Genome-Wide Analysis of C-Repeat Binding Factor Gene Family in Capsicum baccatum and Functional Exploration in Low-Temperature Response
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization Analysis of the CBF Genes in C. baccatum
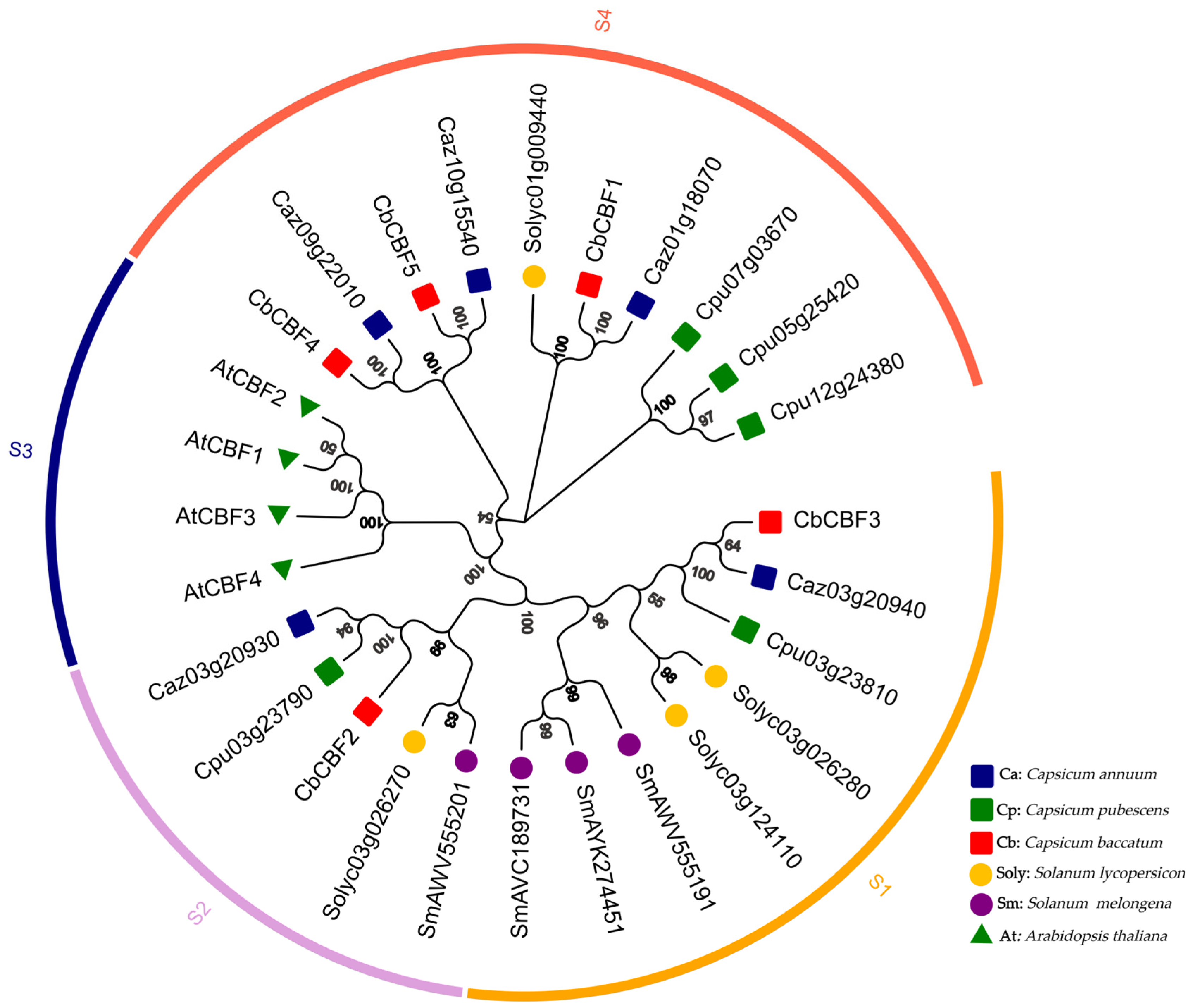
2.2. Evolutionary Analysis of CbCBF Proteins
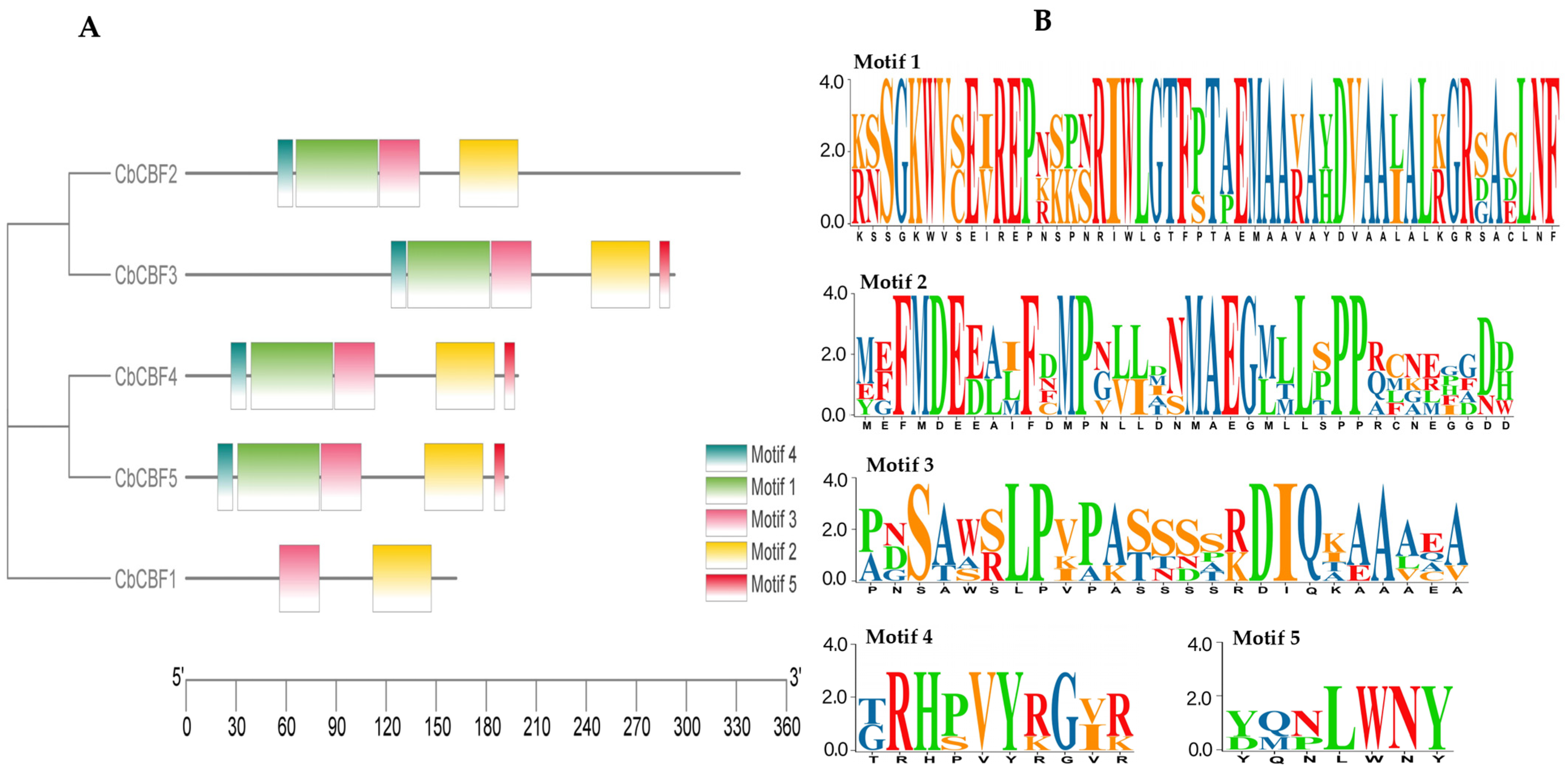
2.3. Conserved Motif Analysis of CbCBF Genes
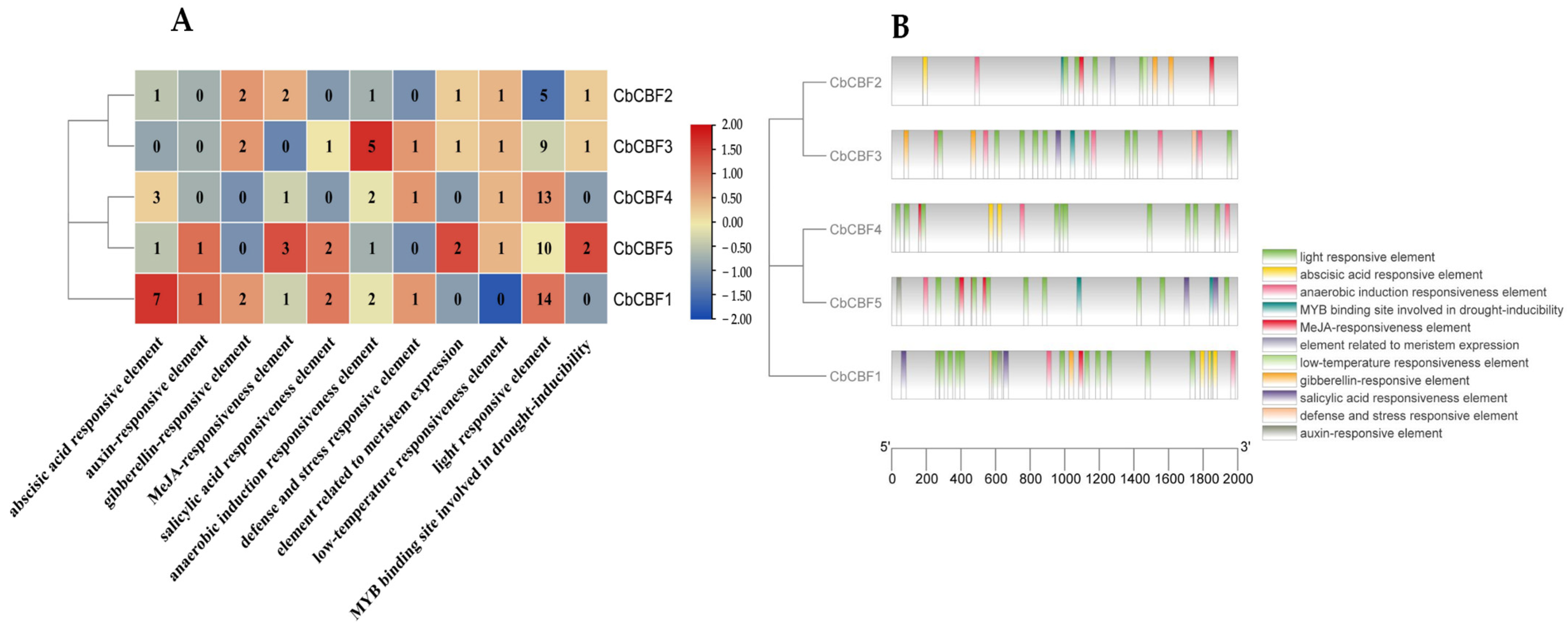
2.4. Cis-Acting Element Analysis of CbCBF Genes
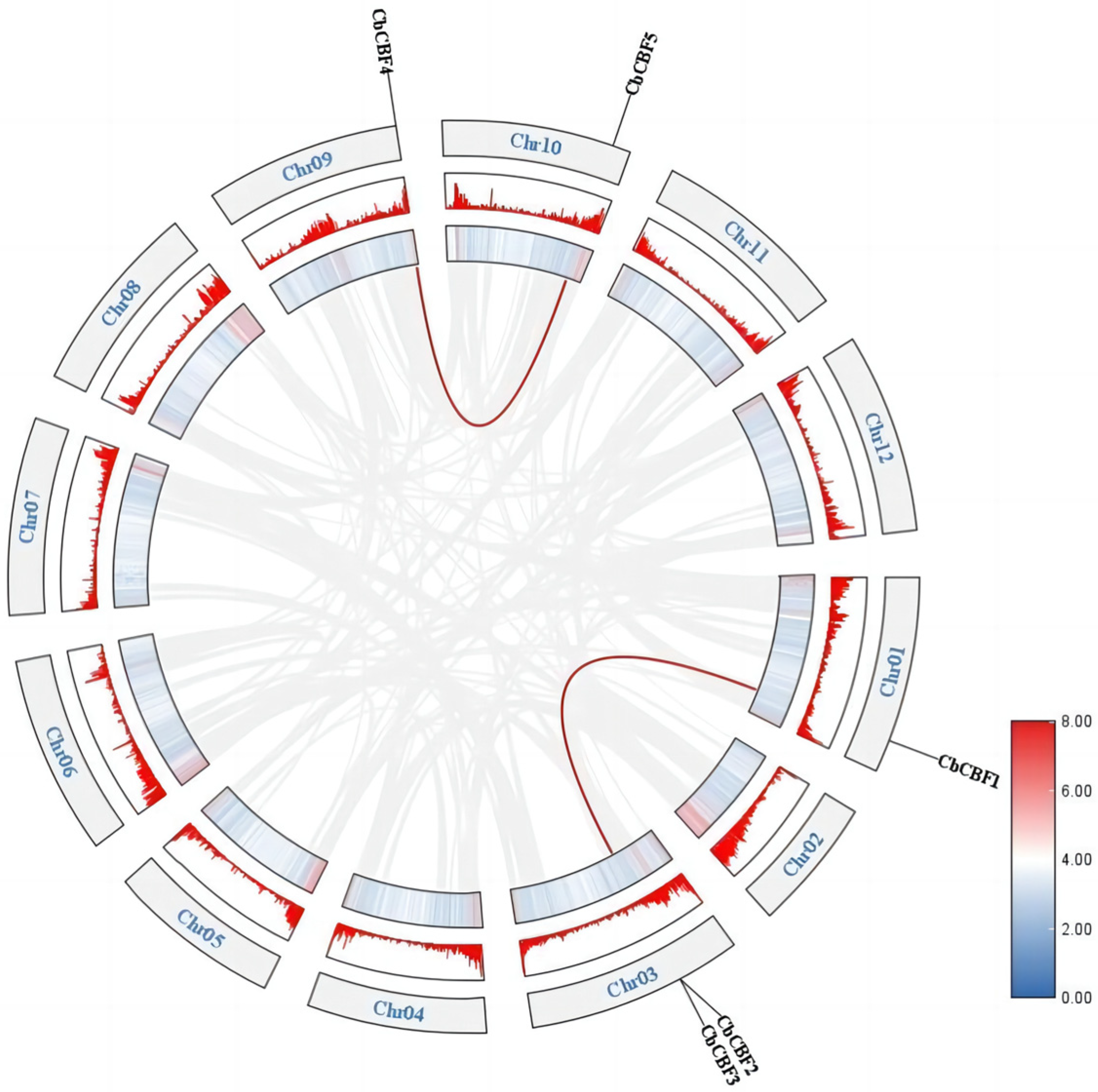
2.5. Collinearity Analysis and Chromosomal Localization of CbCBF Genes
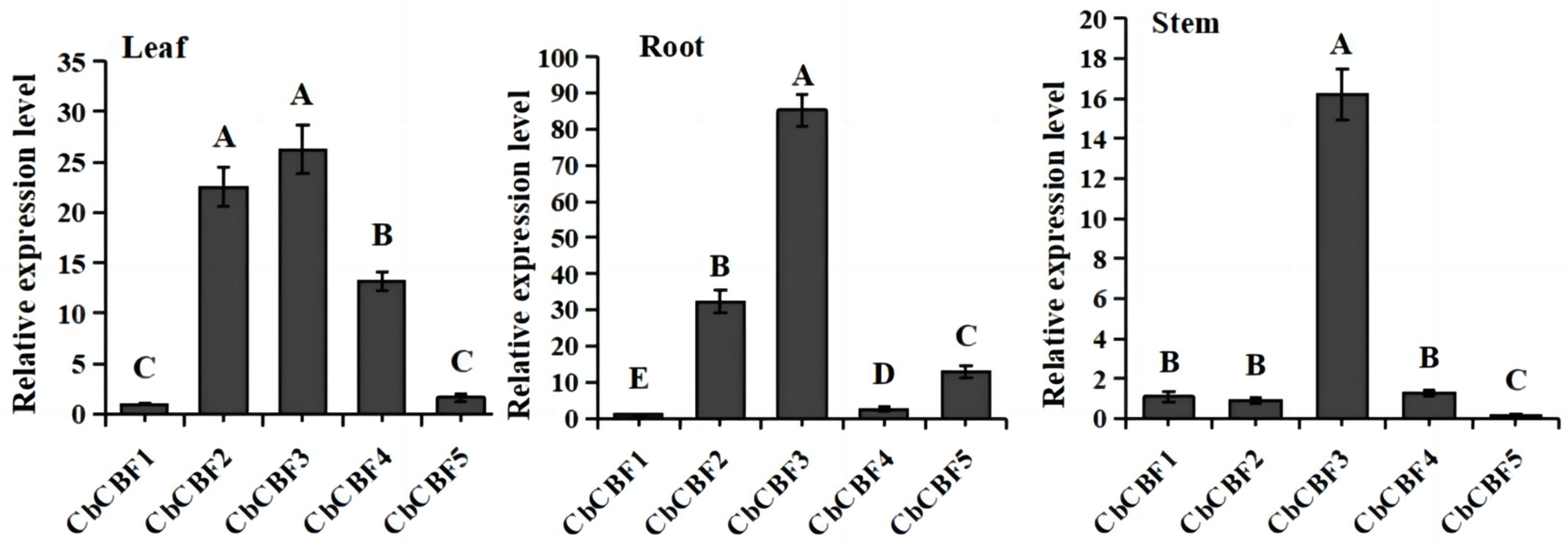
2.6. Tissue-Specific Expression Patterns of CbCBF Genes
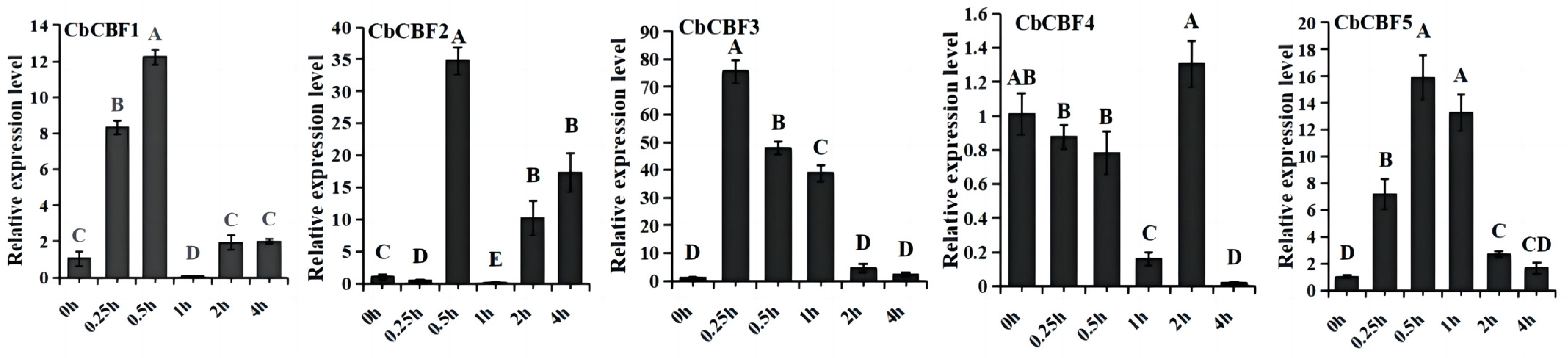
2.7. Expression Patterns of CbCBF Genes in Cold Stress
2.8. Virus-Induced Gene Silencing in C. baccatum
2.9. Silencing the CbCBF3 Gene Reduces Cold Resistance in C. baccatum
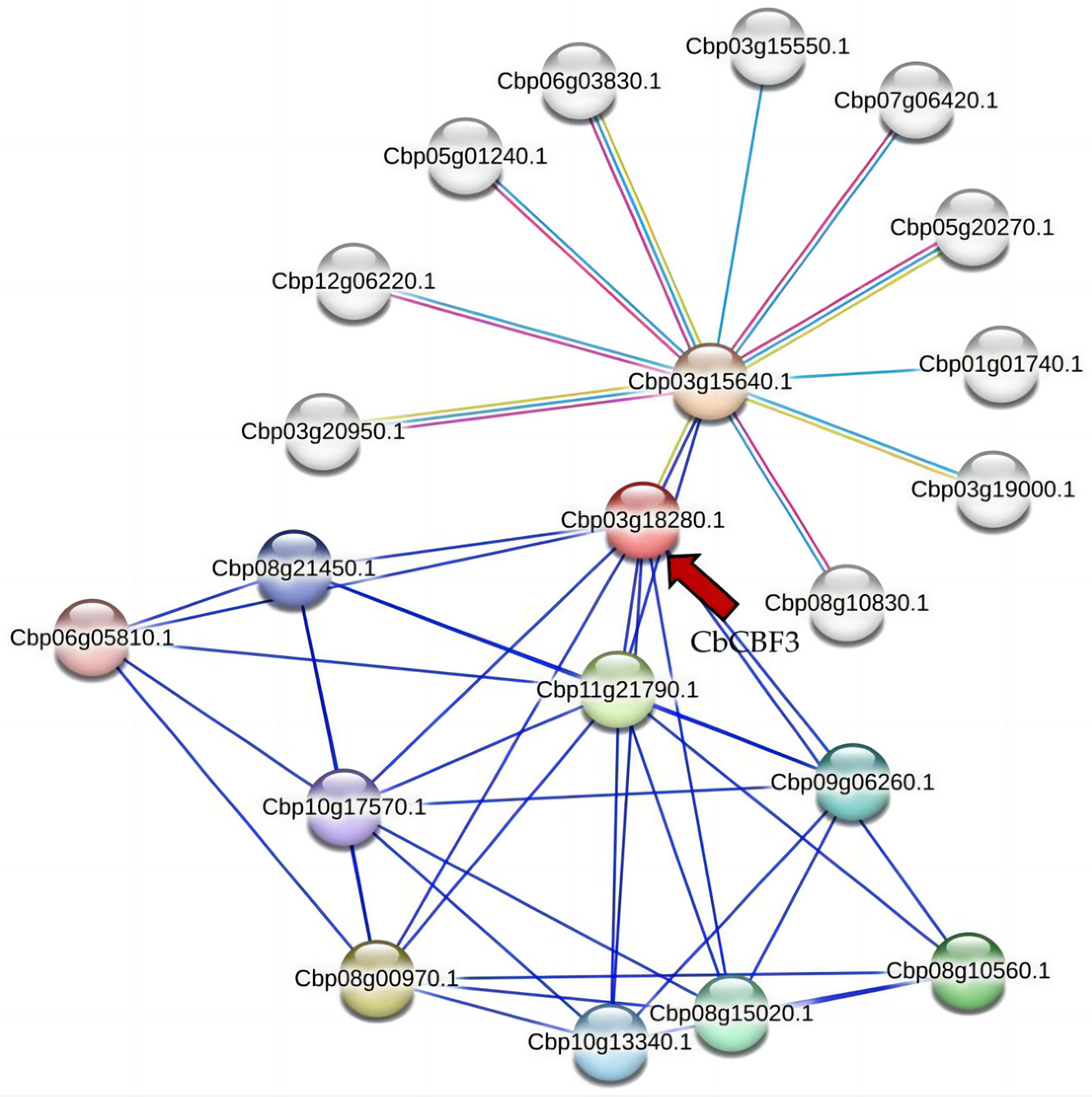
2.10. Protein Interaction Network of CbCBF3 in C. baccatum
3. Discussion
4. Materials and Methods
4.1. Identification of CBF Genes in C. baccatum
4.2. Evolutionary Analysis of CBF Proteins
4.3. Genes Conserved Motifs Analysis
4.4. Cis-Acting Element Prediction
4.5. Collinearity Analysis
4.6. Plant Materials and Treatments
4.7. Functional Analysis of CbCBF3 Based on VIGS
4.8. Virus Vector Construction and qRT-PCR
4.9. Physiological Parameter Determination
4.10. Interaction Network Analysis of CbCBF3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A. Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. In Survival Strategies in Extreme Cold Desiccation: Adaptation Mechanisms Their Applications; Springer: Berlin/Heidelberg, Germany, 2018; pp. 215–232. [Google Scholar] [CrossRef]
- Song, Y.; Feng, L.; Alyafei, M.A.M.; Jaleel, A.; Ren, M. Function of Chloroplasts in Plant Stress Responses. Int. J. Mol. Sci. 2021, 22, 13464. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Hosseinifard, M.; Stefaniak, S.; Ghorbani Javid, M.; Soltani, E.; Wojtyla, Ł.; Garnczarska, M. Contribution of Exogenous Proline to Abiotic Stresses Tolerance in Plants: A Review. Int. J. Mol. Sci. 2022, 23, 5186. [Google Scholar] [CrossRef]
- Eric, R.; Marie Noelle, V.; Alain, Z.; Vaughan, H. Chapter 2 Cold Signalling and Cold Acclimation in Plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar] [CrossRef]
