Functional Characterization of Physcomitrella patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression
Abstract
:1. Introduction
2. Results
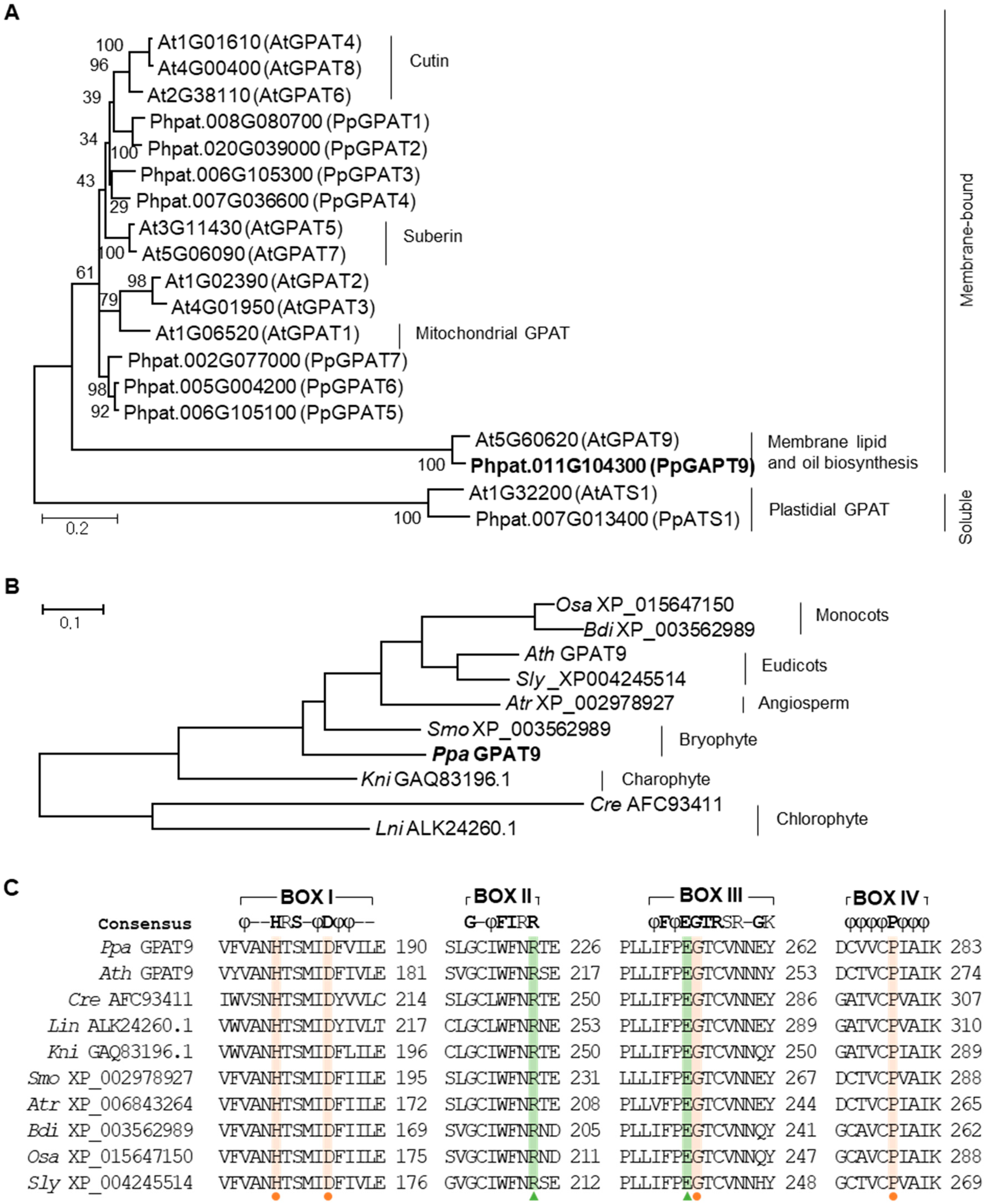
2.1. Isolation of GPAT9 From P. patens
2.2. Expression of PpGPAT9 in the Protonema and Gametophores
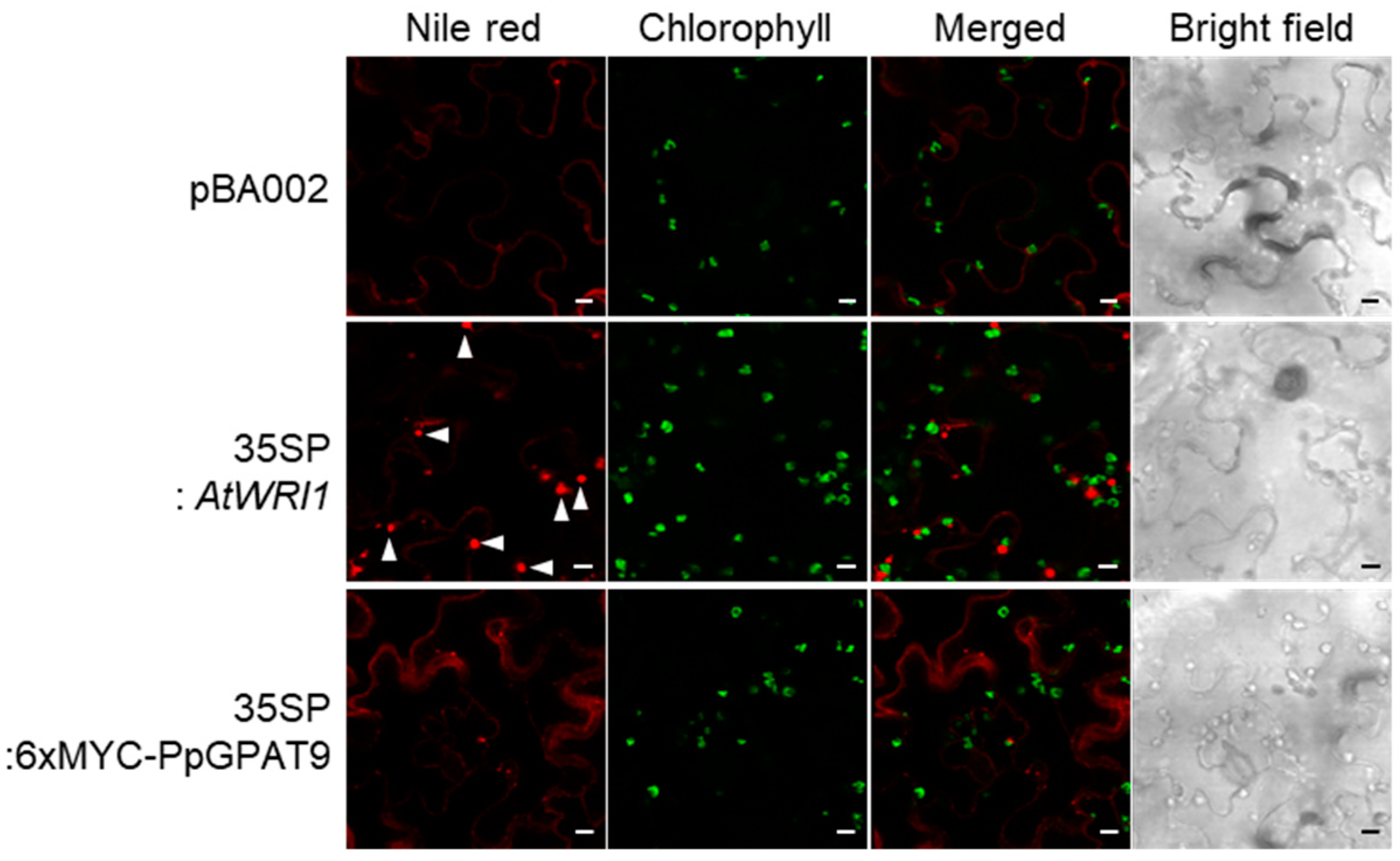
2.3. Subcellular Localization of PpGPAT9
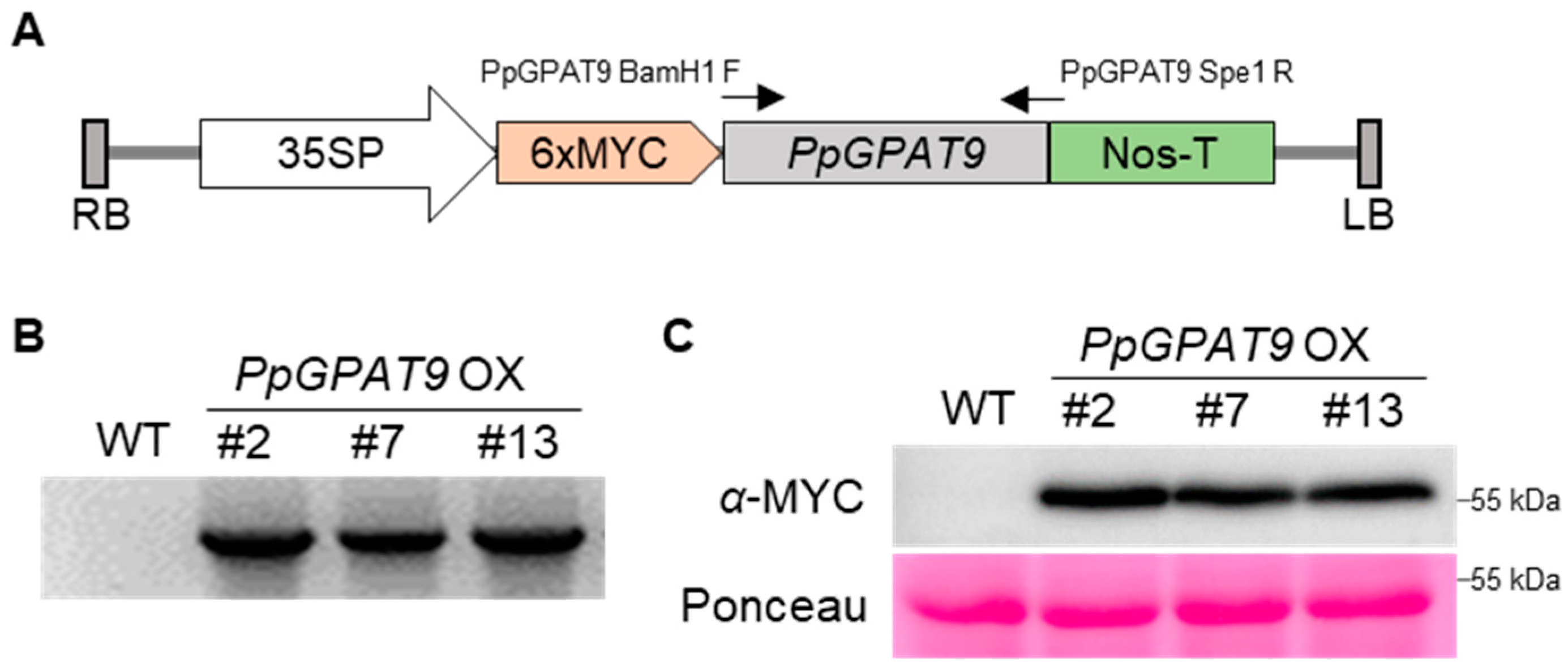
2.4. Generation of Arabidopsis Transgenic Plants Ectopically Expressing PpGPAT9
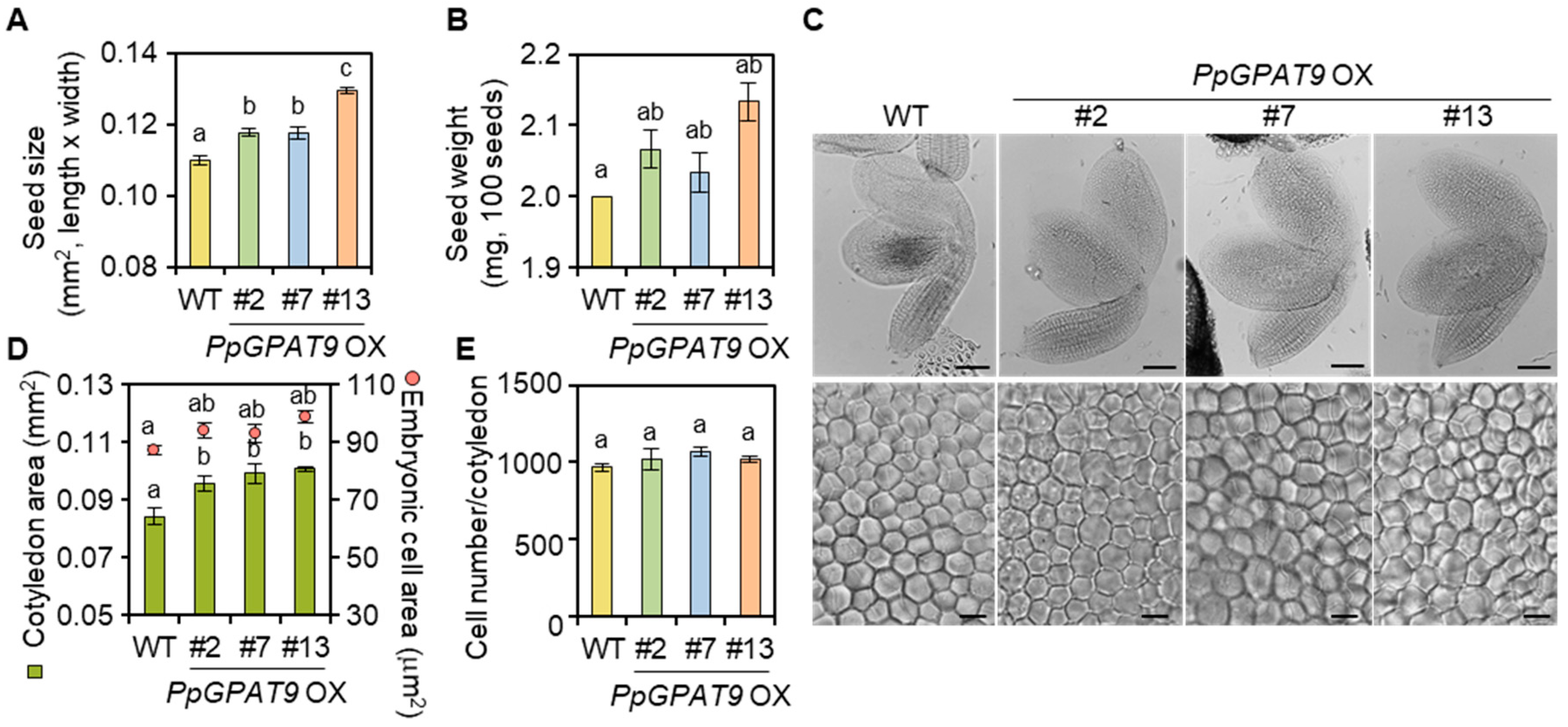
2.5. Morphological and Microscopic Analyses of Transgenic Arabidopsis Seeds Ectopically Expressing PpGPAT9
2.6. Fatty Acid Analysis of Arabidopsis Seeds and Leaves Ectopically Expressing PpGPAT9
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogenetic Tree Construction
4.3. RNA Isolation and Gene Expression Analysis
4.4. Subcellular Localization Using Fluorescent Reporter Proteins
4.5. Ectopic Expression and Transformation
4.6. Protein Extraction and Western Blotting
4.7. Microscopic Analysis of Seeds
4.8. Fatty Acid Analysis
4.9. Transient Expression in N. benthamiana Leaves
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xu, C.; Shanklin, J. Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 2016, 67, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Caldo, K.M.P.; Pal-Nath, D.; Ozga, J.; Lemieux, M.; Weselake, R.J.; Chen, G. Properties and biotechnological applications of Acyl-CoA:diacylglycerol acyltransferase and phospholipid:diacylglycerol acyltransferase from terrestrial plants and microalgae. Lipids 2018, 5, 663–688. [Google Scholar] [CrossRef] [PubMed]
- Biermann, U.; Bornscheuer, U.; Meier, M.A.; Metzger, J.O.; Schafer, H.J. Oils and fats as renewable raw materials in chemistry. Angew. Chem. Int. Ed. Engl. 2011, 50, 3854–3871. [Google Scholar] [CrossRef] [PubMed]
- Foley, P.M.; Beach, E.S.; Zimmerman, J.B. Algae as a source of renewable chemicals: Opportunities and challenges. Green Chem. 2011, 13, 1399–1405. [Google Scholar] [CrossRef]
