Association of Proteomics Changes with Al-Sensitive Root Zones in Switchgrass
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Seedling Plants and Al Treatment
2.2. Physiological Data Collection
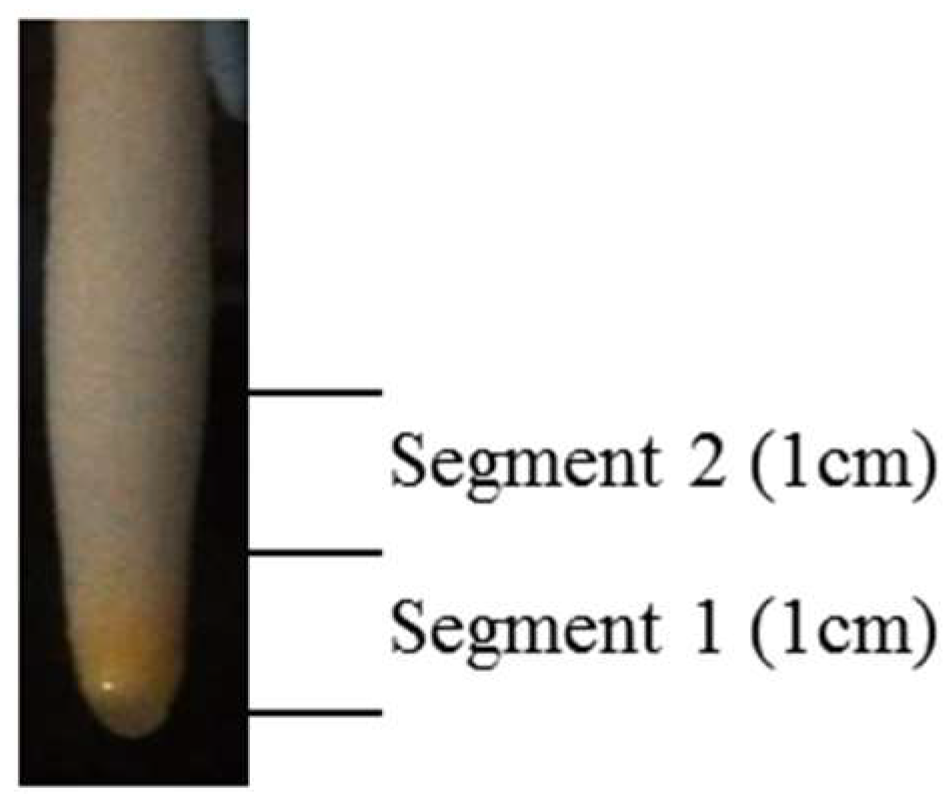
2.3. Tissue Collection and Preparation of Protein Samples
2.4. Tandem Mass Tags (TMT) Labeling and Mass Spectrometry Analysis
2.5. Protein Identification and Quantification, and Statistical Analysis
2.6. Functional Analysis of Identified Proteins
3. Results
3.1. Physiological Changes of Switchgrass Plants under Al Treatments
3.2. Al-Induced Proteome Changes in Different Zones of Root Tips
3.2.1. Total Root Proteome Changes Induced by Al Treatment
3.2.2. Functional Pathways of Al-induced Significantly Changed Proteins
3.2.3. String Interaction Networks among Al-induced Significantly Changed Proteins
- (a)
- STRING pathway analysis of the Segment 1 (the apical 1-cm root tip tissues) root tip proteins
- (b)
- STRING analysis of significantly changed proteins in the Segment 2 (root elongation/maturation zones) root proteins
3.2.4. Other Significantly Changed Proteins in Root Tissues
4. Discussion
- A.
- Mitigation of Al toxicity through regulation of internalization of Al3+ ions and their intercellular sequestration in switchgrass root tips
- B.
- Regulation of cell proliferation and morphological changes in Al-treated roots
- C.
- Modulation of the genome expression system
- D.
- Protein post-translational modifications and protection of protein conformation structures
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Von Uexkull, H.R.; Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 1995, 171, 1–15. [Google Scholar] [CrossRef]
- Foy, C.D. Plant adaptation to acid, aluminum-toxic soils. Commun. Soil Sci. Plant Anal. 1988, 19, 959–987. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Eswaran, H.; Reich, P.; Beinroth, F. Global distribution of soils with acidity. In Plant-Soil Interactions at Low pH; Wright, R.J., Baligar, V.C., Murrmann, R.P., Eds.; Kluwer Academic: Norwell, MA, USA; pp. 159–164.
