1. Introduction
According to the American Cancer Society, breast cancer remains the most commonly diagnosed cancer among women in the United States, predicting over 250,000 new cases in 2017 and accounting for 30% of all new cancer diagnoses [
1]. Breast cancer further ranks second, behind lung cancer, in terms of cancer cell death among women with >40,000 incidences in 2017. While five-year breast cancer survival rates approach 100% in the case of localized tumors (stage 0 or I), the prognosis for metastasized (stage IV) breast cancer is poor, with only 22% survival at 5 years [
2]. Early detection of breast cancer patients is thus crucial to improving recurrence-free survival and quality of life. Markers that distinguish invasive breast cancer phenotypes are urgently required.
Conventional screening tools, such as self-examination, mammography, diagnostic imaging (ultrasound, MRI), genetic screens (e.g., BRCA1), and tissue biopsies [
3,
4], collectively present vital tools for breast cancer detection. As with all tests, limits in sensitivity and specificity will miss some cancers (false-negatives); in other cases, abnormal findings associated with benign disease (false positives) will direct between 55% and 75% of women into unnecessary and potentially toxic chemotherapy [
5,
6,
7]. The next generation of diagnostic and prognostic tests is continuously being sought. In particular, considerable effort is currently devoted to uncovering early molecular indicators released into the blood by cancer cells [
8]. To this end, both DNA from circulating tumor cells [
9] and microRNA released to plasma by cancer cells [
10] have been reported as potential biomarkers. Differentially expressed protein biomarkers have also been observed in plasma [
11]. The presence of high abundance background plasma proteins challenges the detection of low abundance proteins of interest, pointing to a need for more selective and sensitive indicators of breast cancer.
Exosomes are a subclass of extracellular vesicles (EVs), constituting membranous spheroids between 30 and 200 nM released from cells. Initially regarded as “cell dust”, EVs are now unequivocally associated with various types of cancer, and carry specific profiles of proteins, nucleic acids, and lipids that serve to transmit information to other cells. Once secreted by cells into the extracellular medium, EVs retain features of the originating tissue environment [
12]. EVs have also been shown to alter the metabolic activity of neighboring cells [
13], and thus, may have important functional implications for tumor growth and differentiation. Not only are EVs with specific molecular payloads released by cancer cells, but they are secreted in relative higher abundance. Hence, protein profiling of cancer cell-derived exosomes has gained attention as a favorable source of protein biomarkers. Multiple approaches are available to isolate exosomes from the bulk sample. The most common approach is differential ultracentrifugation (UC); although laborious and producing a lower yield, UC remains the “gold standard” in obtaining exosomes in high purity [
14]. Whilst differential centrifugation remains the laboratory method of choice [
15], it is an inefficient and poorly reproducible tool that is unsuited for handling even modest numbers of clinical samples [
16,
17]. Access to ultracentrifugation is also limited in most clinical diagnostic laboratories, demanding robust alternative EV preparations for future translational utility. Alternative approaches, including dialysis, size exclusion chromatography, and polymer- or antibody-induced precipitation, are available [
14].
In an alternative strategy, a synthetic peptide (Vn96) with high affinity for heat shock proteins (HSPs) has recently been shown to selectively isolate EVs by co-precipitation [
18]. Not surprisingly, HSPs are overexpressed in cancer cells [
19], where maintenance of protein homeostasis is at a premium, due to hypoxia, low pH, and limited glucose availability. HSPs are also found in high abundance on the plasma membrane surface of exosomes [
20], thus, the Vn96 pulldown represents an ideal strategy to enrich EVs shed from cancer cells. The Vn96 affinity peptide has previously been shown to recover EV material from urine, plasma, as well as cancer cell lines [
21,
22,
23].
In this study, we demonstrate for the first time the utility of Vn96 in isolating breast cancer EVs for functional proteomic analysis of malignant phenotypes. We employ various breast cancer cells, including the adenocarcinoma models SKBR3, an invasive HER2+ cell and MCF-7, a luminal A cell line model. MCF-10a, a non-tumorigenic breast fibrocystic disease model, is also employed as a control. In-depth proteome analysis by LC-MS/MS, following GELFrEE fractionation of exosomal proteins, reveals numerous differentially expressed proteins, which readily distinguish the various cell lines. The proteins convey unique profiles, particularly in terms of their metabolic and chaperone activity, providing insight into the biological function of breast cancer-derived EVs and potentially pointing a strategy to uncover protein biomarkers for early breast cancer detection.
