electroanalysis 2000, 12, 969–973. [Google Scholar] [CrossRef]
- Jeličová, M.; Metelka, R.; Pejchal, J.; Lierová, A.; Šinkorová, Z. Electrochemical Detection of 8-Hydroxyguanine Using Screen-Printed Carbon Electrodes Modified with Carboxy-Functionalized Multi-Walled Carbon Nanotubes. Mon. Chem. 2019, 150, 1187–1193. [Google Scholar] [CrossRef]
- Lu, N.; Liu, H.; Huang, R.; Gu, Y.; Yan, X.; Zhang, T.; Xu, Z.; Xu, H.; Xing, Y.; Song, Y.; et al. Charge Transfer Platform and Catalytic Amplification of Phenanthroimidazole Derivative: A New Strategy for DNA Bases Recognition. Anal. Chem. 2019, 91, 11938–11945. [Google Scholar] [CrossRef] [PubMed]
- Emran, M.Y.; El-Safty, S.A.; Selim, M.M.; Shenashen, M.A. Selective Monitoring of Ultra-Trace Guanine and Adenine from Hydrolyzed DNA Using Boron-Doped Carbon Electrode Surfaces. Sens. Actuators B Chem. 2021, 329, 129192. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Wei, Y.; Xing, T.; Cao, T.; Wu, S.; Zhu, F. A Copper-Based Metal-Organic Framework/Graphene Nanocomposite for the Sensitive and Stable Electrochemical Detection of DNA Bases. Analyst 2020, 145, 1933–1942. [Google Scholar] [CrossRef]
- Wang, M.; Guo, H.; Xue, R.; Guan, Q.; Zhang, J.; Zhang, T.; Sun, L.; Yang, F.; Yang, W. A Novel Electrochemical Sensor Based on MWCNTs-COOH/Metal-Covalent Organic Frameworks (MCOFs)/Co NPs for Highly Sensitive Determination of DNA Base. Microchem. J. 2021, 167, 106336. [Google Scholar] [CrossRef]
- Goyal, R.N.; Bishnoi, S. Sensitive Voltammetric Sensor for the Determination of Oxidative DNA Damage in Calf Thymus DNA. Biosens. Bioelectron. 2010, 26, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Luais, E.; Gooding, J.J. The Fabrication of Stable Gold Nanoparticle-Modified Interfaces for Electrochemistry. Langmuir 2011, 27, 4176–4183. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Preparation of Aptamer-Linked Gold Nanoparticle Purple Aggregates for Colorimetric Sensing of Analytes. Nat. Protoc. 2006, 1, 246–252. [Google Scholar] [CrossRef]
- Tamano, H.; Nishio, R.; Shakushi, Y.; Sasaki, M.; Koike, Y.; Osawa, M.; Takeda, A. In vitro and in vivo physiology of low nanomolar concentrations of Zn2+ in artificial cerebrospinal fluid. Sci. Rep. 2017, 7, 42897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekanski, A.; Stevanović, J.; Stevanović, R.; Nikolić, B.Ž.; Jovanović, V.M. Glassy Carbon Electrodes I. Characterization and Electrochemical Activation. Carbon 2001, 39, 1195–1205. [Google Scholar] [CrossRef]
- Kwan, W.S.V.; Atanasoska, L.; Miller, L.L. Oligoimide Monolayers Covalently Attached to Gold. Langmuir 1991, 7, 1419–1425. [Google Scholar] [CrossRef]
- Sylvestre, J.P.; Poulin, S.; Kabashin, A.V.; Sacher, E.; Meunier, M.; Luong, J.H.T. Surface Chemistry of Gold Nanoparticles Produced by Laser Ablation in Aqueous Media. J. Phys. Chem. B 2004, 108, 16864–16869. [Google Scholar] [CrossRef]
- Li, S.P.; Kerman, K. Electrochemical Detection of Interaction between Copper(II) and Peptides Related to Pathological α-Synuclein Mutants. Anal. Chem. 2019, 91, 3810–3826. [Google Scholar] [CrossRef]
- Wang, S.; Ferrag, C.; Noroozifar, M.; Kerman, K. Simultaneous Determination of Four DNA Bases at Graphene Oxide/Multi-Walled Carbon Nanotube Nanocomposite-Modified Electrode. Micromachines 2020, 11, 294. [Google Scholar] [CrossRef] [Green Version]
- Bishnoi, S.; Goyal, R.N.; Shim, Y.B. A Novel Nanogold-Single Wall Carbon Nanotube Modified Sensor for the Electrochemical Determination of 8-Hydroxyguanine, a Diabetes Risk Biomarker. Bioelectrochemistry 2014, 99, 24–29. [Google Scholar] [CrossRef]
- Goyal, R.N.; Dryhurst, G. Redox Chemistry of Guanine and 8-Oxyguanine and a Comparison of the Peroxidase-Catalyzed and Electrochemical Oxidation of 8-Oxyguanine. J. Electroanal. Chem. 1982, 135, 75–91. [Google Scholar] [CrossRef]
- Goyal, R.N.; Puri, B.K.; Jain, N. Electrochemical Oxidation of Guanosine-5′-Monophosphate at the Pyrolytic Graphite Electrode. J. Chem. Soc. Perkin Trans. 2 2001, 5, 832–837. [Google Scholar] [CrossRef]
