Enose Lab Made with Vacuum Sampling: Quantitative Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Calibrations with Individual Standard Solutions
2.2. Calibrations with Standard Mixed Solutions
2.3. Lab-Made Enose
2.4. Pre-Procedures before Enose Analysis
2.5. Sample Analysis by Enose
2.6. Statistical Analysis
3. Results
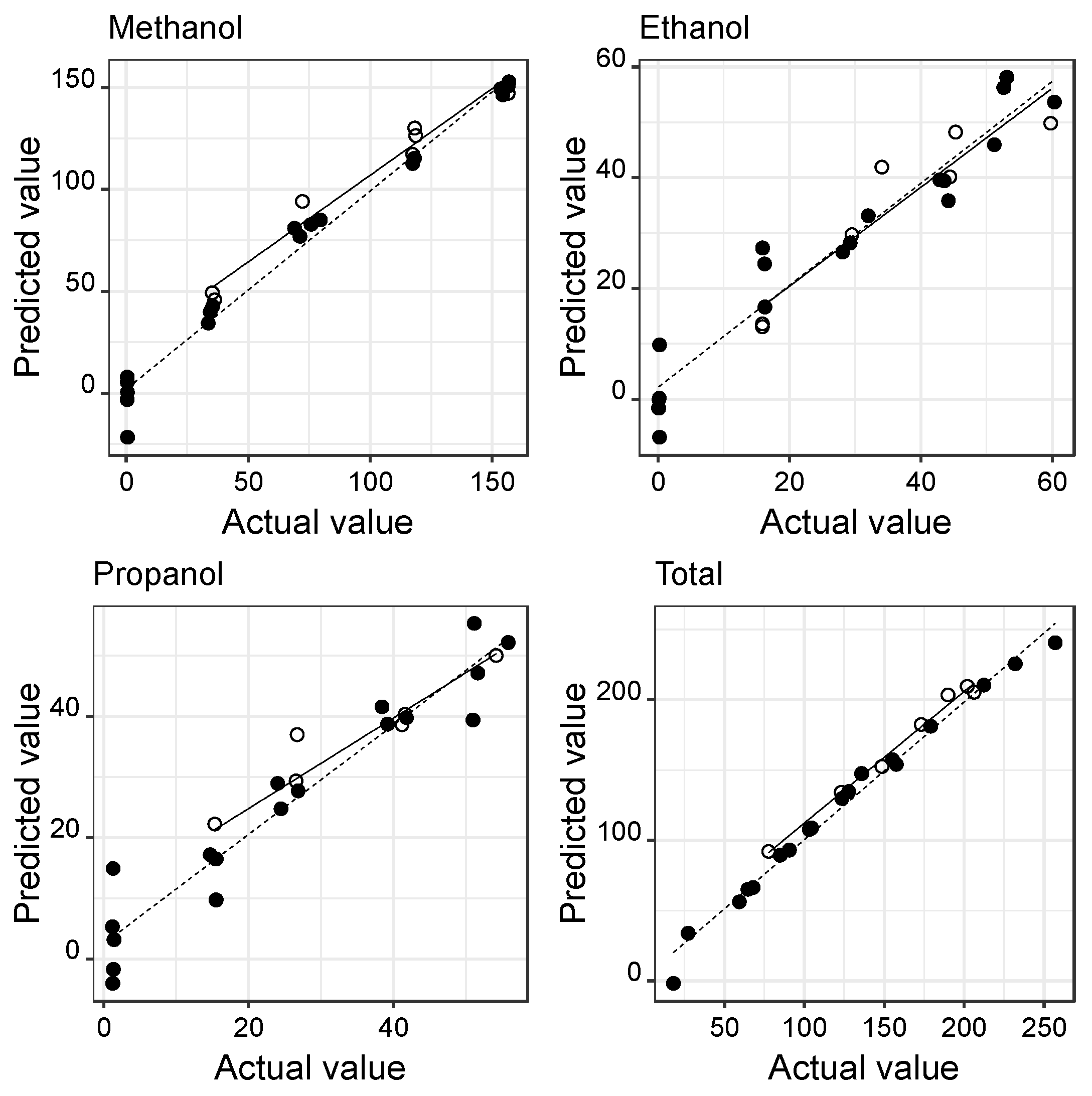
3.1. Calibrations with Individual Standard Solutions
3.2. Calibrations with Standard Mixed Solutions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eduardo, C.; Peñaloza, C. Nariz electrónica: Herramienta para detección de gases empleando redes neuronales artificiales. Electronic nose: Tool for gas detection using Artificial Neural Networks. Rev. Tecnol. Digit. 2018, 8, 39–47. [Google Scholar]
- Boeker, P. On ‘Electronic Nose’ methodology. Sens. Actuators B Chem. 2014, 204, 2–17. [Google Scholar] [CrossRef]
- Jordan Voss, H.G.; Mendes Júnior, J.J.A.; Farinelli, M.E.; Stevan, S.L. A prototype to detect the alcohol content of beers based on an electronic nose. Sensors 2019, 19, 2646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalpana, S.; Lakshmi Baghyam, A. Electronic-nose system for classification of fruits and freshness measurement using K-NN algorithm. Int. J. Innov. Technol. Explor. Eng. 2019, 8, 641–644. [Google Scholar]
- Sanaeifar, A.; ZakiDizaji, H.; Jafari, A.; de la Guardia, M. Early detection of contamination and defect in foodstuffs by electronic nose: A review. Trends Anal. Chem. 2017, 97, 257–271. [Google Scholar] [CrossRef]
- Gliszczyńska-Świgło, A.; Chmielewski, J. Electronic Nose as a Tool for Monitoring the Authenticity of Food. A Review. Food Anal. Methods 2017, 10, 1800–1816. [Google Scholar]
- Dey, A. Semiconductor metal oxide gas sensors: A review. Mater. Sci. Eng. B 2018, 229, 206–217. [Google Scholar] [CrossRef]
- Estakhroyeh, H.R.; Rashedi, E.; Mehran, M. Design and Construction of Electronic Nose for Multi-purpose Applications by Sensor Array Arrangement Using IBGSA. J. Intell. Robot. Syst. Theory Appl. 2018, 92, 205–221. [Google Scholar] [CrossRef]
- Ellis, D.I.; Brewster, V.L.; Dunn, W.B.; Allwood, J.W.; Golovanovc, A.P.; Goodacrea, R. Fingerprinting food: Current technologies for the detection of food adulteration and contamination. Chem. Soc. Rev. 2012, 41, 5706–5727. [Google Scholar] [CrossRef]