3. Results and Discussion
We report a comparative proteome investigation on an in vitro model of breast cancer, examining the extracellular media from SKBR-3 (invasive cancerous cell), MCF-7 (non-invasive), and MCF-10a (immortal but non-cancerous cells). While cells adapted to grow in plastic flasks are regarded as very different from those obtained in vivo, material secreted by cancer cells into the external environment in vitro is likely to produce a similar proteomic profile, reflecting the original growth from which it was derived [
28]. The Vn96 peptide was employed to selectively capture EV material released by cultured cell lines into their growth media. Vn96 targets heat shock proteins (HSPs), overexpressed on the surface of aggressive cancer cells, and by extension, their derivative vesicles [
29]. The Vn96 peptide has been shown to capture exosome-like vesicles containing proteins comparable to EV preparations by traditional ultracentrifugation when analyzed by Western blot [
21].
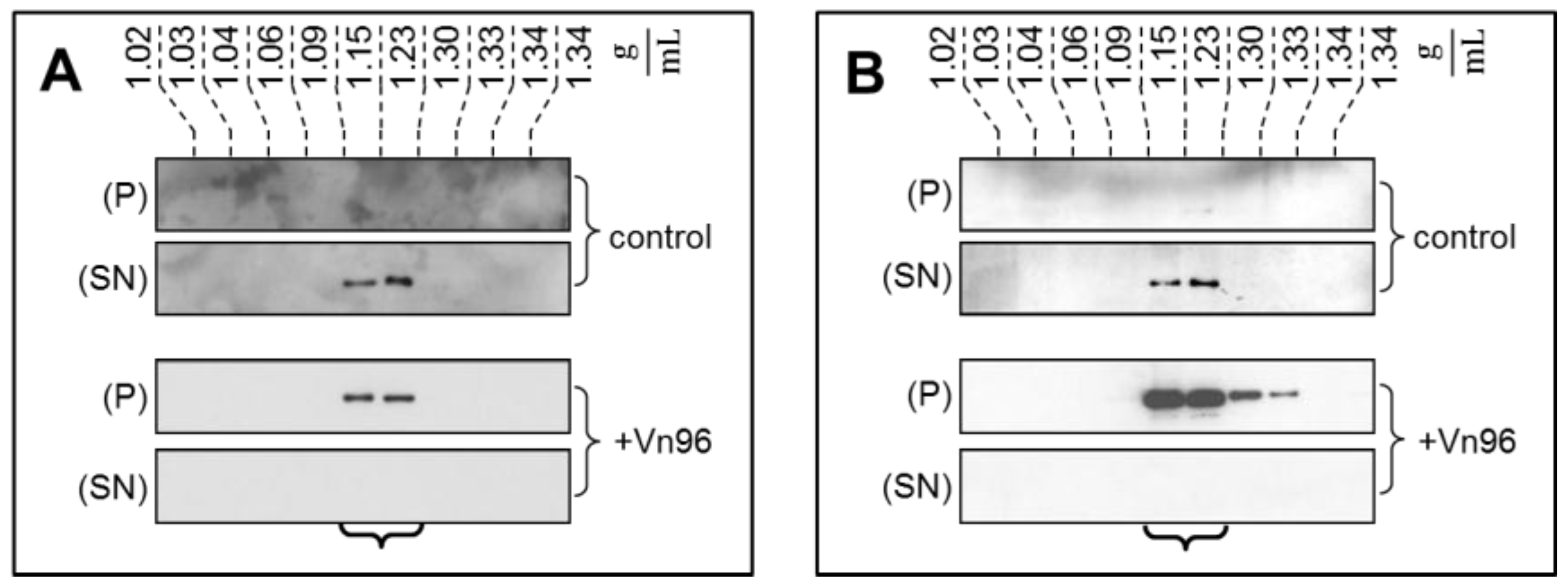
To demonstrate the specificity of Vn96 to pellet EV material, exosomes from SKBR3 were harvested through conventional ultracentrifugation with separation into characteristic flotation zones by sucrose density fractionation. As shown in
Figure 1, the exosomal marker proteins TSG101 and Alix were isolated in the fractions from 1.15 to 1.23 g/mL. Moreover, while these marker proteins remain suspended in the supernatant (SN) following low speed centrifugation, the addition of Vn96 resulted in recovery of exosomal markers in the pellet (P) of the low speed spin. These observations demonstrate the capacity of Vn96 to concentrate proteins associated with vesicular material.
