Fast Regulation of Hormone Metabolism Contributes to Salt Tolerance in Rice (Oryza sativa spp. Japonica, L.) by Inducing Specific Morpho-Physiological Responses
Abstract
:1. Introduction
2. Results and Discussion
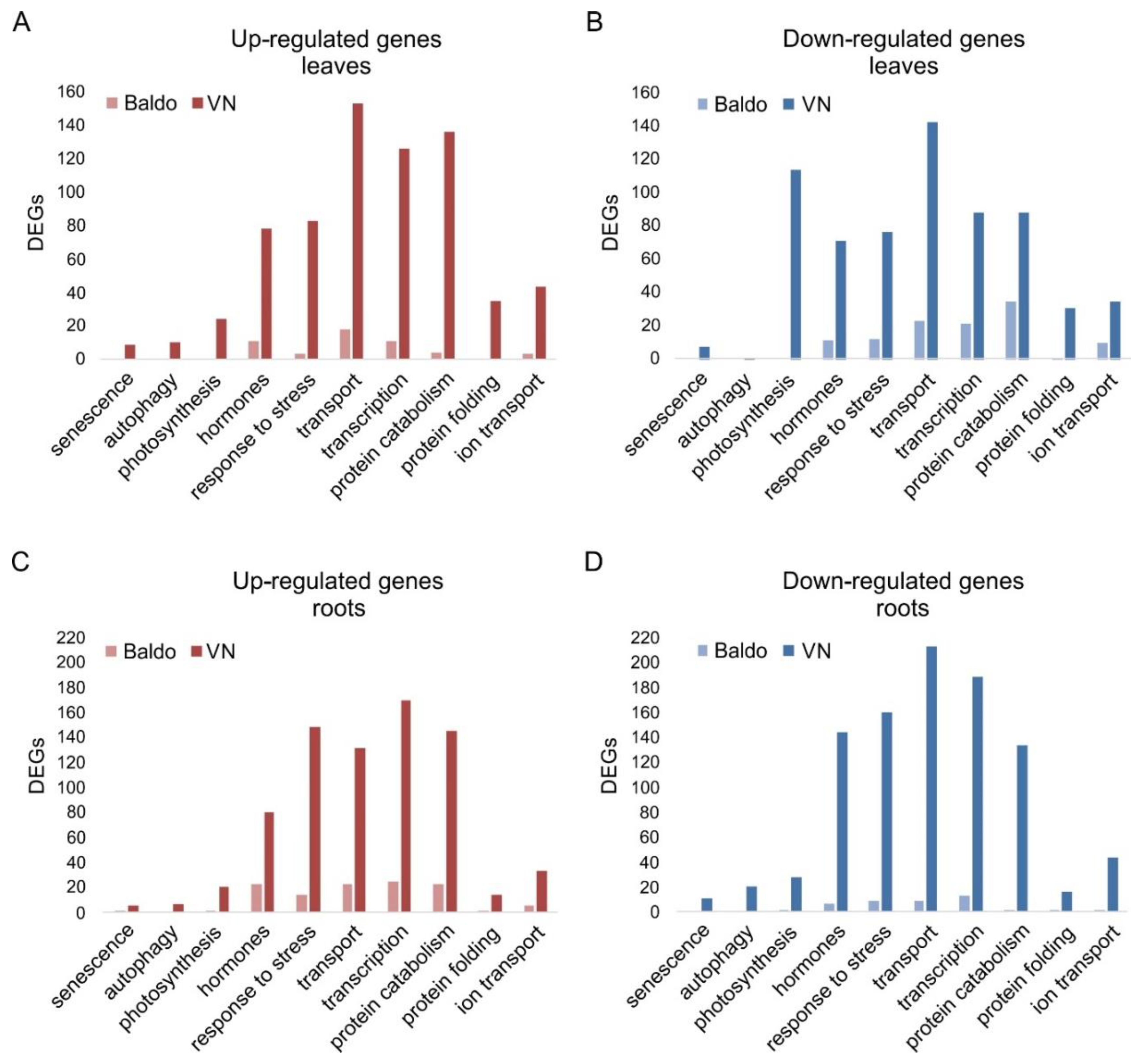
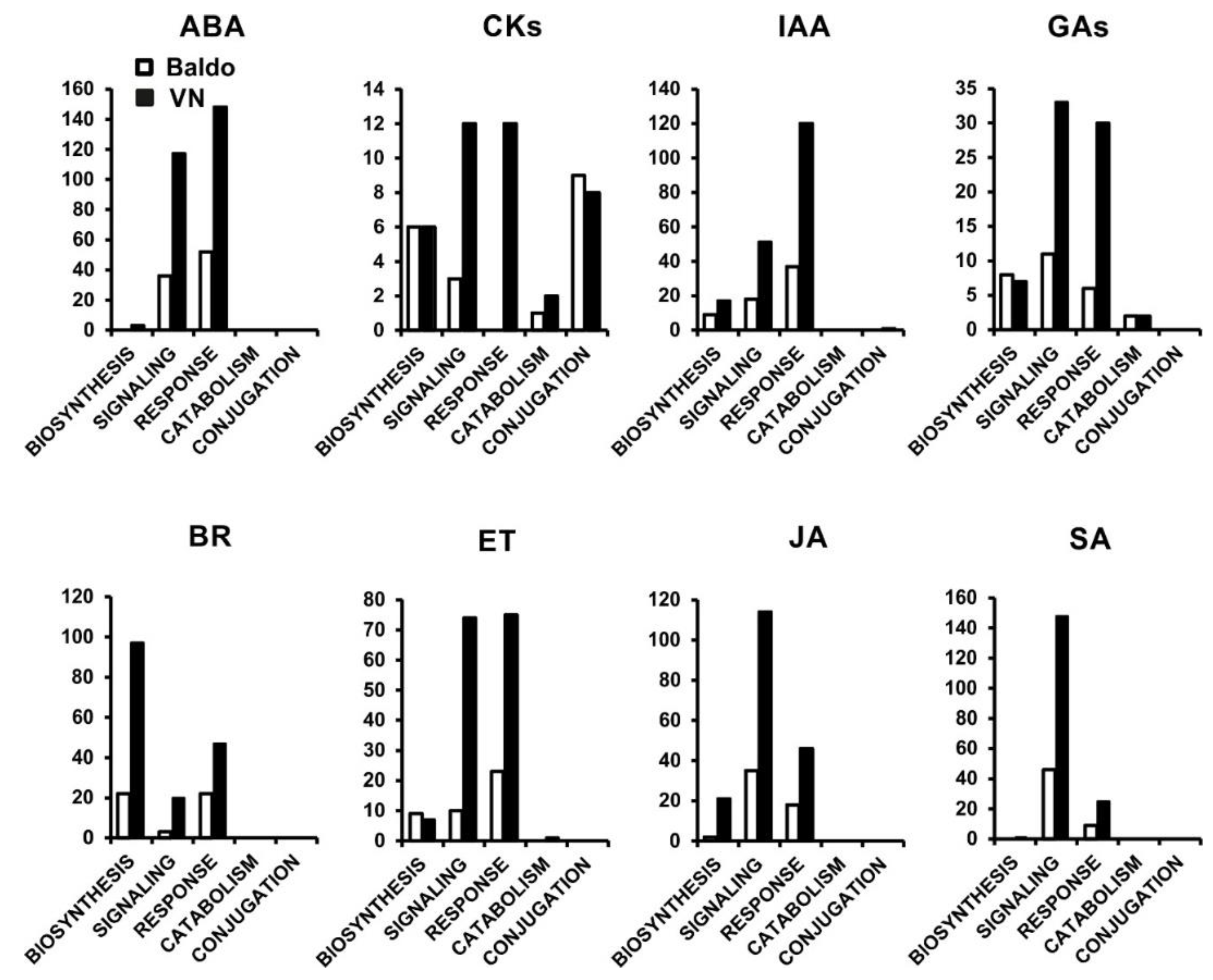
2.1. Transcriptional Changes in Hormone-Related Functional Categories
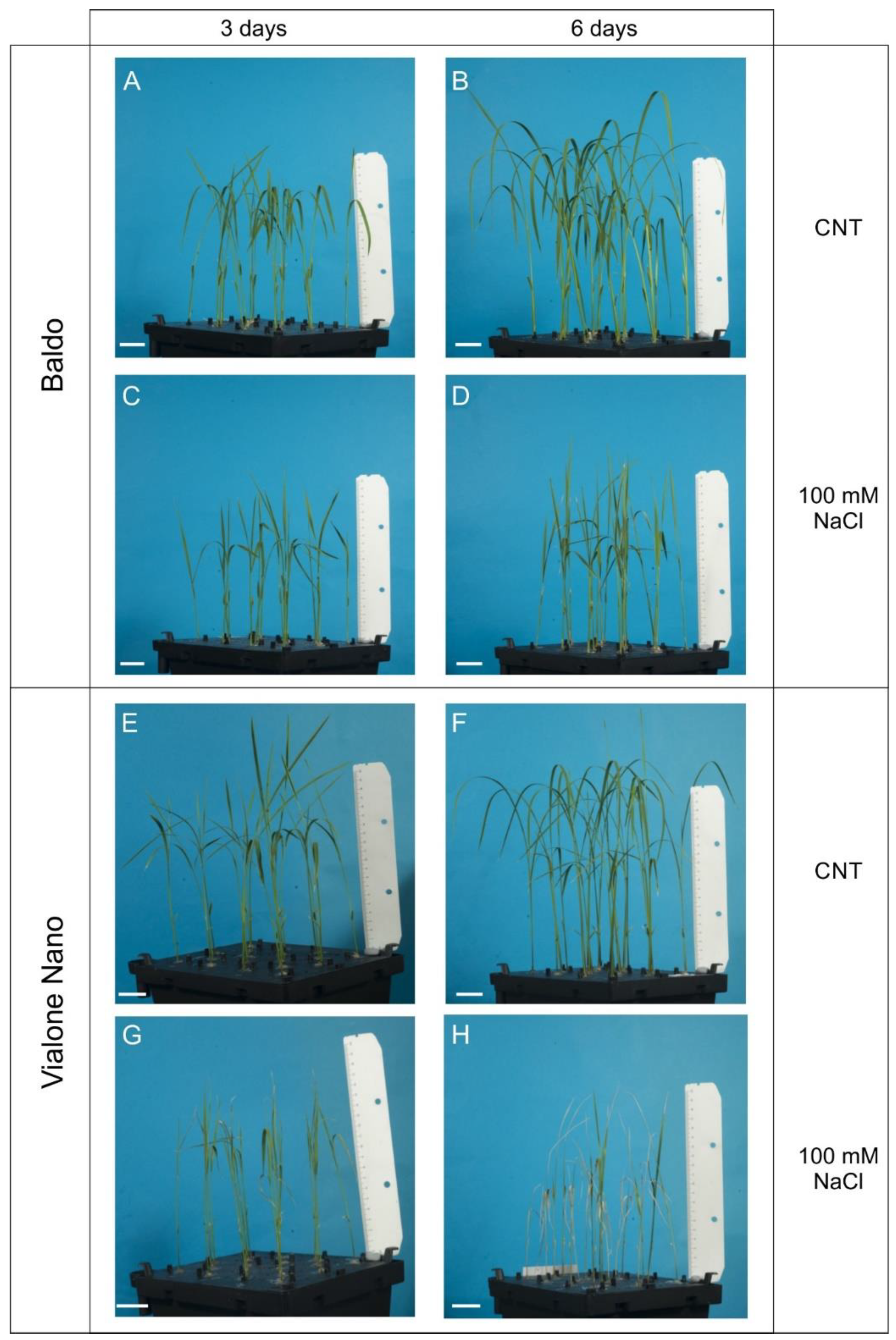
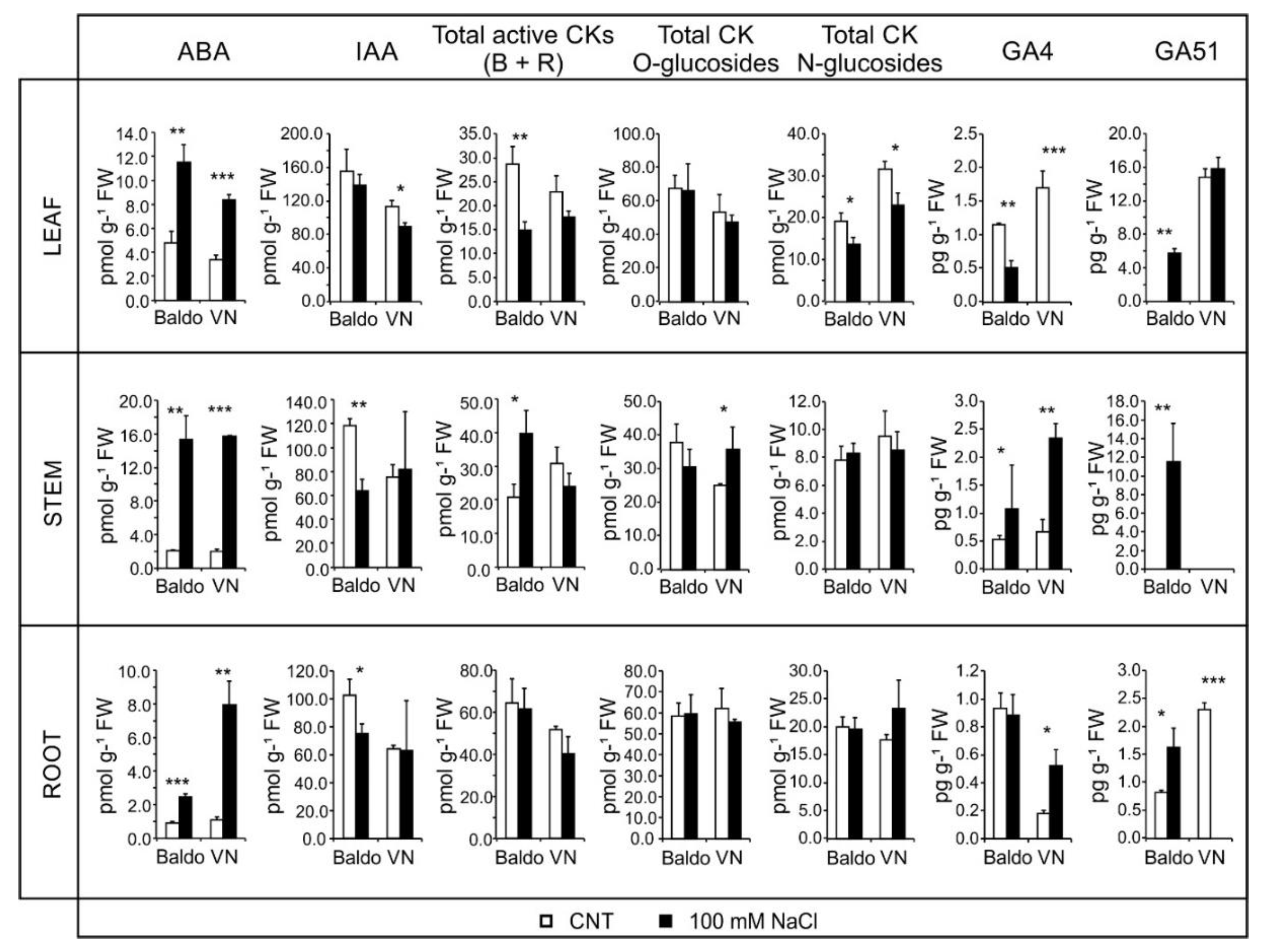
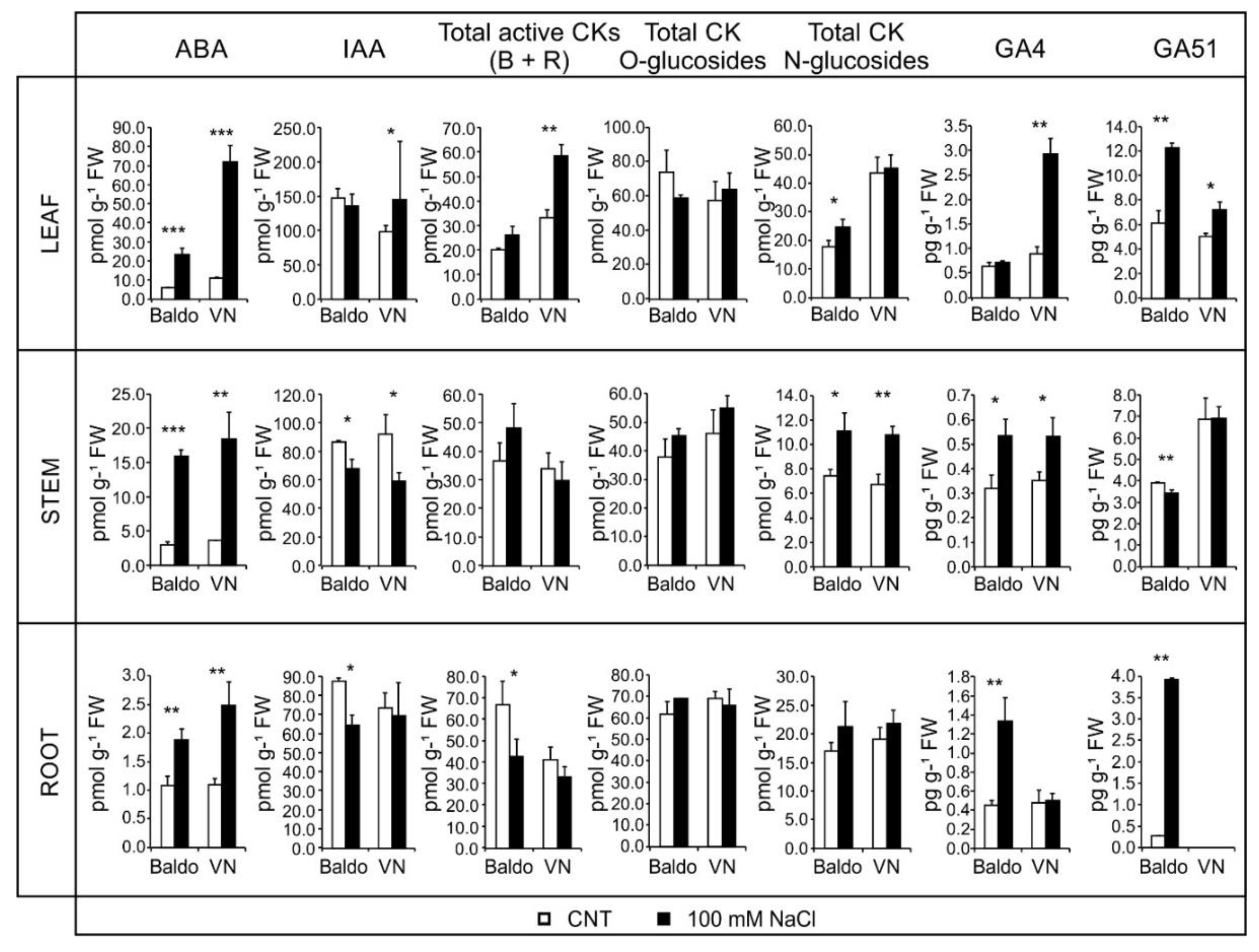
2.2. Changes in Hormonal Contents and Phenotypic Plasticity in Salt-Stressed Plants
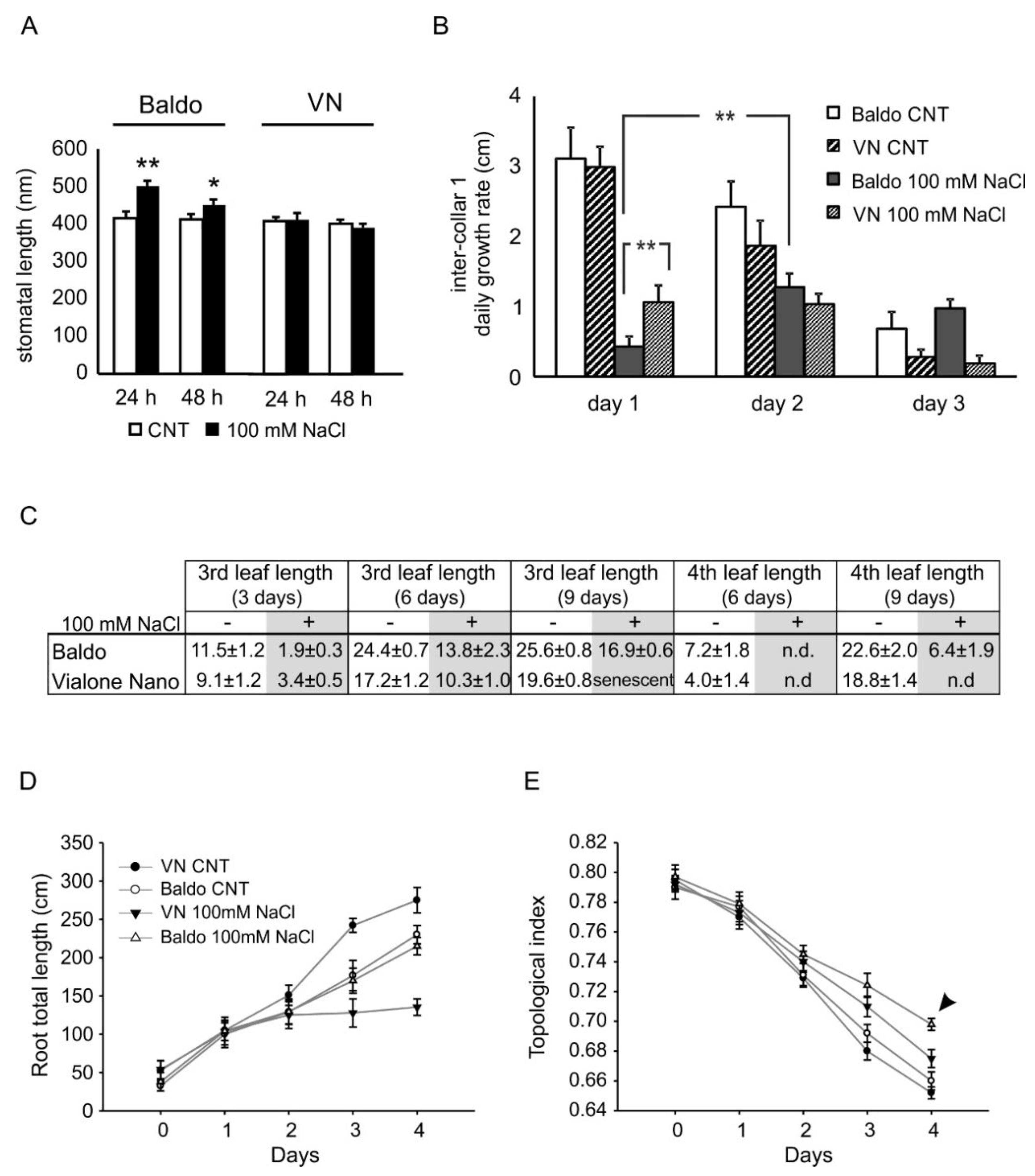
2.2.1. ABA and CKs Levels Control Stomata Aperture
2.2.2. IAA and CKs Levels Control Shoot Growth
2.2.3. ABA, IAA and CKs Crosstalk in Shaping the Root Morphology
2.2.4. Role of GAs in Growth Regulation in Response to Salinity: From Quiescence to Recovery
3. Materials and Methods
3.1. Plant Material
3.2. Transcriptomic Analysis
3.3. Root Morphology Analysis
3.4. Hormones Analysis
3.5. Stomatal Aperture Measurements
3.6. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/soils-portal/soil-management/management-of-some-problem-soils/salt-affected-soils/more-information-on-salt-affected-soils/en/ (accessed on 21 August 2018).
- Das, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; Lv, Q. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of Salt Tolerance-related microRNAs and Their Targets in Maize (Zea mays L.) Using High-throughput Sequencing and Degradome Analysis. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S.-R. Ectopic Expression of OsSta2 Enhances Salt Stress Tolerance in Rice. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Nounjan, N.; Chansongkrow, P.; Charoensawan, V.; Siangliw, J.L.; Toojinda, T.; Chadchawan, S.; Theerakulpisut, P. High Performance of Photosynthesis and Osmotic Adjustment Are Associated With Salt Tolerance Ability in Rice Carrying Drought Tolerance QTL: Physiological and Co-expression Network Analysis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, B.D.; Kumar, A.; Maurya, S.; Krishnan, S.G.; Vinod, K.K.; Singh, M.P.; Ellur, R.K.; Bhowmick, P.K.; Singh, A.K. Marker-Assisted Introgression of Saltol QTL Enhances Seedling Stage Salt Tolerance in the Rice Variety “Pusa Basmati 1”. Available online: https://www.hindawi.com/journals/ijg/2018/8319879/ (accessed on 5 September 2018).
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Pierik, R.; Testerink, C. The Art of Being Flexible: How to Escape from Shade, Salt, and Drought. Plant Physiol. 2014, 166, 5–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierik, R.; de Wit, M. Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. J. Exp. Bot. 2014, 65, 2815–2824. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Voesenek, L.A.C.J. Flooding Stress: Acclimations and Genetic Diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism Is a Response of Plant Roots to Avoid a Saline Environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allu, A.D.; Soja, A.M.; Wu, A.; Szymanski, J.; Balazadeh, S. Salt stress and senescence: Identification of cross-talk regulatory components. J. Exp. Bot. 2014, 65, 3993–4008. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013, 112, 1655–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Jia, L.; Zhang, J. ABA signal in rice under stress conditions. Rice (N. Y.) 2012, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, S.; Kudoyarova, G.R.; Veselov, D.S.; Arkhipova, T.N.; Davies, W.J. Plant hormone interactions: Innovative targets for crop breeding and management. J. Exp. Bot. 2012, 63, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, F.; Parent, B.; Simonneau, T. Control of leaf growth by abscisic acid: Hydraulic or non-hydraulic processes? Plant Cell Environ. 2010, 33, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Davies, W.J.; Wilkinson, S.; Loveys, B. Stomatal control by chemical signalling and the exploitation of this mechanism to increase water use efficiency in agriculture. New Phytol. 2002, 153, 449–460. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, D.; Pang, L.; Kobayashi, A.; Fozard, J.A.; Boudolf, V.; Bhosale, R.; Antoni, R.; Nguyen, T.; Hiratsuka, S.; Fujii, N.; et al. Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat. Plants 2017, 3, 17057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Dietrich, D.; Ng, C.H.; Chan, P.M.Y.; Bhalerao, R.; Bennett, M.J.; Dinneny, J.R. Endodermal ABA Signaling Promotes Lateral Root Quiescence during Salt Stress in Arabidopsis Seedlings. Plant Cell 2013, 25, 324–341. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, S.; Sanguineti, M.C.; Tuberosa, R.; Bellotti, M.; Salvi, S.; Landi, P. Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J. Exp. Bot. 2005, 56, 3061–3070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albacete, A.; Ghanem, M.E.; Martinez-Andujar, C.; Acosta, M.; Sanchez-Bravo, J.; Martinez, V.; Lutts, S.; Dodd, I.C.; Perez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-H.; Liu, Y.