- Ho, D.P.; Ngo, H.H.; Guo, W. A mini review on renewable sources for biofuel. Bioresour. Technol. 2014, 169, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, E.C.; Wilkie, A.C.; Kirst, M.; Rathinasabapathi, B. Metabolic regulation of triacylglycerol accumulation in the green algae: Identification of potential targets for engineering to improve oil yield. Plant Biotechnol. J. 2016, 14, 1649–1660. [Google Scholar] [CrossRef]
- Petrie, J.R.; Shrestha, P.; Belide, S.; Kennedy, Y.; Lester, G.; Liu, Q.; Divi, U.K.; Mulder, R.J.; Mansour, M.P.; Nichols, P.D.; et al. Metabolic engineering Camelina sativa with fish oil-like levels of DHA. PLoS ONE 2014, 9, e85061. [Google Scholar] [CrossRef]
- Ruiz-Lopez, N.; Sayanova, O.; Napier, J.A.; Haslam, R.P. Metabolic engineering of the omega-3 long chain polyunsaturated fatty acid biosynthetic pathway into transgenic plants. J. Exp. Bot. 2012, 63, 2397–2410. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Gimeno, R.E.; Cao, J. Thematic review series: Glycerolipids. Mammalian glycerol-3-phosphate acyltransferases: New genes for an old activity. J. Lipid Res. 2008, 49, 2079–2088. [Google Scholar] [CrossRef]
- Shockey, J.; Regmi, A.; Cotton, K.; Adhikari, N.; Browse, J.; Bates, P.D. Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 2016, 170, 163–179. [Google Scholar] [CrossRef]
- Zou, J.; Wei, Y.; Jako, C.; Kumar, A.; Selvaraj, G.; Taylor, D.C. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 1999, 19, 645–653. [Google Scholar] [CrossRef]
- Kim, H.U.; Li, Y.B.; Huang, A.H.C. Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 2005, 17, 1073–1089. [Google Scholar] [CrossRef]
- Nakamura, Y.; Tsuchiya, M.; Ohta, H. Plastidic phosphatidic acid phosphatases identified in a distinct subfamily of lipid phosphate phosphatases with prokaryotic origin. J. Biol. Chem. 2007, 282, 29013–29021. [Google Scholar] [CrossRef]
- Falcone, D.L.; Gibson, S.; Lemieux, B.; Somerville, C. Identification of a gene that complements an Arabidopsis mutant deficient in chloroplast omega 6 desaturase activity. Plant Physiol. 1994, 106, 1453–1459. [Google Scholar] [CrossRef]