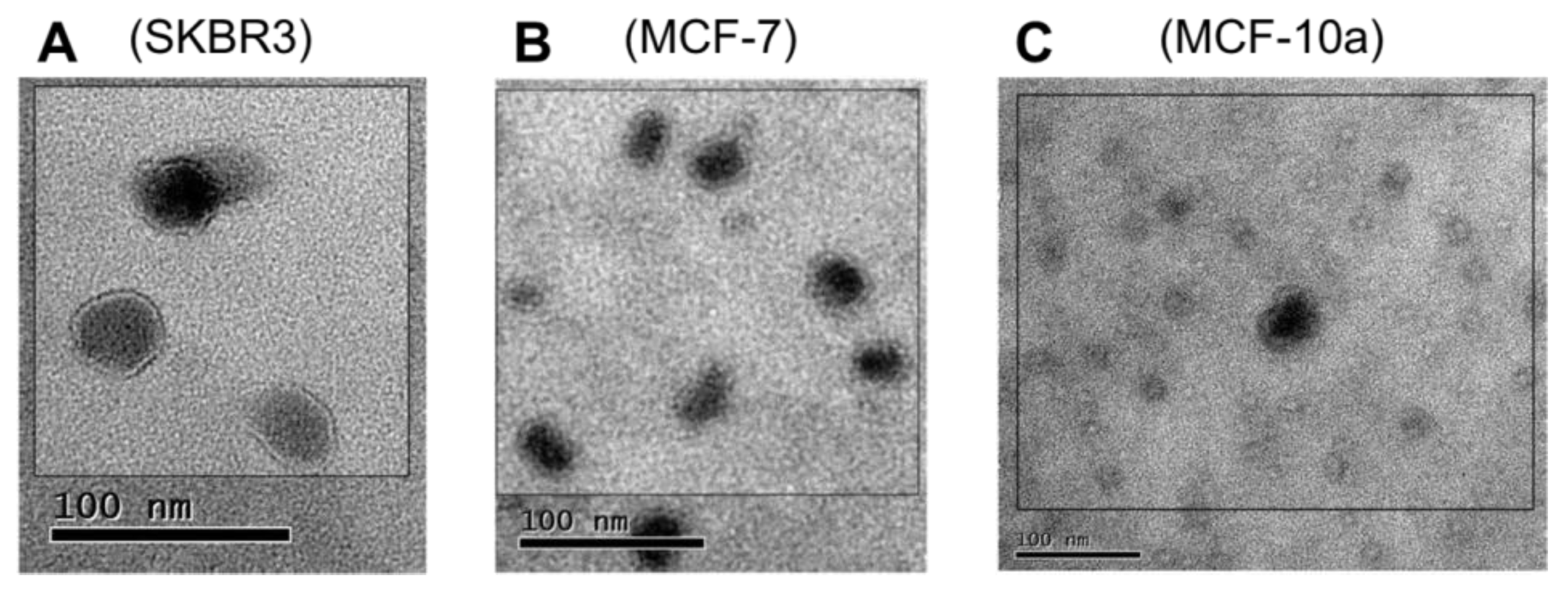
The Vn96 peptide was next employed to directly recover EV materials from filtered bioreactor cell culture media. When observed by TEM
Figure 2, material pulled down by Vn96 generally consisted of bilayer orb structures between 30 and 50 nm in diameter, with electron dense centers. Such features were also evident in the MCF-10a cell line, indicative that HSP-decorated vesicles are also released by these cells. In recent years, the term “small EVs” has been used as an alternative to exosomes [
30]. Vn96 may capture a subset of small EVs, as defined by surface accessorization of HSPs.