-B.; Zhang, X.-S. Auxin–Cytokinin Interaction Regulates Meristem Development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-H.; Liu, Y.-C. Expression of isopentenyl transferase gene (ipt) in leaf and stem delayed leaf senescence without affecting root growth. Plant Cell Rep. 2009, 28, 1759. [Google Scholar] [CrossRef] [PubMed]
- Talla, S.K.; Panigrahy, M.; Kappara, S.; Nirosha, P.; Neelamraju, S.; Ramanan, R. Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. J. Exp. Bot. 2016, 67, 1839–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daszkowska-Golec, A.; Szarejko, I. Open or Close the Gate—Stomata Action Under the Control of Phytohormones in Drought Stress Conditions. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, R.-J.; Han, T.-T.; Cai, W.; Fu, Z.-W.; Lu, Y.-T. Salt Stress Reduces Root Meristem Size by Nitric Oxide-Mediated Modulation of Auxin Accumulation and Signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.-W.; Li, C.-H.; Cao, J.; Zhang, Y.-C.; Zhang, S.-Q.; Xia, Y.-F.; Sun, D.-Y.; Sun, Y. Altered Architecture and Enhanced Drought Tolerance in Rice via the Down-Regulation of Indole-3-Acetic Acid by TLD1/OsGH3.13 Activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and Cell Physiological Analyses in Different Rice Cultivars Provide New Insights Into Adaptive and Salinity Stress Responses. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Formentin, E.; Sudiro, C.; Ronci, M.B.; Locato, V.; Barizza, E.; Stevanato, P.; Ijaz, B.; Zottini, M.; De Gara, L.; Lo Schiavo, F. H2O2 Signature and Innate Antioxidative Profile Make the Difference between Sensitivity and Tolerance to Salt in Rice Cells. Front. Plant Sci. 2018. Accepted for publication. [Google Scholar]
