Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress
Abstract
:1. Introduction
2. Results
2.1. Effects of Waterlogging Stress on Root Growth
2.2. Protein Identification and Quantification
2.3. Differentially Expressed Proteins Response to Waterlogging Stress by iTRAQ
2.4. Gene Onology Annotation of the Differentially Accumulated Proteins
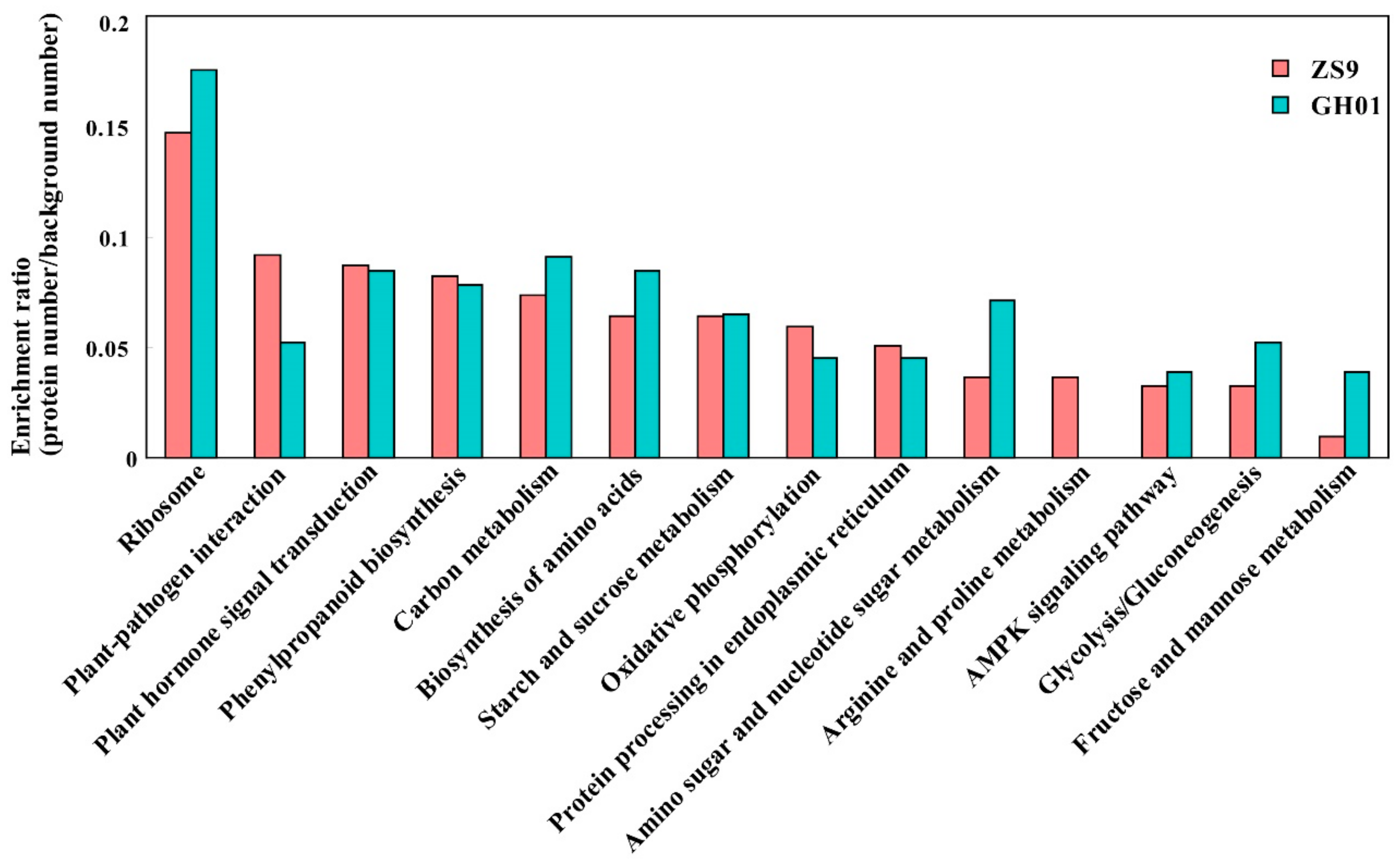
2.5. Pathway Analysis
2.6. Differential Protein Overlapped with ZS9 and GH01
3. Discussion
3.1. Contributions of Proteome Changes to Adapt to Waterlogging Stress
3.2. Waterlogging Stress Affects Proteins Involved in Ethylene Signaling
3.3. Waterlogging Stress Affects Proteins Involved in Protein Phosphorylation
3.4. Waterlogging Stress Affects Proteins Involved in Metabolism
4. Materials and Methods
4.1. Plant Material and Stress Treatments
4.2. Root Morphology and Optical Microscopic Observation
4.3. Protein Extraction
4.4. Protein Digestion and iTRAQ Labeling
4.5. Liquid Chromatography-Tandem Mass Spectrometry
4.6. iTRAQ Analysis
4.7. Bioinformatics Analysis
4.8. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CDPK | calcium-dependent protein kinase |
| ERFs | ethylene response factors |
| GO | Gene Ontology |
| iTRAQ | isobaric tags for relative and absolute quantitation |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
| MAPK | mitogen-activated protein kinase |
| MS | mass spectrometry |
| PTMs | post-translational modifications |
| QTL | quantitative trait locus |
| qRT-PCR | quantitative real time polymerase chain reaction |
| TCA | tricarboxylic acid cycle |
| XTHs | xyloglucan endotransglucosylase/hydrolases |
References
- Jiang, C.; Shi, J.; Li, R.; Long, Y.; Wang, H.; Li, D.; Zhao, J.; Meng, J. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor Appl. Genet. 2014, 127, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Abreu, I.A.; Farinha, A.P.; Negrao, S.; Goncalves, N.; Fonseca, C.; Rodrigues, M.; Batista, R.; Saibo, N.J.; Oliveira, M.M. Coping with abiotic stress: Proteome changes for crop improvement. J. Proteom. 2013, 93, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Xu, J. Abiotic stress responses in plant roots: A proteomics perspective. Front. Plant Sci. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Rampitsch, C.; Bykova, N.V. Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front. Plant Sci. 2015, 6, 209. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Rafii, M.Y.; Ismail, M.R.; Juraimi, A.S.; Rahim, H.A.; Asfaliza, R.; Latif, M.A. Waterlogging tolerance of crops: Breeding, mechanism of tolerance, molecular approaches, and future prospects. Biomed. Res. Int. 2013, 2013, 963525. [Google Scholar] [CrossRef] [PubMed]
- Herzog, M.; Striker, G.G.; Colmer, T.D.; Pedersen, O. Mechanisms of waterlogging tolerance in wheat--a review of root and shoot physiology. Plant Cell Environ. 2016, 39, 1068–1086. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; van Veen, H.; Perata, P. Plant responses to flooding stress. Curr. Opin. Plant Biol. 2016, 33, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.C.; Chou, M.Y.; Chou, S.J.; Li, Y.R.; Peng, H.P.; Shih, M.C. Submergence confers immunity mediated by the wrky22 transcription factor in arabidopsis. Plant Cell 2013, 25, 2699–2713. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteome Res. 2012, 11, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.C.; Shih, M.C. Plant defense after flooding. Plant Signal. Behav. 