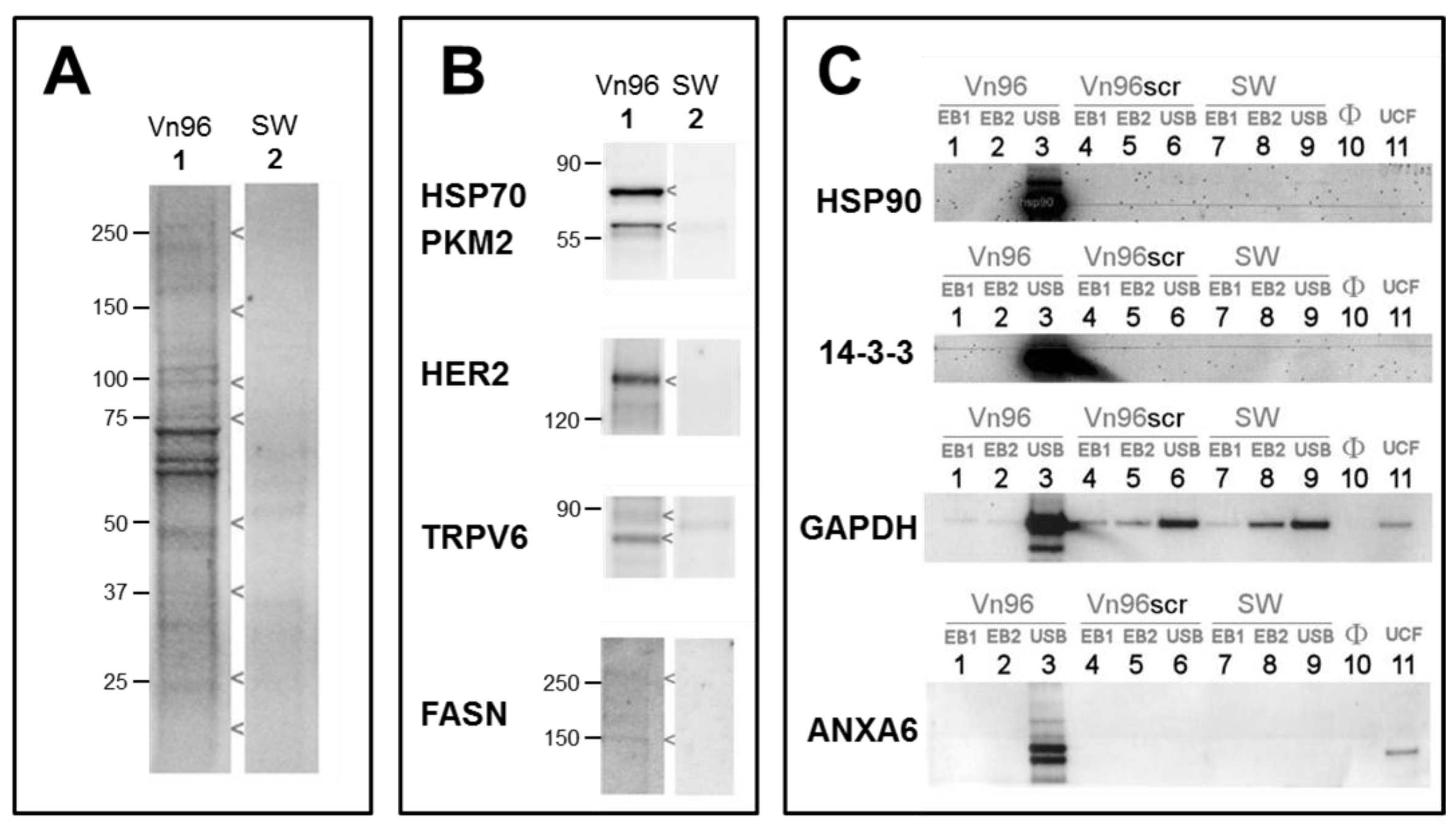
Electrophoretic profiling of Vn96 pull-downs yielded robust total protein profiles (e.g., from SKBR3,
Figure 3A lane 1). Replacement of Vn96 with a random peptide (SW) yielded faint profiles likely to be aggregates of albumin and immunoglobulin from culture medium (
Figure 3A, lane 2). The heat shock proteins (HSP70, HSP90) targeted by Vn96 were depleted from the medium and exclusively recovered in the low centrifuge speed pellet. Employing a random peptide (SW), or scrambled amino acid sequence of Vn96 did not pellet these proteins (
Figure 3B). Western blots of EV material also identified PKM2, receptor kinase HER2, membrane protein TRPV6, and fatty acid synthase; none of which were recovered using the null peptide (
Figure 3B). The stability of EVs isolated by Vn96 from SKBR3 is also evident in
Figure 3C, which employs various commercial detergent washes (components of Millipore’s subcellular proteome extraction kit, S-PEK). As seen in lanes 1 and 2 of
Figure 3C, neither cytosolic Extraction Buffer I (EB1), which contains the mild detergent digitonin, nor membrane and organelle Extraction Buffer II (EB2), containing Triton X-100, would solubilize the EV marker proteins from the Vn96 pellet. Similar results are obtained from the other cell lines. Though EV marker proteins are retained following EB1 and EB2 washes, weakly associated proteins are liberated from the EV pellet by these buffers (see
Supplementary Figure S1). These findings permit incorporation of stringent washes to further purify the EV fraction recovered in the Vn96 pellet. Immunoblot analysis on material recovered from the non-transformed MCF-10a cell culture media, as well as for MCF-7, with the breast cancer exosomal marker CD24 [
31], illustrate that some, but not all, CD24 is released into the supernatant following incubation of the pellet with EB II (results not shown). Thus, we subsequently chose a PBS washing protocol, also under reducing conditions (25 mM TCEP), to achieve a balance of yield and purity. To fully solubilize EV proteins (lane 3 of
Figure 3C), the pellet is boiled in SDS gel-loading buffer, supplemented with 4 M urea (USB). As a consequence, MS analysis demands an SDS-compatible proteomics workflow.