- Ma, B.; Yin, C.-C.; He, S.-J.; Lu, X.; Zhang, W.-K.; Lu, T.-G.; Chen, S.-Y.; Zhang, J.-S. Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice (Oryza sativa L.) Seedlings. PLoS Genet. 2014, 10, e1004701. [Google Scholar] [CrossRef] [PubMed]
- Koeduka, T.; Matsui, K.; Hasegawa, M.; Akakabe, Y.; Kajiwara, T. Rice fatty acid α-dioxygenase is induced by pathogen attack and heavy metal stress: Activation through jasmonate signaling. J. Plant Physiol. 2005, 162, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Nie, S.; Xing, D. Over-expression of Arabidopsis Bax inhibitor-1 delays methyl jasmonate-induced leaf senescence by suppressing the activation of MAP kinase 6. J. Exp. Bot. 2012, 63, 4463–4474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. PNAS 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Counce, P.A.; Keisling, T.C.; Mitchell, A.J. A Uniform, Objective, and Adaptive System for Expressing Rice Development. Crop Sci. 2000, 40, 436. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Ha, C.V.; Nishiyama, R.; Watanabe, Y.; Leyva-González, M.A.; Fujita, Y.; Tran, U.T.; Li, W.; Tanaka, M.; Seki, M.; et al. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. PNAS 2016, 113, 3090–3095. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.E.; Kwon, Y.; Kim, J.H.; Noh, H.; Hong, S.-W.; Lee, H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 2011, 77, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, M.; Matus, J.T.; Francia, P.; Rusconi, F.; Cañón, P.; Medina, C.; Conti, L.; Cominelli, E.; Tonelli, C.; Arce-Johnson, P. The grapevine guard cell-related VvMYB60 transcription factor is involved in the regulation of stomatal activity and is differentially expressed in response to ABA and osmotic stress. BMC Plant Biol. 2011, 11, 142. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-W.; Qi, M.; Yang, Y. A Novel Jasmonic Acid-Inducible Rice myb Gene Associates with Fungal Infection and Host Cell Death. MPMI 2001, 14, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Brütting, C.; Meza-Canales, I.D.; Großkinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis -zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Johnston, R.; Yang, Y.; Gallavotti, A.; Kojima, M.; Travençolo, B.A.; Costa, L.D.; Sakakibara, H.; Jackson, D. Studies of aberrant phyllotaxy1 Mutants of Maize Indicate Complex Interactions between Auxin and Cytokinin Signaling in the Shoot Apical Meristem. Plant Physiol. 2009, 150, 205–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielach, A.; Hrtyan, M.; Tognetti, V.; Bielach, A.; Hrtyan, M.; Tognetti, V.B. Plants under Stress: Involvement of Auxin and Cytokinin. Int. J. Mol. Sci. 2017, 18, 1427. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, L.; Forde, B.G.; Davies, W.J. The Biphasic Root Growth Response to Abscisic Acid in Arabidopsis Involves Interaction with Ethylene and Auxin Signalling Pathways. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; De Smet, I. Localised ABA signalling mediates root growth plasticity. Trends Plant Sci. 2013, 18, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S. Gibberellin Metabolism and its Regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.; Hsing, Y.C.