2013, 8, e26922. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ji, J.; Xu, Q.; Qi, X.; Weng, Y.; Chen, X. The major-effect quantitative trait locus csarn6.1 encodes an aaa atpase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. Plant J. 2018, 93, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M. Root responses to flooding. Curr. Opin. Plant Biol. 2013, 16, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Tuan, P.A.; Mukherjee, S.; Son, S.; Ayele, B.T. Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat. J. Exp. Bot. 2018, 69, 4065–4082. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Kovalev, A.; Gorb, S.N.; Sauter, M. Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 2012, 24, 3296–3306. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Tanaka, A.; Mori, H.; Takamure, I.; Kato, K.; Nakazono, M. Ethylene-dependent aerenchyma formation in adventitious roots is regulated differently in rice and maize. Plant Cell Environ. 2016, 39, 2145–2157. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, N.; Hossain, F.; Mohan, S.; Shiriga, K.; Mittal, S.; Sharma, R.; Singh, R.K.; Gupta, H.S. Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS ONE 2013, 8, e70433. [Google Scholar] [CrossRef] [PubMed]
- Kotula, L.; Clode, P.L.; Striker, G.G.; Pedersen, O.; Lauchli, A.; Shabala, S.; Colmer, T.D. Oxygen deficiency and salinity affect cell-specific ion concentrations in adventitious roots of barley (hordeum vulgare). New Phytol. 2015, 208, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Vwioko, E.; Adinkwu, O.; El-Esawi, M.A. Comparative physiological, biochemical, and genetic responses to prolonged waterlogging stress in okra and maize given exogenous ethylene priming. Front. Physiol 2017, 8, 632. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, M.; Ji, J.; Xu, Q.; Qi, X.; Chen, X. Comparative rna-seq based transcriptome profiling of waterlogging response in cucumber hypocotyls reveals novel insights into the de novo adventitious root primordia initiation. BMC Plant Biol. 2017, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Song, L.; Chen, H.; Valliyodan, B.; Cheng, P.; Ali, L.; Vuong, T.; Wu, C.; Orlowski, J.; Buckley, B.; et al. A major natural genetic variation associated with root system architecture and plasticity improves waterlogging tolerance and yield in soybean. Plant Cell Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Sauter, M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 2009, 21, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Dawood, T.; Yang, X.; Visser, E.J.; Te Beek, T.A.; Kensche, P.R.; Cristescu, S.M.; Lee, S.; Flokova, K.; Nguyen, D.; Mariani, C.; et al. A co-opted hormonal cascade activates dormant adventitious root primordia upon flooding in solanum dulcamara. Plant Physiol. 2016, 170, 2351–2364. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Perata, P.; Voesenek, L.A. Submergence tolerance in rice requires sub1a, an ethylene-response-factor-like gene. Trends Plant Sci. 2007, 12, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. Submergence tolerance conferred by sub1a is mediated by slr1 and slrl1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors snorkel1 and snorkel2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. Hre1 and hre2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Bui, L.T.