- Chang, Y.C.; Yamamoto, Y.; Matsumoto, H. Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 1999, 22, 1009–1017. [Google Scholar] [CrossRef]
- Mossor-Pietraszewska, T.; Kwit, M.; Lêgiewicz, M. The influence of aluminium ions on activity changes of some dehydrogenases and aminotransferases in yellow lupine. Biol. Bull. Pozn. 1997, 34, 4748. [Google Scholar]
- Parrisha, D.J.; Fike, J.H. The biology and agronomy of switchgrass for biofuels. Crit. Rev. Plant Sci. 2005, 24, 423–459. [Google Scholar] [CrossRef]
- Wright, L.L. Historical Perspective on How and Why Switchgrass Was Selected as a “Model” High-Potential Energy Crop; ORNL/TM-2007/109; Oak Ridge National Laboratory: Oak Ridge, TN, USA, 2007.
- Bona, L.; Belesky, D.P. Evaluation of switchgrass entries for acid soil tolerance. Commun. Soil Sci. Plant Anal. 1992, 23, 15–16. [Google Scholar] [CrossRef]
- Babourina, O.; Rengel, Z. Uptake of aluminium into Arabidopsis root cells measured by fluorescent lifetime imaging. Ann. Bot. 2009, 104, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Sivaguru, M.; Horst, W.J. The distal part of the transition zone is the most aluminum-sensitive apical root zone of maize. Plant Physiol. 1998, 116, 155–163. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Pasquali, C.; Appel, R.D.; Ou, K.; Golaz, O.; Sanchez, J.C.; Yan, J.X.; Gooley, A.A.; Hughes, G.; Humphery-Smith, I.; et al. From proteins to proteomes: Large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology 1996, 14, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Xu, J. Abiotic stress responses in plant roots: A proteomics perspective. Front. Plant Sci. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.K.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Barkla, B.J.; Castellanos-Cervantes, T.; de León, J.L.; Matros, A.; Mock, H.P.; Perez-Alfocea, F.; Salekdeh, G.H.; Witzel, K.; Zörb, C. Elucidation of salt stress defense and tolerance mechanisms of crop plants using proteomics—Current achievements and perspectives. Proteomics 2013, 13, 1885–1900. [Google Scholar] [CrossRef] [PubMed]
- Ngara, R.I.; Ndimba, B.K. Understanding the complex nature of salinity and drought-stress response in cereals using proteomics technologies. Proteomics 2014, 14, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Okekeogbu, I.; Ye, Z.; Sangireddy, S.R.; Li, H.; Bhatti, S.; Hui, D.; Zhou, S.; Howe, K.J.; Fish, T.; Yang, Y.; et al. Effect of aluminum treatment on proteomes of radicles of seeds derived from Al-treated tomato plants. Proteomes 2014, 28, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Palmer, M.; Zhou, J.; Bhatti, S.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Differential root proteome expression in tomato genotypes with contrasting drought tolerance exposed to dehydration. J. Am. Soc. Hortic. Sci. 2013, 138, 1–11. [Google Scholar]
- Zhou, S.; Sauve, R.; Fish, T.; Thannhauser, T.W. Salt-induced and salt-suppressed proteins in tomato leaves. J. Am. Soc. Hortic. Sci. 2009, 134, 289–294. [Google Scholar]
- Zhou, S.; Sauvé, R.J.; Liu, Z.; Reddy, S.; Bhatti, S. Heat-induced proteome changes in tomato leaves. J. Am. Soc. Hortic. Sci. 2012, 136, 219–226. [Google Scholar]
- Zhou, S.; Okekeogbu, I.; Sangireddy, S.; Ye, Z.; Li, H.; Bhatti, S.; Hui, D.; Mcdonald, D.W.; Yang, Y.; Giri, S.; et al. Proteome modification in tomato plants upon long-term aluminum treatment. J. Proteom. Res. 2016, 15, 1670–1684. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Sangireddy, S.; Okekeogbu, I.; Zhou, S.; Yu, C.L.; Hui, D.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Drought-induced leaf proteome changes in switchgrass seedlings. Int. J. Mol. Sci. 2016, 17, E1251. [Google Scholar] [CrossRef] [PubMed]
- Magnavaca, R.; Gardner, C.O.; Clark, R. Evaluation of inbred maize lines for aluminum tolerance in nutrient solution. In Genetic Aspects of Plant Mineral Nutrition; Gabelman, H.L.B., Ed.; Martinus Nijhoff: Dordrecht, The Netherlands, 1987; pp. 255–265. [Google Scholar]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- MapMan, Version 3.5.1R2. 2015. Available online: http://MapMan.gabipd.org/web/guest/MapMan (accessed on 10 June 2017).