- Patel, B.R.; Imran, S.; Ye, W.; Weng, H.; Noroozifar, M.; Kerman, K. Simultaneous Voltammetric Detection of Six Biomolecules Using a Nanocomposite of Titanium Dioxide Nanorods with Multi-Walled Carbon Nanotubes. Electrochim. Acta 2020, 362, 137094. [Google Scholar] [CrossRef]
- Zhou, J.; Li, S.; Noroozifar, M.; Kerman, K. Graphene Oxide Nanoribbons in Chitosan for Simultaneous Electrochemical Detection of Guanine, Adenine, Thymine and Cytosine. Biosensors 2020, 10, 30. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Li, B.-Q.; Zheng, J.-B. An Electrochemical Sensor for the Sensitive Determination of Nitrites Based on Pt-PANI-Graphene Nanocomposites. Anal. Methods 2015, 7, 8366–8372. [Google Scholar] [CrossRef]
- Oliveira, S.C.B.; Oliveira-Brett, A.M. Boron Doped Diamond Electrode Pre-Treatments Effect on the Electrochemical Oxidation of dsDNA, DNA Bases, Nucleobases, Homopolynucleobases and Biomarker 8-Oxoguanine. J. Electroanal. Chem. 2010, 648, 60–66. [Google Scholar] [CrossRef]
- Yin, H.; Zhou, Y.; Ma, Q.; Ai, S.; Ju, P.; Zhu, L.; Lu, L. Electrochemical Oxidation Behavior of Guanine and Adenine on Graphene-Nafion Composite Film Modified Glassy Carbon Electrode and the Simultaneous Determination. Process. Biochem. 2010, 45, 1707–1712. [Google Scholar] [CrossRef]
- González-Meza, O.A.; Larios-Durán, E.R.; Gutiérrez-Becerra, A.; Casillas, N.; Escalante, J.I.; Bárcena-Soto, M. Development of a Randles-Ševčík-like Equation to Predict the Peak Current of Cyclic Voltammetry for Solid Metal Hexacyanoferrates. J. Solid State Electrochem. 2019, 23, 3123–3133. [Google Scholar] [CrossRef]
- Moldenhauer, J.; Meier, M.; Paul, D.W. Rapid and Direct Determination of Diffusion Coefficients Using Microelectrode Arrays. J. Electrochem. Soc. 2016, 163, H672–H678. [Google Scholar] [CrossRef]
| Platform | Method | Nucleobases | LOD (μM) | Linear Range (μM) | Ref. |
|---|---|---|---|---|---|
| Au nanoparticle-SWCNT-modified edge-plane pyrolytic graphite electrode | SWV | G | 0.005 | 0.1–2.0 | [40] |
| 8-OH-G | 0.001 | 2.5 × 10−4–6.25 × 10−3 | |||
| Screen-printed carbon electrode modified with MWCNT-COOH | DPV | 8-OH-G | 0.57 | 0.3–12.0 | [26] |
| Boron-doped diamond electrode | DPV | 8-OH-G | 1.0 | 0–11.4 | [46] |
| Bare GCE | DPV | 8-OH-G | 0.8 | 6.0–28.0 | [25] |
| Graphene-Nafion-modified GCE | DPV | G | 0.58 | 2–120 | [47] |
| GO-MWCNT hybrid with chitosan-modified GCE | DPV | G | 0.11 | 1.0–78.0 | [39] |
| GO nanoribbons in chitosan-modified GCE | DPV | G | 0.0018 | 0.013–256 | [44] |
| Copper metal-organic framework with reduced graphene oxide-modified GCE | DPV | G | 0.012 | 0.02–100 | [29] |
| MWCNT-COOH with a Cu -porphyrin metal covalent organic framework with Co nanoparticles on GCE | DPV | G | 0.0055 | 0.04–130 | [30] |
| GCE-Ph-S-AuNPs | DPV | 8-OH-G | 0.020 | 1.3–12.0 & 12.0–125.0 | This work |
| G | 0.021 | 0.3–12.0 & 12.0–60.0 |
| Matrix | Analyte | Detected (µM) | Spiked (µM) | Found (µM) ± SD | Relative Recovery (%) ± SD |
|---|---|---|---|---|---|
| FS-DNA | 8-OH-G | - | 2.0 | 1.97 ± 0.08 | 98.5 ± 0.08 |
| G | - | 2.0 | 2.04 ± 0.06 | 102.0 ± 0.06 | |
| 8-OH-G | - | 15.0 | 15.5 ± 0.10 | 103.3 ± 0.10 | |
| G | - | 15.0 | 14.8 ± 0.12 | 98.7 ± 0.12 | |
| ACSF | 8-OH-G | - | 6.0 | 6.2 ± 0.18 | 103.3 ± 0.18 |
| G | - | 10.0 | 9.6 ± 0.07 | 96.0 ± 0.07 | |
| 8-OH-G | - | 40.0 | 42.7 ± 0.21 | 106.7 ± 0.21 | |
| G | - | 45.0 | 46.3 ± 0.16 | 102.8 ± 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltani, N.; Hassan, Q.; Noroozifar, M.; Kerman, K. Au Nanoparticles on 4-Thiophenol-Electrodeposited Carbon Surfaces for the Simultaneous Detection of 8-Hydroxyguanine and Guanine. Chemosensors 2023, 11, 326. https://doi.org/10.3390/chemosensors11060326
Soltani N, Hassan Q, Noroozifar M, Kerman K. Au Nanoparticles on 4-Thiophenol-Electrodeposited Carbon Surfaces for the Simultaneous Detection of 8-Hydroxyguanine and Guanine. Chemosensors. 2023; 11(6):326. https://doi.org/10.3390/chemosensors11060326
Chicago/Turabian StyleSoltani, Niloufar, Qusai Hassan, Meissam Noroozifar, and Kagan Kerman. 2023. "Au Nanoparticles on 4-Thiophenol-Electrodeposited Carbon Surfaces for the Simultaneous Detection of 8-Hydroxyguanine and Guanine" Chemosensors 11, no. 6: 326. https://doi.org/10.3390/chemosensors11060326