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Licausi, F. Constitutively expressed erf-vii transcription factors redundantly activate the core anaerobic response in arabidopsis thaliana. Plant Sci. 2015, 236, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Gasch, P.; Fundinger, M.; Muller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant erf-vii transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Tan, X.; Hu, C.; Zeng, L.; Lu, G.; Fu, G.; Cheng, Y.; Zhang, X. The transcriptome of Brassica napus L. Roots under waterlogging at the seedling stage. Int. J. Mol. Sci. 2013, 14, 2637–2651. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Hu, C.; Zeng, L.; Cheng, Y.; Xu, M.; Zhang, X. A comparison of screening methods to identify waterlogging tolerance in the field in Brassica napus L. During plant ontogeny. PLoS ONE 2014, 9, e89731. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Kim, K.S.; Jang, Y.S.; Hwang, J.H.; Lee, D.H.; Choi, I.H. Global gene expression responses to waterlogging in leaves of rape seedlings. Plant Cell Rep. 2014, 33, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Marmagne, A.; Brabant, P.; Thiellement, H.; Alix, K. Analysis of gene expression in resynthesized Brassica napus allotetraploids: Transcriptional changes do not explain differential protein regulation. New Phytol. 2010, 186, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Gu, F.; Liu, D.; Yin, C.; Zhao, S.; Chen, H.; Zhang, J.; Yang, C.; Zhan, X.; Zhang, M. Proteomic analysis of elite soybean jidou17 and its parents using itraq-based quantitative approaches. Proteome Sci. 2013, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Han, X.; Geng, C.; Zhao, Y.; Zhang, Z.; Qiu, F. Comparative proteomic analysis revealing the complex network associated with waterlogging stress in maize (Zea mays L.) seedling root cells. Proteomics 2015, 15, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomic 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Jorrin, J.V.; Maldonado, A.M.; Castillejo, M.A. Plant proteome analysis: A 2006 update. Proteomics 2007, 7, 2947–2962. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Sun, Y.; Zhao, X.; Xu, P. Recent advance in high accuracy itraq for quantitative proteomics. Sheng Wu Gong Cheng Xue Bao 2014, 30, 1073–1082. [Google Scholar] [PubMed]
- Xu, X.; Ji, J.; Ma, X.; Xu, Q.; Qi, X.; Chen, X. Comparative proteomic analysis provides insight into the key proteins involved in cucumber (Cucumis sativus L.) adventitious root emergence under waterlogging stress. Front. Plant Sci. 2016, 7, 1515. [Google Scholar] [CrossRef] [PubMed]
- Bandehagh, A.; Salekdeh, G.H.; Toorchi, M.; Mohammadi, A.; Komatsu, S. Comparative proteomic analysis of canola leaves under salinity stress. Proteomics 2011, 11, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Shao, M.; He, Y.; Guan, R.; Chu, P.; Jiang, H. Proteome dynamics and physiological responses to short-term salt stress in Brassica napus leaves. PLoS ONE 2015, 10, e0144808. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhao, Y.; Wang, Y.; Shi, Z.; Zhang, P.; Zhang, Y.; Song, W.; Zhao, J. Comparative proteomics of contrasting maize genotypes provides insights into salt-stress tolerance mechanisms. J. Proteome Res. 2018, 17, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, X.; Niu, F.; Sun, X.; Hu, Z.; Zhang, H. Itraq-based quantitative proteomic analysis of wheat roots in response to salt stress. Proteomics 2017, 17, 1600265. [Google Scholar] [CrossRef] [PubMed]