- Mumyakmaz, B.; Karabacak, K. An E-Nose-based indoor air quality monitoring system: Prediction of combustible and toxic gas concentrations. Turk. J. Electr. Eng. Comput. Sci. 2015, 23, 729–740. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, H.; Wang, X.; Zheng, H.; Chen, Z.; Meng, C. Development of Electronic Nose for Qualitative and Quantitative Monitoring of Volatile Flammable Liquids. Sensors 2020, 20, 1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aagód, G.; Guz, A.; Sabba, F.; Sobczuk, H. Detection of Wastewater Treatment Process Disturbances in Bioreactors Using the E-Nose Technology. Ecol. Chem. Eng. S 2018, 25, 405–418. [Google Scholar]
- System, E.; Dur, C. Concentration Detection of the E. coli Bacteria in Drinking Water Treatment Plants through an E-Nose. Water 2019, 11, 774. [Google Scholar]
- Viejo, C.G.; Fuentes, S.; Godbole, A.; Widdicombe, B.; Unnithan, R.R. Development of a low-cost e-nose to assess aroma profiles: An artificial intelligence application to assess beer quality. Sens. Actuators B Chem. 2020, 308, 127688. [Google Scholar] [CrossRef]
- Buratti, S.; Benedetti, S. Chapter 28—Alcoholic Fermentation Using Electronic Nose and Electronic Tongue; Electronic Noses and Tongues in Food Science; Academic Press: San Diego, CA, USA, 2016. [Google Scholar]
- Cui, S.; Wu, J.; Wang, J.; Wang, X. Discrimination of American ginseng and Asian ginseng using electronic nose and gas chromatography-mass spectrometry coupled with chemometrics. J. Ginseng Res. 2017, 41, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieganowski, A.; Jaromin-Glen, K.; Guz, Ł.; Łagód, G.; Jozefaciuk, G.; Franus, W.; Suchorab, Z.; Sobczuk, H. Evaluating soil moisture status using an e-nose. Sensors 2016, 16, 886. [Google Scholar] [CrossRef] [Green Version]
- Majchrzak, T.; Wojnowski, W.; Dymerski, T.; Jacek, G. Electronic noses in classification and quality control of edible oils: A review. Food Chem. 2018, 246, 192–201. [Google Scholar] [CrossRef]
- Teixeira, G.G.; Dias, L.G.; Rodrigues, N.; Marx, Í.M.G.; Veloso, A.C.A.; Pereira, J.A.; Peres, A.M. Application of a lab-made electronic nose for extra virgin olive oils commercial classification according to the perceived fruitiness intensity. Talanta 2021, 226, 122122. [Google Scholar] [CrossRef]