- Abdullah, S.N.A.; Azzeme, A.M.; Yousefi, K. Fine-Tuning Cold Stress Response Through Regulated Cellular Abundance and Mechanistic Actions of Transcription Factors. Front. Plant Sci. 2022, 13, 850216. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, H.; Liu, Z.; Zhai, H.; Liu, Q. The sweet potato transcription factor IbbHLH33 enhances chilling tolerance in transgenic tobacco. Czech J. Genet. Plant Breed. 2022, 58, 210–222. [Google Scholar] [CrossRef]
- Han, J.; Li, X.; Li, W.; Yao, A.; Niu, C.; Hou, R.; Liu, W.; Wang, Y.; Zhang, L.; Han, D. Overexpression of Malus baccata WRKY40 (MbWRKY40) enhances stress tolerance in Arabidopsis subjected to cold and drought. Plant Stress 2023, 10, 100209. [Google Scholar] [CrossRef]
- Zhang, S.; Cao, K.; Wei, Y.; Jiang, S.; Ye, J.; Xu, F.; Chen, Y.; Shao, X. PpBZR1, a BES/BZR transcription factor, enhances cold stress tolerance by suppressing sucrose degradation in peach fruit. Res. Sq. 2023, 202, 107972. [Google Scholar] [CrossRef]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-Repeat/Dehydration-Responsive Element Binding Factor Cold-Response Pathway Are Conserved in Brassica napus and Other Plant Species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef]
- Medina, J.; Bargues, M.; Terol, J.; Pérez-Alonso, M.; Salinas, J. The Arabidopsis CBF Gene Family Is Composed of Three Genes Encoding AP2 Domain-Containing Proteins Whose Expression Is Regulated by Low Temperature but Not by Abscisic Acid or Dehydration. Plant Physiol. 1999, 119, 463–470. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Li, J.; Liu, Q.; Li, Y.-Q.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Cloning and Functional Analysis of a Novel DREB1/CBF Transcription Factor Involved in Cold-Responsive Gene Expression in Zea mays L. Plant Cell Physiol. 2004, 45, 1042–1052. [Google Scholar] [CrossRef]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004, 39, 905–919. [Google Scholar] [CrossRef]
- Fang, X.; Lin, Y.; Chen, C.; Pervaiz, T.; Wang, X.; Luo, H.; Fang, J.; Shangguan, L. Whole genome identification of CBF gene families and expression analysis in Vitis vinifera L. Czech J. Genet. Plant Breed. 2023, 59, 119–132. [Google Scholar] [CrossRef]