- Gao, J.; Ajjawi, I.; Manoli, A.; Sawin, A.; Xu, C.; Froehlich, J.E.; Last, R.L.; Benning, C. FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 2009, 60, 832–839. [Google Scholar] [CrossRef]
- Heilmann, I.; Mekhedov, S.; King, B.; Shanklin, J. Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 2004, 136, 4237–4245. [Google Scholar] [CrossRef]
- Iba, K.; Gibson, S.; Nishiuchi, T.; Fuse, T.; Nishimura, M.; Arondel, V.; Hugly, S.; Somerville, C. A gene encoding a chloroplast omega-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J. Biol. Chem. 1993, 268, 24099–24105. [Google Scholar]
- McConn, M.; Hugly, S.; Somerville, C. A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast [omega]-3 desaturase. Plant Physiol. 1994, 106, 1609–1614. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Quettier, A.L.; Kroon, J.T.M.; Craddock, C.; Adams, N.; Slabas, A.R. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 2010, 22, 2796–2811. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Peisker, H.; Weth, A.; Baumgartner, W.; Dormann, P.; Frentzen, M. Extraplastidial cytidinediphosphate diacylglycerol synthase activity is required for vegetative development in Arabidopsis thaliana. Plant J. 2013, 75, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Browse, J.; McConn, M.; James, D., Jr.; Miquel, M. Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate: Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 1993, 268, 16345–16351. [Google Scholar] [PubMed]
- Stahl, U.; Carlsson, A.S.; Lenman, M.; Dahlqvist, A.; Huang, B.Q.; Banas, W.; Banas, A.; Stymne, S. Cloning and functional characterization of a Phospholipid:Diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 2004, 135, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Kunst, L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997, 12, 121–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornberg, A.; Pricer, W.E., Jr. Enzymatic esterification of alpha-glycerophosphate by long chain fatty acids. J. Biol. Chem. 1953, 204, 345–357. [Google Scholar] [PubMed]