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I.; Hoagland, D.R. A Comparison of Water Culture and Soil as Media for Crop Production. Science 1939, 89, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Falda, M.; Toppo, S.; Pescarolo, A.; Lavezzo, E.; Di Camillo, B.; Facchinetti, A.; Cilia, E.; Velasco, R.; Fontana, P. Argot2: A large scale function prediction tool relying on semantic similarity of weighted Gene Ontology terms. BMC Bioinform. 2012, 13, S14. [Google Scholar] [CrossRef] [PubMed]
- Fontana, P.; Cestaro, A.; Velasco, R.; Formentin, E.; Toppo, S. Rapid Annotation of Anonymous Sequences from Genome Projects Using Semantic Similarities and a Weighting Scheme in Gene Ontology. PLoS ONE 2009, 4, e4619. [Google Scholar] [CrossRef] [PubMed]
- Lavezzo, E.; Falda, M.; Fontana, P.; Bianco, L.; Toppo, S. Enhancing protein function prediction with taxonomic constraints—The Argot2.5 web server. Methods 2016, 93, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar]
- Fitter, A.H.; Stickland, T.R. Architectural analysis of plant root systems 2. Influence of nutrient supply on architecture in contrasting plant species. New Phytol. 1991, 118, 383–389. [Google Scholar] [CrossRef] [Green Version]
- Fitter, A.H.; Stickland, T.R.; Harvey, M.L.; Wilson, G.W. Architectural analysis of plant root systems 1. Architectural correlates of exploitation efficiency. New Phytol. 1991, 118, 375–382. [Google Scholar] [CrossRef] [Green Version]
- Floková, K.; Tarkowská, D.; Miersch, O.; Strnad, M.; Wasternack, C.; Novák, O. UHPLC–MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 2014, 105, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, T.; Tarkowská, D.; Novák, O.; Hedden, P.; Strnad, M. Analysis of gibberellins as free