- Hu, Z.; Ban, Q.; Hao, J.; Zhu, X.; Cheng, Y.; Mao, J.; Lin, M.; Xia, E.; Li, Y. Genome-Wide Characterization of the C-repeat Binding Factor (CBF) Gene Family Involved in the Response to Abiotic Stresses in Tea Plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 921. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, J.; Wang, R.; Liu, W.; Chen, S.; Wang, Y.; Yu, Y.; Qu, G.; Chen, S. Genome-Wide Identification and Expression Profiles of C-Repeat Binding Factor Transcription Factors in Betula platyphylla under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 10573. [Google Scholar] [CrossRef]
- Than, P.P.; Prihastuti, H.; Phoulivong, S.; Taylor, P.W.; Hyde, K.D. Chilli anthracnose disease caused by Colletotrichum species. J. Zhejiang Univ. Sci. B 2008, 9, 764–778. [Google Scholar] [CrossRef]
- Kim, H.-G.; Bae, J.-H.; Jastrzebski, Z.; Cherkas, A.; Heo, B.-G.; Gorinstein, S.; Ku, Y.-G. Binding, Antioxidant and Anti-proliferative Properties of Bioactive Compounds of Sweet Paprika (Capsicum annuum L.). Plant Foods Hum. Nutr. 2016, 71, 129–136. [Google Scholar] [CrossRef]
- Surh, Y.-J. More Than Spice: Capsaicin in Hot Chili Peppers Makes Tumor Cells Commit Suicide. J. Natl. Cancer Inst. 2002, 94, 1263–1265. [Google Scholar] [CrossRef]
- Perry, L.; Dickau, R.; Zarrillo, S.; Holst, I.; Pearsall, D.M.; Piperno, D.R.; Berman, M.J.; Cooke, R.G.; Rademaker, K.; Ranere, A.J. Starch Fossils and the Domestication and Dispersal of Chili Peppers (Capsicum spp. L.) in the Americas. Science 2007, 315, 986–988. [Google Scholar] [CrossRef]
- Kraft, K.H.; Brown, C.H.; Nabhan, G.P.; Luedeling, E.; Luna Ruiz, J.d.J.; Coppens d’Eeckenbrugge, G.; Hijmans, R.J.; Gepts, P. Multiple lines of evidence for the origin of domesticated chili pepper, Capsicum annuum, in Mexico. Proc. Natl. Acad. Sci. USA 2014, 111, 6165–6170. [Google Scholar] [CrossRef]
- Barboza, G.E.; Carrizo García, C.; Leiva González, S.; Scaldaferro, M.; Reyes, X. Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS ONE 2019, 14, e0209792. [Google Scholar] [CrossRef]
- Dias, G.; Gomes, V.; Moraes, T.; Zottich, U.; Rabelo, G.; Carvalho, A.; Moulin, M.; Gonçalves, L.; Rodrigues, R.; Da Cunha, M. Characterization of Capsicum species using anatomical and molecular data. Genet. Mol. Res. 2013, 12, 6488–6501. [Google Scholar] [CrossRef] [PubMed]
- Barboza, G.E.; García, C.C.; de Bem Bianchetti, L.; Romero, M.V.; Scaldaferro, M. Monograph of wild and cultivated chili peppers (Capsicum L., Solanaceae). PhytoKeys 2022, 200, 1. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, E.; Zhang, D.; Mays, A.D.; Saftner, R.A.; Stommel, J.R. Genetic diversity in Capsicum baccatumis significantly influenced by its ecogeographical distribution. BMC Genet. 2012, 13, 68. [Google Scholar] [CrossRef]