- Barron, E.J.; Stumpf, P.K. Fat metabolism in higher plants. XIX. The biosynthesis of triglycerides by avocado-mesocarp enzymes. Biochim. Biophys. Acta 1962, 60, 329–337. [Google Scholar] [CrossRef]
- Gao, Q.; Shang, Y.; Huang, W.; Wang, C. Glycerol-3-phosphate acyltransferase contributes to triacylglycerol biosynthesis, lipid droplet formation, and host invasion in Metarhizium robertsii. Appl. Environ. Microbiol. 2013, 79, 7646–7653. [Google Scholar] [CrossRef]
- Niu, Y.F.; Wang, X.; Hu, D.X.; Balamurugan, S.; Li, D.W.; Yang, W.D.; Liu, J.S.; Li, H.Y. Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum. Biotechnol. Biofuels 2016, 9, 60. [Google Scholar] [CrossRef]
- Röttig, A.; Steinbüchel, A. Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 277–321. [Google Scholar] [CrossRef]
- Waschburger, E.; Kulcheski, F.R.; Veto, N.M.; Margis, R.; Margis-Pinheiro, M.; Turchetto-Zolet, A.C. Genome-wide analysis of the glycerol-3-phosphate acyltransferase (GPAT) gene family reveals the evolution and diversification of plant GPATs. Genet. Mol. Biol. 2018, 41, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J.; Knight, C.D. The Moss Physcomitrella patens, a model system with potential for the study of plant reproduction. Plant Cell 1993, 5, 1483–1488. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.D.; Chen, G.; Mietkiewska, E.; Tomasi, P.; Jayawardhane, K.; Dyer, J.M.; Weselake, R.J. Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids. J. Exp. Bot. 2016, 67, 4627–4638. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Hirasawa, E.; Takemura, T.; Takahashi, S.; Chokshi, K.; Pancha, I.; Tanaka, K.; Imamura, S. Accelerated triacylglycerol production without growth inhibition by overexpression of a glycerol-3-phosphate acyltransferase in the unicellular red alga Cyanidioschyzon merolae. Sci. Rep. 2018, 8, 12410. [Google Scholar] [CrossRef] [PubMed]