- Loutfi, A.; Coradeschi, S.; Mani, G.K.; Shankar, P.; Rayappan, J.B.B. Electronic noses for food quality: A review. J. Food Eng. 2015, 144, 103–111. [Google Scholar] [CrossRef]
- Chen, Y. Reference-related component analysis: A new method inheriting the advantages of PLS and PCA for separating interesting information and reducing data dimension. Chemom. Intell. Lab. Syst. 2016, 156, 196–202. [Google Scholar] [CrossRef]
- Ma, H.; Wang, T.; Li, B.; Cao, W.; Zeng, M.; Yang, J.; Su, Y.; Hu, N.; Zhou, Z.; Yang, Z. A low-cost and efficient electronic nose system for quantification of multiple indoor air contaminants utilizing HC and PLSR. Sens. Actuators B. Chem. 2022, 350, 130768. [Google Scholar] [CrossRef]
- Wijaya d r Afianti, F.; Arifianto, A.; Rahmawati, D.; Kodogiannis, V.S. Ensemble machine learning approach for electronic nose signal processing. Sens. Bio-Sens. Res. 2022, 36, 100495. [Google Scholar] [CrossRef]
- Kuhn, M. Caret: Classification and Regression Training; R Package Version 6.0-86; Astrophysics Source Code Library: Cambridge, MA, USA, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Saleh, A.K.M.E.; Arashi, M.; Golam, B.M. Theory of Ridge Regression Estimation with Applications; Wiley Series in Probability and Statistics Book 285; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019. [Google Scholar]
- Roig, B.; Thomas, O. Rapid estimation of global sugars by UV photodegradation and UV spectrophotometry. Anal. Chim. Acta 2003, 477, 325–329. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Costa Arca, V.; Peres, A.M.; Machado, A.A.S.C.; Bona, E.; Dias, L.G. Sugars’ Quantifications Using a Potentiometric Electronic Tongue with Cross-Selective Sensors: Influence of an Ionic Background. Chemosensors 2019, 7, 43. [Google Scholar] [CrossRef] [Green Version]




| Coded Levels | |||
|---|---|---|---|
| Assay No. | Compound 1 | Compound 2 | Compound 3 |
| 1 | 0 | 0 | 0 |
| 2 | 0 | −2 | −1 |
| 3 | −2 | −1 | −2 |
| 4 | −1 | −2 | 2 |
| 5 | −2 | 2 | 2 |
| 6 | 2 | 2 | 0 |
| 7 | 2 | 0 | −1 |
| 8 | 0 | −1 | 2 |
| 9 | −1 | 2 | −1 |
| 10 | 2 | −1 | 1 |
| 11 | −1 | 1 | 1 |
| 12 | 1 | 1 | 0 |
| 13 | 1 | 0 | 2 |
| 14 | 0 | 2 | 1 |