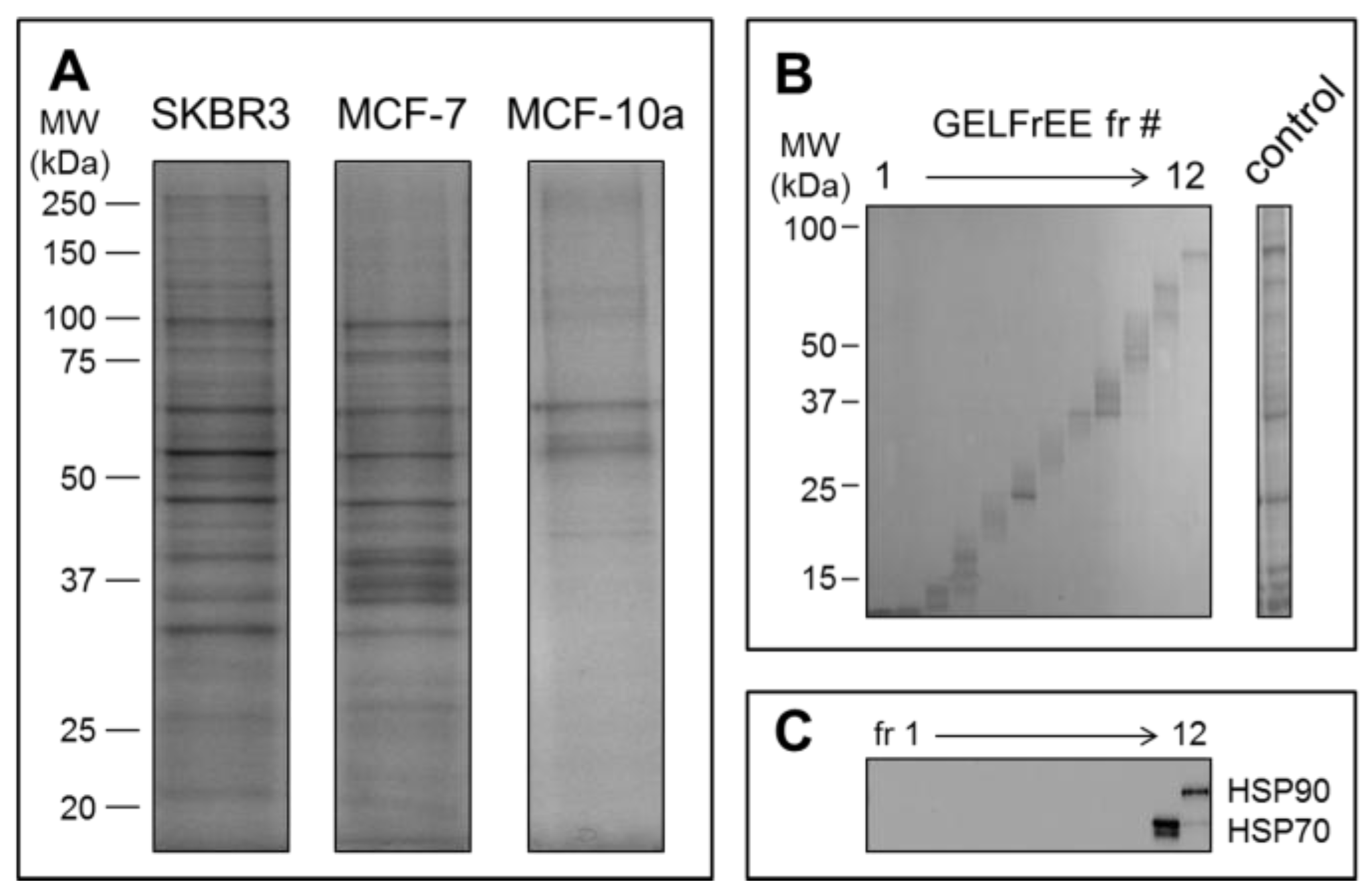
Mass spectrometry detection followed GELFrEE fractionation of the solubilized EV proteomes. GELFrEE enables recovery of proteins in an SDS-containing buffer, with separation according to molecular weight. SDS depletion via organic solvent precipitation permits LC/MS of trypsin-digested proteins. As demonstrated in
Figure 4A, a higher abundance of EV proteins was recovered from the SKBR3 and MCF-7 cell lines, relative to the non-cancerous MCF-10a, in support of the theory that aggressive cancer cells will overexpress EV materials. The decreased abundance of proteins recovered by Vn96 from the MCF-10a cell media may also reflect a lower number of Vn96 binding opportunities, due to lack of surface expressed HSP/chaperones. Nonetheless, it is clear that a greater concentration of proteins is recovered from the tumorigenic cell lines. GELFrEE resolved the proteins over a mass range extending to ~100 kDa
Figure 4B, isolating proteins in discrete fractions according to molecular weight (
Figure 4C). A detailed listing of the identified proteins and peptides from each of the three cell lines are provided as
supplementary files (Tables S1–S3).