- Strotbek, C.; Krinninger, S.; Frank, W. The moss Physcomitrella patens: Methods and tools from cultivation to targeted analysis of gene function. Int. J. Dev. Biol. 2013, 57, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Beike, A.K.; Jaeger, C.; Zink, F.; Decker, E.L.; Reski, R. High contents of very long-chain polyunsaturated fatty acids in different moss species. Plant Cell Rep. 2014, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Grimsley, N.H.; Grimsley, J.M.; Hartmann, E. Fatty acid composition of mutants of the moss Physcomitrella patens. Phytochemistry 1981, 20, 1519–1524. [Google Scholar] [CrossRef]
- Resemann, H.C.; Lewandowska, M.; Gomann, J.; Feussner, I. Membrane lipids, waxes and oxylipins in the moss model organism Physcomitrella patens. Plant Cell Physiol. 2019, 60, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Beisson, F.; Li, Y.; Bonaventure, G.; Pollard, M.; Ohlrogge, J.B. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 2007, 19, 351–368. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Pollard, M.; Sauveplane, V.; Pinot, F.; Ohlrogge, J.; Beisson, F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. USA 2009, 106, 22008–22013. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Beisson, F.; Koo, A.J.K.; Molina, I.; Pollard, M.; Ohlrogge, J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. USA 2007, 104, 18339–18344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Simpson, J.P.; Li-Beisson, Y.; Beisson, F.; Pollard, M.; Ohlrogge, J.B. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: Substrate specificity, sn-2 preference, and evolution. Plant Physiol. 2012, 160, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Xia, Q.; Dauk, M.; Shen, W.; Selvaraj, G.; Zou, J. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell. 2003, 15, 1872–1887. [Google Scholar] [CrossRef] [PubMed]
- Lewin, T.M.; Wang, P.; Coleman, R.A. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 1999, 38, 5764–5771. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Kim, H.; Go, Y.S.; Lee, S.B.; Hur, C.G.; Kim, H.U.; Suh, M.C. Identification of functional BrFAD2-1 gene encoding microsomal delta-12 fatty acid desaturase from Brassica rapa and development of Brassica napus containing high oleic acid contents. Plant Cell Rep. 2011, 30, 1881–1892. [Google Scholar] [CrossRef] [PubMed]