acids by ultra performance liquid chromatography–tandem mass spectrometry. Talanta 2013, 112, 85–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Hosek, P.; Soudek, P.; Knirsch, V.; Vankova, R. Hormonal dynamics during salt stress responses of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea. Plant Sci. 2017, 264, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.G.; Roberts, J.; Jordan, W.R.; Drew, M.C. Growth, Water Relations, and Accumulation of Organic and Inorganic Solutes in Roots of Maize Seedlings during Salt Stress. Plant Physiol. 1997, 113, 881–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Formentin, E.; Barizza, E.; Stevanato, P.; Falda, M.; Massa, F.; Tarkowskà, D.; Novák, O.; Lo Schiavo, F. Fast Regulation of Hormone Metabolism Contributes to Salt Tolerance in Rice (Oryza sativa spp. Japonica, L.) by Inducing Specific Morpho-Physiological Responses. Plants 2018, 7, 75. https://doi.org/10.3390/plants7030075
Formentin E, Barizza E, Stevanato P, Falda M, Massa F, Tarkowskà D, Novák O, Lo Schiavo F. Fast Regulation of Hormone Metabolism Contributes to Salt Tolerance in Rice (Oryza sativa spp. Japonica, L.) by Inducing Specific Morpho-Physiological Responses. Plants. 2018; 7(3):75. https://doi.org/10.3390/plants7030075
Chicago/Turabian StyleFormentin, Elide, Elisabetta Barizza, Piergiorgio Stevanato, Marco Falda, Federica Massa, Danuše Tarkowskà, Ondřej Novák, and Fiorella Lo Schiavo. 2018. "Fast Regulation of Hormone Metabolism Contributes to Salt Tolerance in Rice (Oryza sativa spp. Japonica, L.) by Inducing Specific Morpho-Physiological Responses" Plants 7, no. 3: 75. https://doi.org/10.3390/plants7030075