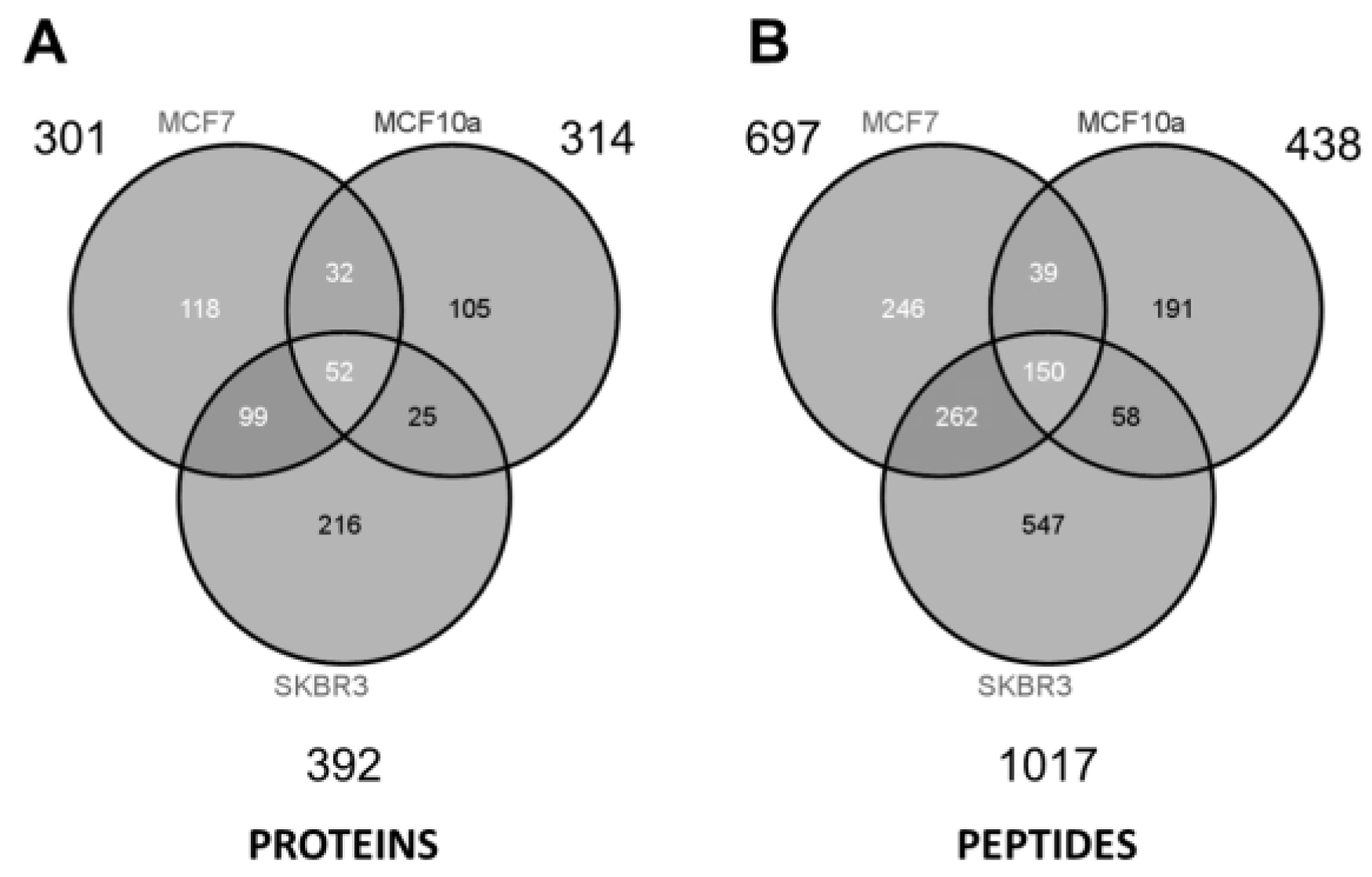
As summarized in the Venn diagrams of
Figure 5, some ~300 to 400 unique proteins were identified from each cell type (minimum 2 peptides per protein). The largest number of identified proteins (392) was seen from the most aggressive cancer cell line (SKBR3), and might be a reflection of the increased prominence of HSPs expressed on the EV surface. Considering all three samples and technical replicates (three GELFrEE runs per sample type), MS collectively identified 647 unique proteins across the various cell lines. Despite marked differences in protein concentration across the three cell lines, similar numbers of proteins were identified through MS. This may reflect on the limitations of an MS platform which favors detection of the most abundant components. While greater differences are reflected at the peptide level
Figure 5, comparative analysis of the discrete proteomes is best presented through changes in the protein abundance. Here, spectral counting is employed as a means of assessing relative protein abundance across the three cell types. In total, we obtained 7400 peptide spectral matches (PSMs) from SKBR3; 4584 PSMs from MCF-7; and 2381 PSMs from MCF-10a, being indicative of the greater concentration of EV proteins recovered in the most invasive phenotype.