- García-Oliveira, A.L.; Benito, C.; Prieto, P.; de Andrade Menezes, R.; Rodrigues-Pousada, C.; Guedes-Pinto, H.; Martins-Lopes, P. Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and proton (H+) toxicity in bread wheat (Triticum aestivum L.). BMC Plant Biol. 2013, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, A.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef] [PubMed]
- Izbiańska, K.; Arasimowicz-Jelonek, M.; Deckert, J. Phenylpropanoid pathway metabolites promote tolerance response of lupine roots to lead stress. Ecotoxicol. Environ. Saf. 2014, 110, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.R.; Rikiishi, S.; Matsumoto, H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 2002, 128, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Ezaki, B.; Gardner, R.C.; Ezaki, Y.; Matsumoto, H. Expression of aluminium-induced genes in transgenic Arabidopsis plants can ameliorate aluminium stress and/or oxidative stress. Plant Physiol. 2000, 122, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Sauve, R.; Boone, B.; Levy, S. Identification of genes associated with aluminium toxicity in tomato roots using cDNA microarrays. Plant Stress 2008, 2, 113–120. [Google Scholar]
- Richards, K.D.; Schott, E.J.; Sharma, Y.K.; Davis, K.R.; Gardner, R.C. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998, 116, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Wan, J.X.; Sun, Y.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Xyloglucan endotransglucosylase-hydrolase17 interacts with xyloglucan endotransglucosylase-hydrolase31 to confer xyloglucan endotransglucosylase action and affect aluminum sensitivity in Arabidopsis. Plant Physiol. 2014, 165, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Péscsvéradi, A.; Nagy, Z.; Varga, A.; Vashegyi, A.; Labadi, I.; Galbacs, G.; Zsoldos, F. Chloroplastic glutamine synthetase is activated by direct binding of aluminium. Physiol. Plant. 2009, 135, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Arenhart, R.A.; Lima, J.C.; Pedron, M.; Carvalho, F.E.; Silveira, J.A.; Rosa, S.B.; Caverzan, A.; Andrade, C.M.; Schünemann, M.; Margis, R.; et al. Involvement of ASR genes in aluminium tolerance mechanisms in rice. Plant Cell Environ. 2013, 36, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Arenhart, R.A.; Bai, Y.; de Oliveira, L.F.; Neto, L.B.; Schunemann, M.; Maraschin Fdos, S.; Mariath, J.; Silverio, A.; Sachetto-Martins, G.; Margis, R.; et al. New insights into aluminum tolerance in rice: The ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol. Plant 2014, 7, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Ohyama, Y.; Kobayashi, Y.; Ito, H.; Iuchi, S.; Fujita, M.; Zhao, C.R.; Tanveer, T.; Ganesan, M.; Kobayashi, M.; et al. STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 2014, 7, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jurandir, V.M.; Jon, S.; Leon, V.K. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Maron, L.G.; Piñeros, M.A.; Guimarães, C.T.; Magalhaes, J.V.; Pleiman, J.K.; Mao, C.; Shaff, J.; Belicuas, S.N.; Kochian, L.V. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010, 61, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Maron, L.G.; Guimarães, C.T.; Kirst, M.; Albert, P.S.; Birchler, J.A.; Bradbury, P.J.; Buckler, E.S.; Coluccio, A.E.; Danilova, T.V.; Kudrna, D.; et al. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 2013, 110, 5241–5246. [Google Scholar] [CrossRef] [PubMed]