- Momo, J.; Kumar, A.; Islam, K.; Ahmad, I.; Rawoof, A.; Ramchiary, N. A comprehensive update on Capsicum proteomics: Advances and future prospects. J. Proteom. 2022, 261, 104578. [Google Scholar] [CrossRef]
- Meghvansi, M.; Siddiqui, S.; Khan, M.H.; Gupta, V.; Vairale, M.; Gogoi, H.; Singh, L. Naga chilli: A potential source of capsaicinoids with broad-spectrum ethnopharmacological applications. J. Ethnopharmacol. 2010, 132, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Saleh, B.K.; Omer, A.; Teweldemedhin, B. Medicinal uses and health benefits of chili pepper (Capsicum spp.): A review. MOJ Food Process. Technol. 2018, 6, 325–328. [Google Scholar] [CrossRef]
- Govindarajan, V.; Salzer, U.J. Capsicum—production, technology, chemistry, and quality—part II. Processed products, standards, world production and trade. Crit. Rev. Food Sci. Nutr. 1986, 23, 207–288. [Google Scholar] [CrossRef]
- Zimmer, A.R.; Leonardi, B.; Miron, D.; Schapoval, E.; de Oliveira, J.R.; Gosmann, G. Antioxidant and anti-inflammatory properties of Capsicum baccatum: From traditional use to scientific approach. J. Ethnopharmacol. 2012, 139, 228–233. [Google Scholar] [CrossRef]
- Rodríguez-Burruezo, A.; Prohens, J.; Raigón, M.D.; Nuez, F. Variation for bioactive compounds in ají (Capsicum baccatum L.) and rocoto (C. pubescens R. & P.) and implications for breeding. Euphytica 2009, 170, 169–181. [Google Scholar] [CrossRef]
- Mahasuk, P.; Taylor, P.; Mongkolporn, O. Identification of Two New Genes Conferring Resistance to Colletotrichum acutatum in Capsicum baccatum. Phytopathology 2009, 99, 1100–1104. [Google Scholar] [CrossRef]
- Yoon, J.B.; Park, H.G. Trispecies bridge crosses,(Capsicum annuum× C. chinense) × C. baccatum, as an alternative for introgression of anthracnose resistance from C. baccatum into C. annuum. Hortic. Environ. Biotechnol. 2005, 46, 5–9. [Google Scholar]
- Manzur, J.P.; Fita, A.; Prohens, J.; Rodríguez-Burruezo, A. Successful Wide Hybridization and Introgression Breeding in a Diverse Set of Common Peppers (Capsicum annuum) Using Different Cultivated Ají (C. baccatum) Accessions as Donor Parents. PLoS ONE 2015, 10, e0144142. [Google Scholar] [CrossRef]
- Aune, D.; Giovannucci, E.; Boffetta, P.; Fadnes, L.T.; Keum, N.; Norat, T.; Greenwood, D.C.; Riboli, E.; Vatten, L.J.; Tonstad, S. Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality-a systematic review and dose-response meta-analysis of prospective studies. Int. J. Epidemiol. 2017, 46, 1029–1056. [Google Scholar] [CrossRef]