- Echevarria-Zomeno, S.; Fernandez-Calvino, L.; Castro-Sanz, A.B.; Lopez, J.A.; Vazquez, J.; Castellano, M.M. Dissecting the proteome dynamics of the early heat stress response leading to plant survival or death in arabidopsis. Plant Cell Environ. 2016, 39, 1264–1278. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. Kobas 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, W.; Hashiguchi, A.; Nishimura, M.; Tian, J.; Komatsu, S. Metabolic profiles of flooding-tolerant mechanism in early-stage soybean responding to initial stress. Plant Mol. Biol. 2017, 94, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.P.; Liu, H.; Argueso, C.T.; Pereira, A.; Vera Cruz, C.; Verdier, V.; Leach, J.E. Rna-seq analysis reveals insight into enhanced rice xa7-mediated bacterial blight resistance at high temperature. PLoS ONE 2017, 12, e0187625. [Google Scholar] [CrossRef] [PubMed]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef] [PubMed]
- Valerio, L.; De Meyer, M.; Penel, C.; Dunand, C. Expression analysis of the arabidopsis peroxidase multigenic family. Phytochemistry 2004, 65, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Koiwa, H.; Cushman, M.A.; Ray, A.; Bufford, D.; Kore-eda, S.; Matsumoto, T.K.; Zhu, J.; Cushman, J.C.; Bressan, R.A.; et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001, 126, 363–375. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Zhang, Z.G.; Yan, D.Q.; Zhang, J.S.; Chen, S.Y. A salt-responsive receptor-like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor. Appl. Genet. 2004, 109, 377–383. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, A.; Vuylsteke, M.; Van Hummelen, P.; Zabeau, M.; Van Der Straeten, D. Transcriptional profiling by cdna-aflp and microarray analysis reveals novel insights into the early response to ethylene in arabidopsis. Plant J. 2004, 39, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.; Kim, M.S.; Sicher, R.C.; Bae, H.J.; Bailey, B.A. Necrosis- and ethylene-inducing peptide from fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in arabidopsis. Plant Physiol. 2006, 141, 1056–1067. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, X.; Chen, L.; Li, J. A rapid screening methods to identify waterlogging tolerance in Brassica napus L. Chin. J. Oil Crop sci. 2006, 28, 138–143. [Google Scholar]
- Muller, M.; Munne-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Eysholdt-Derzso, E.; Sauter, M. Hypoxia and the erfvii transcription factor hre2 promote adventitious root elongation in arabidopsis. Plant Biol. (Stuttg.) 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Li, P.F.; Chen, M.K.; Lee, Y.I.; Yang, C.H. Forever young flower negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control arabidopsis floral organ senescence and abscission. Plant Physiol. 2015, 168, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Mase, K.; Ishihama, N.; Mori, H.; Takahashi, H.; Kaminaka, H.; Kodama, M.; Yoshioka, H. Ethylene-responsive ap2/erf transcription factor macd1 participates in phytotoxin-triggered programmed cell death. Mol. Plant Microbe Interact. 2013, 26, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Burgos, M.S.; Messmer, M.M.; Stamp, P.; Schmid, J.E. Flooding tolerance of spelt (Triticum spelta L.) compared to wheat (Triticum aestivum L.)—A physiological and genetic approach. Euphytica 2001, 122, 287–295. [Google Scholar] [CrossRef]