- Pineros, M.; Kochian, L.V. Novel properties of the wheat aluminum tolerance organic acid transporter (TaALMT1) revealed by electrophysiological characterization in Xenopus oocytes: Functional and structural implications. Plant Physiol. 2008, 147, 2131–2146. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Liu, J.; Dong, D.; Jia, X.; Mccouch, S.; Kochian, L.V. Natural variation underlies alterations in NRAT1 expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Shen, R.F.; Nagao, S.; Tanimoto, E. Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 2004, 45, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Geisler, M.J.B.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminum tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Zhang, W.; Ryan, P.; Tyerman, S. Malate-permeable channels and cation channels activated by aluminum in the apical cells of wheat root roots. Plant Physiol. 2001, 125, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Hoekenga, O.A.; Pineros, M.A. How do crop plants tolerate acid soils?—Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Hawes, M.C.; Gunawardena, U.; Miyasaka, S.; Zhao, X. The role of root border cells in plant defense. Trends Plant Sci. 2000, 5, 128–133. [Google Scholar] [CrossRef]
- Miyasaka, S.C.; Hawes, M.C. Possible role of root border cells in detection and avoidance of aluminum toxicity. Plant Physiol. 2001, 125, 1978–1987. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wang, N.; Xing, C.; Wang, F.; Wu, K.; Du, X. Immobilization of aluminum with mucilage secreted by root cap and root border cells is related to aluminum resistance in Glycine max L. Environ. Sci. Pollut. Res. Int. 2013, 20, 8924–8933. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Liu, J.Y.; Fang, J.; Tao, L.; Shen, R.F.; Li, Y.L.; Xiao, H.D.; Feng, Y.M.; Wen, H.X.; Guan, J.H.; et al. Boron supply enhances aluminum tolerance in root border cells of Pea (Pisum sativum) by interacting with cell wall pectins. Front. Plant Sci. 2017, 8, 742. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Hoekenga, O.A.; Itoh, H.; Nakashima, M.; Saito, S.; Shaff, J.E.; Maron, L.G.; Piñeros, M.A.; Kochian, L.V.; Koyama, H. Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 2007, 145, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Kopittke, P.M.; McKenna, B.A.; Karunakaran, C.; Dynes, J.J.; Arthur, Z.; Gianoncelli, A.; Kourousias, G.; Menzies, N.W.; Ryan, P.R.; Wang, P.; et al. Aluminum complexation with malate within the root apoplast Differs between aluminum resistant and sensitive wheat lines. Front Plant Sci. 2017, 8, 1377. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Ryan, P.R.; Randall, P.J. Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol. 1993, 103, 695–762. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, P.B.; Tai, C.Y.; Kochian, L.V.; Howell, S.H. Arabidopsis mutants with increased sensitivity to aluminum. Plant Physiol. 1996, 110, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Belal, R.; Tang, R.; Li, Y.; Mabrouk, Y.; Badr, E.; Luan, S. An ABC transporter complex encoded by Aluminum Sensitive 3 and NAP3 is required for phosphate deficiency responses in Arabidopsis. Biochem. Biophys. Res. Commun. 2015, 463, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Zelinová, V.; Halušková, L.; Huttová, J.; Illéš, P.; Mistrík, I.; Valentovičová, K.; Tamás, L. Short-term aluminium-induced changes in barley root tips. Protoplasma 2011, 248, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Kirik, V.; Grini, P.E.; Mathur, J.; Klinkhammer, I.; Adler, K.; Bechtold, N.; Herzog, M.; Bonneville, J.M.; Hülskamp, M. The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the alpha/beta-tubulin monomer balance. Plant Cell 2002, 14, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.L.; Rahman, A.; Baskin, T.I.; Kieber, J.J. Two leucine-rich repeat receptor kinases mediate signaling linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 2008, 20, 3065–3079. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Q.; Liu, H.; Shen, S. Phylogenetic and functional analysis of the basic transcription factor gene BTF3 from Jatropha curcas. Plant Growth Regul. 2017, 82, 247–257. [Google Scholar] [CrossRef]
- Smykowski, A.; Zimmermann, P.; Zentgraf, U. G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol. 2010, 153, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Krainer, A.R.; Conway, G.C.; Kozak, D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 1990, 62, 35–42. [Google Scholar] [CrossRef]