- Oyebode, O.; Gordon-Dseagu, V.; Walker, A.; Mindell, J.S. Fruit and vegetable consumption and all-cause, cancer and CVD mortality: Analysis of Health Survey for England data. Epidemiol. Community Health 2014, 68, 856–862. [Google Scholar] [CrossRef]
- Hu, D.; Huang, J.; Wang, Y.; Zhang, D.; Qu, Y. Fruits and Vegetables Consumption and Risk of Stroke. Stroke 2014, 45, 1613–1619. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.; Visser, R.G.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- López, P.; Gorzalczany, S.; Acevedo, C.; Alonso, R.; Ferraro, G. Chemical study and anti-inflammatory activity of Capsicum chacoense and C. baccatum. Rev. Bras. Farmacogn. 2012, 22, 455–458. [Google Scholar] [CrossRef]
- Guillen, N.G.; Tito, R.; Mendoza, N.G. Capsaicinoids and pungency in Capsicum chinense and Capsicum baccatum fruits. Pesqui. Agropecuária Trop. 2018, 48, 237–244. [Google Scholar] [CrossRef]
- Albrecht, E.; Zhang, D.; Saftner, R.A.; Stommel, J.R. Genetic diversity and population structure of Capsicum baccatum genetic resources. Genet. Resour. Crop Evol. 2012, 59, 517–538. [Google Scholar] [CrossRef]
- Rácz, A.; Czégény, G.; Kutyáncsánin, D.; Nagy, N.; Hideg, É.; Csepregi, K. Fight against cold: Photosynthetic and antioxidant responses of different bell pepper cultivars (Capsicum annuum L.) to cold stress. Biol. Futur. 2023, 74, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Leite, P.; Rodrigues, R.; Silva, R.; Pimenta, S.; Medeiros, A.; Bento, C.; Gonçalves, L. Molecular and agronomic analysis of intraspecific variability in Capsicum baccatum var. pendulum accessions. Genet. Mol. Res. 2016, 15, 1–16. [Google Scholar] [CrossRef]
- Hussein, H.; Mekki, B.; Abd El-Sadek, M.; El Lateef, E. Effect of L-Ornithine application on improving drought tolerance in sugar beet plants. Heliyon 2019, 5, e02631. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.C.; Yeap, W.C.; Appleton, D.R.; Ho, C.-L.; Kulaveerasingam, H. Identification of drought responsive Elaeis guineensis WRKY transcription factors with sensitivity to other abiotic stresses and hormone treatments. BMC Genom. 2022, 23, 164. [Google Scholar] [CrossRef]
- Goyal, P.; Manzoor, M.M.; Vishwakarma, R.A.; Sharma, D.; Dhar, M.K.; Gupta, S. A Comprehensive Transcriptome-Wide Identification and Screening of WRKY Gene Family Engaged in Abiotic Stress in Glycyrrhiza glabra. Sci. Rep. 2020, 10, 373. [Google Scholar] [CrossRef]