| 15 | 2 | 1 | 2 |
| 16 | 1 | 2 | −2 |
| 17 | 2 | −2 | −2 |
| 18 | −2 | −2 | 0 |
| 19 | −2 | 0 | 1 |
| 20 | 0 | 1 | −2 |
| 21 | 1 | −2 | 1 |
| 22 | −2 | 1 | −1 |
| 23 | 1 | −1 | −1 |
| 24 | −1 | −1 | 0 |
| 25 | −1 | 0 | −2 |
| Code | Sensor | Tested Gases |
|---|---|---|
| S1 | TGS 2600 B00 | Methane, CO, isobutane, ethanol, and H2 |
| S2 | TGS 2602 | H2, NH3, ethanol, H2S, and toluene |
| S3 | TGS 2610 C00 | Ethanol, isobutane, and H2 |
| S4 | TGS 2611 C00 | Ethanol, H2, isobutane, and methane |
| S5 | TGS 2610 D00 | Ethanol, H2, and isobutane |
| S6 | TGS 2611 E00 | Ethanol, isobutane, H2, and methane |
| S7 | TGS 2612 | Ethanol, methane, isobutane, and propane |
| S8 | TGS 826 A00 | Isobutane, H2, ammonia, and ethanol |
| S9 | TGS 823 C12N | Methane, CO, isobutane, n-hexane, benzene, ethanol, and acetone |
| Compound | Boiling Point, °C | Cmin, g/L | Cmax, g/L | Resistance Interval, Ohm |
|---|---|---|---|---|
| Methanol | 64.7 | 0.10 | 315.2 | [186; 33,374] |
| Ethanol | 78.4 | 0.14 | 307.5 | [194; 33,170] |
| Propanol | 97.0 | 0.14 | 306.9 | [225; 33,252] |
| Acetaldehyde | 20.2 | 0.018 | 56.4 | [106; 28,072] |
| Ethyl acetate | 77.1 | 0.024 | 34.8 | [75; 29,022] |
| Compound | Lambda | RMSE | MAE | R2 |
|---|---|---|---|---|
| Mixture of 3 alcohols | ||||
| Methanol | 0.0028 | 17.37 ± 15.33 | 14.08 ± 11.29 | 0.98 ± 0.05 |
| Ethanol | 0.0028 | 10.49 ± 5.04 | 8.93 ± 3.94 | 0.84 ± 0.26 |
| Propanol | 0.0028 | 10.72 ± 7.49 | 8.93 ± 5.64 | 0.88 ± 0.18 |
| Total | 0.0028 | 15.37 ± 15.06 | 12.33 ± 11.42 | 0.97 ± 0.06 |
| Mixture of 3 organic compounds with different functional groups | ||||
| Acetaldehyde | 1.99 | 0.19 ± 0.07 | 0.17 ± 0.06 | 0.54 ± 0.41 |
| Ethanol | 0.0028 | 9.43 ± 4.33 | 8.36 ± 3.98 | 0.90 ± 0.18 |
| Ethyl acetate | 0.0028 | 2.88 ± 1.38 | 2.59 ± 1.22 | 0.96 ± 0.08 |
| Total | 0.0028 | 7.98 ± 3.57 | 6.99 ± 3.33 | 0.93 ± 0.12 |
| Training Data Set | Testing Data Set | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | RSE | R2 | p-Value | Slope (p-Value) | Intercept (p-Value) | RSE | R2 | p-Value | Slope (p-Value) | Intercept (p-Value) |
| Mixture of 3 alcohols | ||||||||||
| Methanol | 7.90 | 0.993 | <0.001 | 0.99 ± 0.02 (<0.001) | Ns (0.528) | 8.05 | 0.966 | <0.001 | 0.85 ± 0.07 (<0.001) | 21.88 ± 7.29 (0.030) |