- Gidda, S.K.; Shockey, J.M.; Rothstein, S.J.; Dyer, J.M.; Mullen, R.T. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: Functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 2009, 47, 867–879. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–896. [Google Scholar] [CrossRef]
- Li, M.; Welti, R.; Wang, X. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 2006, 142, 750–761. [Google Scholar] [CrossRef]
- Yang, Z.; Ohlrogge, J.B. Turnover of fatty acids during natural senescence of Arabidopsis, Brachypodium, and switchgrass and in Arabidopsis beta-oxidation mutants. Plant Physiol. 2009, 150, 1981–1989. [Google Scholar] [CrossRef]
- Sanjaya; Miller, R.; Durrett, T.P.; Kosma, D.K.; Lydic, T.A.; Muthan, B.; Koo, A.J.; Bukhman, Y.V.; Reid, G.E.; Howe, G.A.; et al. Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell 2013, 25, 677–693. [Google Scholar] [CrossRef]
- An, D.; Suh, M.C. Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 2015, 9, 137–148. [Google Scholar] [CrossRef]
- Huang, C.Y.; Chung, C.I.; Lin, Y.C.; Hsing, Y.I.; Huang, A.H. Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol. 2009, 150, 1192–1203. [Google Scholar] [CrossRef]
- Nishida, I.; Tasaka, Y.; Shiraishi, H.; Murata, N. The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate acyltransferase of Arabidopsis thaliana. Plant Mol. Biol. 1993, 21, 267–277. [Google Scholar] [CrossRef]
- Yang, W.; Pollard, M.; Li-Beisson, Y.; Beisson, F.; Feig, M.; Ohlrogge, J. A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc. Natl. Acad. Sci. USA 2010, 107, 12040–12045. [Google Scholar] [CrossRef]
- Lee, S.B.; Yang, S.U.; Pandey, G.; Kim, M.-S.; Hyoung, S.; Choi, D.; Shin, J.S.; Suh, M.C. Occurrence of land-plant-specific glycerol-3-phosphate acyltransferases enabled cuticle formation and gametophore development in Physcomitrella patens. New Phytol. 2019. (In press) [Google Scholar]