- Poot-Poot, W.; Rodas-Junco, B.A.; Muñoz-Sánchez, J.A.; Hernández-Sotomayor, S.M.T. Protoplasts: A friendly tool to study aluminum toxicity and coffee cell viability. Springerplus 2016, 5, 1452. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Chang, J.G.; Hendriks, I.A.; Sigurðsson, J.O.; Olsen, J.V.; Vertegaal, A.C. System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel small ubiquitin-like modified target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteom. 2015, 14, 1419–1434. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.H.M.; Parveen, A.; Verma, A.K.; Ahmad, I.; Arshad, M.; Mahdi, A.A. Aluminum induced endoplasmic reticulum stress mediated cell death in SH-SY5Y neuroblastoma cell line is independent of p53. PLoS ONE. 2014, 9, e98409. [Google Scholar]
- Panda, S.K.; Baluska, F.; Matsumoto, H. Aluminum stress signaling in plants. Plant Signal Behav. 2009, 4, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant. Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
| Treatment | Segment 1 (Apical Root Tissues) | Segment 2 (Elongation/Maturation Zone) | ||
|---|---|---|---|---|
| Replicate | Labelling Tag | Replicate | Labelling Tag | |
| Control | Control-1 | 129N | Control-1 | 128N |
| Control-2 | 129C | Control-2 | 126 | |
| Control-3 | 128N | Control-3 | 129N | |
| 400 µM | 400 µM-1 | 126 | 400 µM-1 | 129C |
| 400 µM-2 | 127C | 400 µM-2 | 127N | |
| 400 µM-3 | 131 | 400 µM-3 | 128C | |
| Physiological Measurements | Control | Al-treated |
|---|---|---|
| Photosynthetic rate (μmol CO2/m2/s) | 23.43 ± 0.20A | 18.07 ± 2.76B |
| Conductance (mol H2O/m2/s) | 0.20 ± 0.01A | 0.18 ± 0.04A |
| Transpiration rate (mmol H2O/m−2/s) | 5.47 ± 0.77A | 3.79 ± 0.90B |
| Water use efficiency (WUE) (μmol CO2/mmol H2O) | 4.41 ± 0.23A | 5.97 ± 2.66A |
| Types of Protein Data | Segment 1 (Apical 1-cm Root-Tip) | Segment 2 (Elongation/Maturation Zone) |
|---|---|---|
| Number of identified proteins | 6309 | 7288 |
| Number of proteins identified with 2 or more peptides and quantified | 4130 | 4636 |
| Number of significantly changed proteins | 164 (3.9% of quantified proteins) | 52 (1.1% of quantified proteins) |
| Functional Pathway | Segment 1 (Apical-1 cm Root-Tip) | Segment 2 (Elongation/Maturation Zone) | ||
|---|---|---|---|---|
| Higher abundance | Lower abundance | Higher abundance | Lower abundance | |
| Amino acid metabolism | 1 | 1 | ||
| Cell cycle | 3 | |||
| Cell organization | 5 | 4 | ||
| Cell vesicle transport | 2 | |||
| Cell wall synthesis and modification proteins | 3 | 1 | 1 | |
| Development | 1 | 1 | 1 | |
| DNA metabolism | 2 | 1 | 1 | |
| Enzyme families | 2 | 1 | 5 | |
| Lipid metabolism | 2 | 1 | ||
| Major CHO metabolism | 2 | 1 | ||
| Mitochondrial electron transport | 5 | 2 | ||
| N-metabolism | 1 | |||
| Energy metabolism | 5 | |||
| Nucleotide metabolism | 1 | |||
| Phyto hormone metabolism | 2 | 1 | ||
| Protein degradation | 5 | 3 | ||
| Protein synthesis | 23 | 1 | 2 | 4 |
| Protein targeting | 4 | 1 | ||
| Protein post translational modification | 5 | |||
| Redox | 3 | 1 | ||
| RNA metabolism | 14 | 1 | ||
| Secondary metabolism | 3 | 1 | ||
| Signaling | 4 | 3 | ||
| Stress proteins | 10 | 3 | 1 | 7 |
| Transport | 3 | 2 | 1 | |
| Unknown and others | 43 | 2 | 7 | |
| Total number of proteins | 145 | 19 | 17 | 35 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangu, M.; Ye, Z.; Bhatti, S.; Zhou, S.; Yang, Y.; Fish, T.; Thannhauser, T.W. Association of Proteomics Changes with Al-Sensitive Root Zones in Switchgrass. Proteomes 2018, 6, 15. https://doi.org/10.3390/proteomes6020015
Rangu M, Ye Z, Bhatti S, Zhou S, Yang Y, Fish T, Thannhauser TW. Association of Proteomics Changes with Al-Sensitive Root Zones in Switchgrass. Proteomes. 2018; 6(2):15. https://doi.org/10.3390/proteomes6020015
Chicago/Turabian StyleRangu, Mahesh, Zhujia Ye, Sarabjit Bhatti, Suping Zhou, Yong Yang, Tara Fish, and Theodore W. Thannhauser. 2018. "Association of Proteomics Changes with Al-Sensitive Root Zones in Switchgrass" Proteomes 6, no. 2: 15. https://doi.org/10.3390/proteomes6020015