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Wang, H.; Xing, H.; Zhao, J.; Guo, N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae. BMC Plant Biol. 2019, 19, 598. [Google Scholar] [CrossRef]
- Barrero-Gil, J.; Salinas, J. Gene Regulatory Networks Mediating Cold Acclimation: The CBF Pathway. In Survival Strategies in Extreme Cold and Desiccation; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–22. [Google Scholar] [CrossRef]
- Kothari, S.; Joshi, A.; Kachhwaha, S.; Ochoa-Alejo, N. Chilli peppers—A review on tissue culture and transgenesis. Biotechnol. Adv. 2010, 28, 35–48. [Google Scholar] [CrossRef]
- Li, W.; Chen, Y.; Ye, M.; Lu, H.; Wang, D.; Chen, Q. Evolutionary history of the C-repeat binding factor/dehydration-responsive element-binding 1 (CBF/DREB1) protein family in 43 plant species and characterization of CBF/DREB1 proteins in Solanum tuberosum. BMC Evol. Biol. 2020, 20, 142. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Quan, X.; Shan, Q.; Wang, W.; Yin, N.; Wang, S.; Wang, Z.; He, W. Comprehensive analysis of cucumber C-repeat/dehydration-responsive element binding factor family genes and their potential roles in cold tolerance of cucumber. BMC Plant Biol. 2022, 22, 270. [Google Scholar] [CrossRef]
- Zhou, H.; Ma, J.; Liu, H.; Zhao, P. Genome-Wide Identification of the CBF Gene Family and ICE Transcription Factors in Walnuts and Expression Profiles under Cold Conditions. Int. J. Mol. Sci. 2023, 25, 25. [Google Scholar] [CrossRef]
- Liu, J.; Magwanga, R.O.; Xu, Y.; Wei, T.; Kirungu, J.N.; Zheng, J.; Hou, Y.; Wang, Y.; Agong, S.G.; Okuto, E. Functional Characterization of Cotton C-Repeat Binding Factor Genes Reveal Their Potential Role in Cold Stress Tolerance. Front. Plant Sci. 2021, 12, 766130. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef]
- Mohseni, S.; Che, H.; Djillali, Z.; Dumont, E.; Nankeu, J.; Danyluk, J. Wheat CBF gene family: Identification of polymorphisms in the CBF coding sequence. Genome 2012, 55, 865–881. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription Factor CBF4 Is a Regulator of Drought Adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Guo, L.; Cui, F.; Shen, Y.; Ye, X.; Deng, D.; Wang, S.; Zhu, J.; Wu, W. Innovations and stepwise evolution of CBFs/DREB1s and their regulatory networks in angiosperms. J. Integr. Plant Biol. 2022, 64, 2111–2125. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Baulcombe, D.C. Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 1999, 2, 109–113. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; Della-Cioppa, G.; Harvey, D.; Hanley, K.; Grill, L. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef]