- McCartney, A.W.; Dyer, J.M.; Dhanoa, P.K.; Kim, P.K.; Andrews, D.W.; McNew, J.A.; Mullen, R.T. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 2004, 37, 156–173. [Google Scholar] [CrossRef]
- Vanhercke, T.; El Tahchy, A.; Liu, Q.; Zhou, X.R.; Shrestha, P.; Divi, U.K.; Ral, J.P.; Mansour, M.P.; Nichols, P.D.; James, C.N.; et al. Metabolic engineering of biomass for high energy density: Oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 2014, 12, 231–239. [Google Scholar] [CrossRef]
- An, D.; Kim, H.; Ju, S.; Go, Y.S.; Kim, H.U.; Suh, M.C. Expression of Camelina WRINKLED1 isoforms rescue the seed phenotype of the Arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 2017, 8, 34. [Google Scholar] [CrossRef]
- Ivarson, E.; Leiva-Eriksson, N.; Ahlman, A.; Kanagarajan, S.; Bulow, L.; Zhu, L.H. Effects of overexpression of WRI1 and hemoglobin genes on the seed oil content of Lepidium campestre. Front. Plant Sci. 2016, 7, 2032. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Banaś, W.; Carlsson, A.S.; Banaś, A. Effect of overexpression of PDAT gene on Arabidopsis growth rate and seed oil content. J. Agric. Sci. 2014, 6, 65–79. [Google Scholar] [CrossRef]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef]
- Kelly, A.A.; van Erp, H.; Quettier, A.L.; Shaw, E.; Menard, G.; Kurup, S.; Eastmond, P.J. The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 2013, 162, 1282–1289. [Google Scholar] [CrossRef]
- van Erp, H.; Kelly, A.A.; Menard, G.; Eastmond, P.J. Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiol. 2014, 165, 30–36. [Google Scholar] [CrossRef]
- Kim, H.; Park, J.H.; Kim, D.J.; Kim, A.Y.; Suh, M.C. Functional analysis of diacylglycerol acyltransferase1 genes from Camelina sativa and effects of CsDGAT1B overexpression on seed mass and storage oil content in C. sativa. Plant Biotechnol. Rep. 2016, 10, 141–153. [Google Scholar] [CrossRef]
- Wu, X.L.; Liu, Z.H.; Hu, Z.H.; Huang, R.Z. BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J. Integr. Plant Biol. 2014, 56, 582–593. [Google Scholar] [CrossRef]