| Ethanol | 5.61 | 0.974 | <0.001 | 0.97 ± 0.04 (<0.001) | Ns (0.342) | 5.47 | 0.981 | <0.001 | 0.95 ± 0.05 (<0.001) | Ns (0.674) |
| Propanol | 5.33 | 0.972 | <0.001 | 0.96 ± 0.04 (<0.001) | Ns (0.231) | 5.58 | 0.978 | <0.001 | 1.01 ± 0.06 (<0.001) | Ns (0.052) |
| Total | 7.91 | 0.997 | <0.001 | 1.00 ± 0.01 (<0.001) | Ns (0.587) | 6.91 | 0.999 | <0.001 | 1.04 ± 0.02 (<0.001) | Ns (0.050) |
| Mixture of 3 organic compounds with different functional groups | ||||||||||
| Acetaldehyde | 0.06 | 0.07 | 0.303 | 0.10 ± 0.09 (0.303) | 0.16 ± 0.02 (<0.001) | 0.04 | 0.02 | 0.786 | −0.03 ± 0.09 (0.786) | 0.23 ± 0.03 (<0.001) |
| Ethanol | 4.05 | 0.957 | <0.001 | 0.84 ± 0.04 (<0.001) | 4.42 ± 1.54 (0.011) | 2.01 | 0.973 | <0.001 | 0.72 ± 0.05 (<0.001) | 5.41 ± 1.90 (0.036) |
| Ethyl acetate | 1.56 | 0.995 | <0.001 | 0.99 ± 0.02 (<0.001) | Ns (0.591) | 1.14 | 0.998 | <0.001 | 1.07 ± 0.02 (<0.001) | Ns (0.411) |
| Total | 4.36 | 0.966 | <0.001 | 0.89 ± 0.04 (<0.001) | 4.72 ± 2.10 (0.039) | 2.26 | 0.986 | <0.001 | 0.83 ± 0.04 (<0.001) | 6.57 ± 2.52 (0.048) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, G.G.; Peres, A.M.; Estevinho, L.; Geraldes, P.; Garcia-Cabezon, C.; Martin-Pedrosa, F.; Rodriguez-Mendez, M.L.; Dias, L.G. Enose Lab Made with Vacuum Sampling: Quantitative Applications. Chemosensors 2022, 10, 261. https://doi.org/10.3390/chemosensors10070261
Teixeira GG, Peres AM, Estevinho L, Geraldes P, Garcia-Cabezon C, Martin-Pedrosa F, Rodriguez-Mendez ML, Dias LG. Enose Lab Made with Vacuum Sampling: Quantitative Applications. Chemosensors. 2022; 10(7):261. https://doi.org/10.3390/chemosensors10070261
Chicago/Turabian StyleTeixeira, Guilherme G., António M. Peres, Letícia Estevinho, Pedro Geraldes, Cristina Garcia-Cabezon, Fernando Martin-Pedrosa, Maria Luz Rodriguez-Mendez, and Luís G. Dias. 2022. "Enose Lab Made with Vacuum Sampling: Quantitative Applications" Chemosensors 10, no. 7: 261. https://doi.org/10.3390/chemosensors10070261
APA StyleTeixeira, G. G., Peres, A. M., Estevinho, L., Geraldes, P., Garcia-Cabezon, C., Martin-Pedrosa, F., Rodriguez-Mendez, M. L., & Dias, L. G. (2022). Enose Lab Made with Vacuum Sampling: Quantitative Applications. Chemosensors, 10(7), 261. https://doi.org/10.3390/chemosensors10070261