- Hashiguchi, A.; Komatsu, S. Impact of post-translational modifications of crop proteins under abiotic stress. Proteomes 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.G.; Barrett-Wilt, G.A.; Sussman, M.R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 15986–15991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonhomme, L.; Valot, B.; Tardieu, F.; Zivy, M. Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol. Cell. Proteomics 2012, 11, 957–972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lv, D.; Ge, P.; Bian, Y.; Chen, G.; Zhu, G.; Li, X.; Yan, Y. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteomic 2014, 109, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.D.; Zhu, T.; Verstraeten, I.; van de Cotte, B.; International Wheat Genome Sequencing Consortium; Gevaert, K.; De Smet, I. Temperature-induced changes in the wheat phosphoproteome reveal temperature-regulated interconversion of phosphoforms. J. Exp. Bot. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. Tasnrk2.4, an snf1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in arabidopsis. J. Exp. Bot. 2010, 61, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.G. Phosphorylation of rack1 in plants. Plant Signal. Behav. 2015, 10, e1022013. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Hayashi, N.; Kikuchi, S.; Ohsugi, R. Cdpk-mediated abiotic stress signaling. Plant Signal. Behav. 2012, 7, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, W.Z.; Zhang, Y.; Deng, M.; Niu, F.; Yang, B.; Wang, X.; Wang, B.; Liang, W.; Deyholos, M.K.; et al. Identification, expression and interaction analyses of calcium-dependent protein kinase (cpk) genes in canola (Brassica napus L.). BMC Genom. 2014, 15, 211. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Moon, J.H.; Murata, Y.; Kuchitsu, K.; Leonhardt, N.; DeLong, A.; Schroeder, J.I. Disruption of a guard cell-expressed protein phosphatase 2a regulatory subunit, rcn1, confers abscisic acid insensitivity in arabidopsis. Plant Cell 2002, 14, 2849–2861. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.K.; Willmann, R.; Kolb, D.; Grefen, L.; Lajunen, H.M.; Bethke, G.; Lee, J.; Nurnberger, T.; Gust, A.A. The arabidopsis mitogen-activated protein kinase phosphatase pp2c5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 2010, 153, 1098–1111. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants (Basel) 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Munns, R.; Burton, R.A.; Gilliham, M.; Wege, S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018, 269, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Van Sandt, V.S.; Suslov, D.; Verbelen, J.P.; Vissenberg, K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Maris, A.; Kaewthai, N.; Eklof, J.M.; Miller, J.G.; Brumer, H.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Differences in enzymic properties of five recombinant xyloglucan endotransglucosylase/hydrolase (xth) proteins of arabidopsis thaliana. J. Exp. Bot. 2011, 62, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Peschke, V.M.; Sachs, M.M. Characterization and expression of transcripts induced by oxygen deprivation in maize (Zea mays L.). Plant Physiol. 1994, 104, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Song, T.; Chu, M.; Yu, F.; Kumar, S.; Karunakaran, C.; Peng, G. Evaluating changes in cell-wall components associated with clubroot resistance using fourier transform infrared spectroscopy and RT-PCR. Int. J. Mol. Sci. 2017, 18, 2058. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cosgrove, D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000, 51, 1543–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christianson, J.A.; Llewellyn, D.J.; Dennis, E.S.; Wilson, I.W. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.J.; Oyanagi, A.; Komatsu, S. Cell wall proteome of wheat roots under flooding stress using gel-based and lc ms/ms-based proteomics approaches. Biochim. Biophys. Acta 2010, 1804, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, Y.; Nakamura, T.; Komatsu, S. Identification of indicator proteins associated with flooding injury in soybean seedlings using label-free quantitative proteomics. J. Proteome Res. 2013, 12, 4785–4798. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- James, E.K.; Olivares, F.L.; de Oliveira, A.L.; dos Reis, F.B., Jr.; da Silva, L.G.; Reis, V.M. Further observations on the interaction between sugar cane and gluconacetobacter diazotrophicus under laboratory and greenhouse conditions. J. Exp. Bot 2001, 52, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Sheoran, I.S.; Olson, D.J.; Ross, A.R.; Sawhney, V.K. Proteome analysis of embryo and endosperm from germinating tomato seeds. Proteomics 2005, 5, 3752–3764. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.; Lindell, G.; Kallstrom, B.N.; Branca, R.M.; Danielsson, K.G.; Dahlberg, M.; Larson, B.; Forshed, J.; Lehtio, J. Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes. Mol. Cell. Proteomic 2012, 11, M112.016998. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids. Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids. Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Hu, X.N.; Zhang, Y.Q.; Miao, Z.Y.; Xie, C.; Meng, X.Z.; Deng, J.; Wen, J.Q.; Mysore, K.S.; Frugier, F.; et al. Opposing control by transcription factors myb61 and myb3 increases freezing tolerance by relieving c-repeat binding factor suppression. Plant Physiol. 2016, 172, 1306–1323. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Xiao, Z.; Jian, H.; Peng, L.; Qu, C.; Fu, M.; He, B.; Tie, L.; Liang, Y.; Xu, X.; et al. A combination of genome-wide association and transcriptome analysis reveals candidate genes controlling harvest index-related traits in Brassica napus. Sci. Rep. 2016, 6, 36452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-wide identification and structural analysis of bzip transcription factor genes in Brassica napus. Genes (Basel) 2017, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chang, W.; Fan, Y.; Sun, W.; Qu, C.; Zhang, K.; Liu, L.; Xu, X.; Tang, Z.; Li, J.; et al. Genome-wide identification and characterization of nodule-inception-like protein (nlp) family genes in Brassica napus. Int. J. Mol. Sci. 2018, 19, 2270. [Google Scholar] [CrossRef] [PubMed]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Qiao, X.; Tian, Z.; Zhang, X.; Zou, X.; Cheng, Y.; Lu, G.; Zeng, L.; Fu, G.; Ding, X.; et al. Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress. Plants 2018, 7, 71. https://doi.org/10.3390/plants7030071
Xu J, Qiao X, Tian Z, Zhang X, Zou X, Cheng Y, Lu G, Zeng L, Fu G, Ding X, et al. Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress. Plants. 2018; 7(3):71. https://doi.org/10.3390/plants7030071
Chicago/Turabian StyleXu, Jinsong, Xing Qiao, Zhitao Tian, Xuekun Zhang, Xiling Zou, Yong Cheng, Guangyuan Lu, Liu Zeng, Guiping Fu, Xiaoyu Ding, and et al. 2018. "Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress" Plants 7, no. 3: 71. https://doi.org/10.3390/plants7030071