- Li, H.; Zhang, D.; Xie, K.; Wang, Y.; Liao, Q.; Hong, Y.; Liu, Y. Efficient and high-throughput pseudorecombinant-chimeric Cucumber mosaic virus-based VIGS in maize. Plant Physiol. 2021, 187, 2865–2876. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Chakravarthy, S.; Martin, G.B. Virus-induced Gene Silencing (VIGS) in Nicotiana benthamiana and Tomato. J. Vis. Exp. 2009, e1292. [Google Scholar] [CrossRef]
- Qi, X.; Mo, Q.; Li, J.; Zi, Z.; Xu, M.; Yue, S.; Zhao, H.; Zhu, H.; Wang, G. Establishment of virus-induced gene silencing (VIGS) system in Luffa acutangula using Phytoene desaturase (PDS) and tendril synthesis related gene (TEN). Plant Methods 2023, 19, 94. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Kang, J.-y.; Choi, H.-i.; Im, M.-y.; Kim, S.Y. Arabidopsis Basic Leucine Zipper Proteins That Mediate Stress-Responsive Abscisic Acid Signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Zarka, D.G.; Okamoto, H.; Thomashow, M.F.; Knight, M.R. Abscisic Acid Induces CBF Gene Transcription and Subsequent Induction of Cold-Regulated Genes via the CRT Promoter Element. Plant Physiol. 2004, 135, 1710–1717. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 Kinase Modulates Freezing Tolerance by Enhancing ICE1 Stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Gao, C.; Mumtaz, M.A.; Zhou, Y.; Yang, Z.; Shu, H.; Zhu, J.; Bao, W.; Cheng, S.; Yin, L.; Huang, J. Integrated Transcriptomic and Metabolomic Analyses of Cold-Tolerant and Cold-Sensitive Pepper Species Reveal Key Genes and Essential Metabolic Pathways Involved in Response to Cold Stress. Int. J. Mol. Sci. 2022, 23, 6683. [Google Scholar] [CrossRef]
- Yang, Y.; Guang, Y.; Wang, F.; Chen, Y.; Yang, W.; Xiao, X.; Luo, S.; Zhou, Y. Characterization of Phytochrome-Interacting Factor Genes in Pepper and Functional Analysis of CaPIF8 in Cold and Salt Stress. Front. Plant Sci. 2021, 12, 746517. [Google Scholar] [CrossRef]