Supplementary Table S4 details a full comparative assessment of the spectral matches observed per protein across the three cell lines.
Supplementary Table S5 compares the identified proteins to the ExoCarta protein database [
32], indicating that the majority of proteins detected (509 of 575 gene products) were previously found to be associated with exosomal material.
Reflecting the origin of EVs as they are produced by the cell, and considering the cellular distribution of proteins identified from the EV fractions (
Supplementary Figure S2), it is not surprising that several of the identified proteins were associated with the membrane (19%), or cellular surface (12%). The large number of extracellular proteins is reflective of the heat shock proteins along with other proteins normally released by cells.
Table 1,
Table 2 and
Table 3 summarize the top 25 proteins (excluding probable contaminants), as categorized according to primary function (metabolism; chaperone and protein binding; skeletal and assembly).
Multiple cytoskeleton proteins (keratins, actin, myosin, etc.) dominated the list of identified proteins (see
Supplementary Tables S1–S3). Keratins are frequently encountered as unavoidable contaminant proteins during sample collection and processing. As such, suspected keratin contaminants (keratin type 1, 2, 5, 9, 14, 16) were among the identified proteins. These proteins exhibit minimal statistical difference in peptide spectral counts among the three cell types. However, such contaminants are easily distinguishable from bona fide cancer markers, such as cytokeratin 8, 18, and 19. These molecules are found at the surface of cancer cells, and are thus either incorporated as bystanders, or as part of the EV assembly and embarkation process. CK8 and CK18 are well documented as secreted cancer biomarkers, and may be detected in serum of patients with breast cancer receiving chemotherapy [
33]. These markers were shown to resist extraction by wash buffers EB1 and EB2 (
Supplementary Figure S1), and are thus likely to be strongly associated or embedded within the extracellular vesicles. CK19 has been suggested to be likely involved in driving the more aggressive tumor proliferation, invasion, and metastasis associated with HER-2/neu-positive tumors [
34]. By their PSMs, each of these marker proteins were highly elevated in the cancer cell lines relative to MCF-10a (
Table 1,
Table 2 and
Table 3). Thus, these proteins are included in the lists of relevant EV components.
Considering the full list of proteins collectively identified in the EV material, a broad range of biological function is conveyed (
Supplementary Figure S2). The largest group of proteins identified (18%) were associated with metabolic activity. The chaperone/binding proteins were also prominent among the identified groups. A preliminary comparison of the molecular pathways associated with the identified proteins is seen through Ingenuity Pathway Analysis (IPA), for which the top 12 pathways of each cell type are depicted in
Supplementary Figure S3. The most distinguishing feature was the glycolysis/gluconeogenesis and the pentose phosphate pathways, represented at the top of IPA pathways represented in SKBR3 (second and fourth for MCF-7). By sharp contrast, metabolic enzymes constitutive of these pathways were essentially absent from MCF-10a.
Cancers are dominated by metabolic pathways that vary from normative physiology with emphasis on accelerated uptake of glucose and glutamine, aerobic glycolysis, decreased mitochondrial activity, and enhanced lipogenesis.
Table 4 provides a quantitative comparison of proteins involved in metabolic pathways. Perhaps the most well-known characteristic, the Warburg effect [
35], refers to the avid consumption of glucose for direction into a glycolytic pathway with the accumulation of lactate, rather than incorporation of pyruvate into the tricarboxylic acid (TCA) cycle for oxidative phosphorylation. Nine of the ten canonical enzymes of glycolysis were represented in the Vn96 captured EVs of both SKBR3 and MCF-7, though only five were observed in MCF-10a. Some elements were exclusively found in the invasive and non-invasive VNEs, such as isoforms of pyruvate kinase and lactate dehydrogenase. Lactate dehydrogenase is perhaps the most widely recognized secreted enzyme of glycolysis contributing to the acidification paradigm of the Warburg effect [
36]. Metabolism is thus closely linked to cancer progression, because the ability for a cell to proliferate is dependent upon the availability of nutrients to build new cells. Glycolytic enzymes also protect cancer cells from stress by inhibiting apoptosis, and correlate well with resistance to radio- and chemotherapy [
37,
38]. Early diversions are promoted toward the pentose phosphate pathway as the means to generate nucleotide and amino acids. Of relevance was the detection of phosphogluconate dehydrogenase in higher abundance for the cancerous cell lines.