- Weber, H.; Borisjuk, L.; Wobus, U. Sugar import and metabolism during seed development. Trends Plant Sci. 1997, 2, 169–174. [Google Scholar] [CrossRef]
- Paya-Milans, M.; Aznar-Moreno, J.A.; Balbuena, T.S.; Haslam, R.P.; Gidda, S.K.; Perez-Hormaeche, J.; Mullen, R.T.; Thelen, J.J.; Napier, J.A.; Salas, J.J.; et al. Sunflower HaGPAT9-1 is the predominant GPAT during seed development. Plant Sci. 2016, 252, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Ashton, N.W.; Cove, D.J. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol. Gen. Genet. 1977, 154, 87–95. [Google Scholar] [CrossRef]
- Nishiyama, T.; Hiwatashi, Y.; Sakakibara, I.; Kato, M.; Hasebe, M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 2000, 7, 9–17. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Buda, G.J.; Barnes, W.J.; Fich, E.A.; Park, S.; Yeats, T.H.; Zhao, L.; Domozych, D.S.; Rose, J.K. An ATP binding cassette transporter is required for cuticular wax deposition and desiccation tolerance in the moss Physcomitrella patens. Plant Cell 2013, 25, 4000–4013. [Google Scholar] [CrossRef]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Martinez-Trujillo, M.; Limones-Briones, V.; Cabrera-Ponce, J.L. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 2004, 22, 63–70. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.U.; Kim, J.; Kim, H.; Suh, M.C. Functional Characterization of Physcomitrella patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression. Plants 2019, 8, 284. https://doi.org/10.3390/plants8080284
Yang SU, Kim J, Kim H, Suh MC. Functional Characterization of Physcomitrella patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression. Plants. 2019; 8(8):284. https://doi.org/10.3390/plants8080284
Chicago/Turabian StyleYang, Sun Ui, Juyoung Kim, Hyojin Kim, and Mi Chung Suh. 2019. "Functional Characterization of Physcomitrella patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression" Plants 8, no. 8: 284. https://doi.org/10.3390/plants8080284