- Wang, L.; Shen, X.; Chen, X.; Ouyang, Q.; Tan, X.; Tao, N. Exogenous Application of Melatonin to Green Horn Pepper Fruit Reduces Chilling Injury during Postharvest Cold Storage by Regulating Enzymatic Activities in the Antioxidant System. Plants 2022, 11, 2367. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Harmut, A. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–383. [Google Scholar] [CrossRef]
- Tikoria, R.; Kaur, A.; Ohri, P. Physiological, biochemical and structural changes in tomato plants by vermicompost application in different exposure periods under glass house conditions. Plant Physiol. Biochem. 2023, 197, 107656. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| CbCBF1 | Cbp01g28680 | 162 | 18,402.47 | 4.63 | 55.57 | −0.701 | Nucleus |
| CbCBF2 | Cbp03g18270 | 332 | 36,252.64 | 5.96 | 62.57 | −0.534 | Cytoplasm, Nucleus |
| CbCBF3 | Cbp03g18280 | 293 | 32,782.27 | 5.03 | 56.14 | −0.33 | Cytoplasm. |
| CbCBF4 | Cbp09g26670 | 199 | 21,643.08 | 5.4 | 39.53 | −0.476 | Cytoplasm, Nucleus |
| CbCBF5 | Cbp10g16750 | 193 | 20,203.37 | 5.68 | 50.39 | −0.462 | Nucleus |
| Primer Name | Gene ID | Forward Primer Sequence (5′ –> 3′) | Reverse Primer Sequence (5′ –> 3′) |
|---|---|---|---|
| CbCBF1 | Cbp01g28680 | CAATTTAAGAGGAGGGCAGCTC | TGAAGAGCCGCGATTTGGAT |
| CbCBF2 | Cbp03g18270 | ATGGCGGAAGGGCTAATGTT | TACCGCTTCCGGGACAAAC |
| CbCBF3 | Cbp03g18280 | TCCCTACTGCTGAAATGGCG | ACTCTGATGGTCGGAAAGCC |
| CbCBF4 | Cbp09g26670 | ATGCGGATCTCAACTTCCCG | ACTGCTTCTTTACGTGCGGA |
| CbCBF5 | Cbp10g16750 | AGGCTGGAAATATGGCTGGG | TGCAGGAAGGTTGAGACGTG |
| PDS | Cbp05g00330 | GGCTAAGGATTTCCGGCCTT | GACAAACCACCCAAACCTGC |
| Actin | Cbp03g21510 | GGTCGGAATGGGACAGAAGG | GGTGCCTCCGTTAGGAGAAC |
| TRV | None | TGGGAGATGATACGCTGTT | CCTAAAACTTCAGACACG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Cai, Q.; Luo, L.; Sun, Z.; Li, L. Genome-Wide Analysis of C-Repeat Binding Factor Gene Family in Capsicum baccatum and Functional Exploration in Low-Temperature Response. Plants 2024, 13, 549. https://doi.org/10.3390/plants13040549
Yang Y, Cai Q, Luo L, Sun Z, Li L. Genome-Wide Analysis of C-Repeat Binding Factor Gene Family in Capsicum baccatum and Functional Exploration in Low-Temperature Response. Plants. 2024; 13(4):549. https://doi.org/10.3390/plants13040549
Chicago/Turabian StyleYang, Yanbo, Qihang Cai, Li Luo, Zhenghai Sun, and Liping Li. 2024. "Genome-Wide Analysis of C-Repeat Binding Factor Gene Family in Capsicum baccatum and Functional Exploration in Low-Temperature Response" Plants 13, no. 4: 549. https://doi.org/10.3390/plants13040549