Beyond glycolysis, the provision of raw materials into peripheral biosynthetic pathways is crucial to cancer survival and colonization of areas distal to the primary tumor [
39]. For example, glycolysis channels raw material into lipid biosynthesis for membrane expansion and vesicle production [
40,
41]. Lipogenesis, required for membrane expansion and vesicle production, and typical of the aggressive cancer, would benefit from the donation of precursors. As seen in
Table 4, a high proportion of peptides originate from fatty acid synthase (FASN) in the invasive SKBR3. Similarly, tumor protein D52 was also exclusively observed in SKBR3, being implicated in increased capacity for storing lipid typical of invasive cancer cells [
42]. The significance of FASN abundance in an invasive phenotype is likely associated with the promotion of membrane biogenesis. FASN is frequently associated with invasive cancer, and has been proposed as a therapeutic target [
43]. It is normally expressed in low levels when dietary sources are sufficient. However, FASN expression and activity in cancer cells can be very high, and becomes associated with lipid rafts following cell signaling events [
44]. Serum levels of FASN have been found extracellularly in breast cancer [
45], and are predictive of colorectal cancer stage [
46]. FASN has previously been found in exosomes from multiple cancer cells [
32]. Accordingly, it is possible that proportional representation of FASN in exosomes is a prospective biomarker of invasive phenotype [
47].
As ligands for the Vn96 affinity peptide, individual canonical heat shock proteins are listed in
Table 5. HSP60 was the most abundantly represented heat shock protein among the three cell lines, and most highly expressed in SKBR3. HSP60 is a known surface-displayed molecule, secreted by cancer cells, and an important marker of cancer-derived exosomes [
48]. Isoforms of HSP90 were more extensively represented in SKBR3, though also observed in the non-cancerous cell line MCF7. HSP90-alpha constitutes the extracellular isoform, and is particularly characteristic of invasive cancer [
49]. Although location of HSP90 isoforms was not determined, this family of proteins is frequently found on the cell surface of cancer cells, and by extension, on derivative vesicles [
50]. HSP90 is imperative as a stabilizing chaperone of a broad range of aberrantly overactive receptors and kinases imperative to cancer. Transient HSP–multi protein complexes, coined as the “epichaperome” and found in high prevalence on numerous cancers, have been shown to play important roles in facilitating cell regulation and survival [
51]. Thus, while smaller chaperones, HSP10 or HSP27, were observed in higher abundance in the cancerous cell lines, it is unknown if these are independently capable of engaging Vn96. However, HSP10 and 27 have been implicated in multifunctional chaperone networks in invasive breast cancer [
52]. HSP chaperone complexes not only present biomarkers for cancer diagnostics, but have been identified as targets for drug therapy [
51].
The functional elements of a metastatic cascade reside in secreted proteins invested in HSP-decorated EVs. Cumulatively, these proteins enable cells to detach, invade tissue, and access circulation, while buffering against toxicity. Proteome profiling of the EV materials from malignant vs non-invasive phenotypes reveals multiple features conducive to malignancy, some of which have only been appreciated in the last year. A short selection is provided in
Table 6. The role of these proteins in cancer progression is described in the table with reference to literature. For example, protein disulfide isomerases are multifunctional chaperones instrumental in the breakage and rearrangement of disulfide bonds of extracellular matrix proteins, being required for detachment, extravasation, and intravasation at secondary sites, particularly with regard to cellular matrix remodeling. Other proteins may protect or preserve aspects crucial to metabolism (e.g., selenium-binding protein 1), while other proteins may promote expression of specific proteins or enable function to maintain the malignant phenotype (14-3-3 zeta). The Vn96 protocol to isolate EV materials uncovers multiple distinguishing protein features, which collectively constitute candidate biomarkers of breast cancer.
Previous investigations on breast cancer cell lines [
66], or of their secreted exosomes [
67], have highlighted proteomic profiles indicative of cancer cell proliferation and mobility, which was also apparent from our study. Our observations also corroborate a proteomic study that identified proteins involved in metabolic and detoxification pathways as highly expressed in HER-2/neu-positive breast cancer [
68].