Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants
Abstract
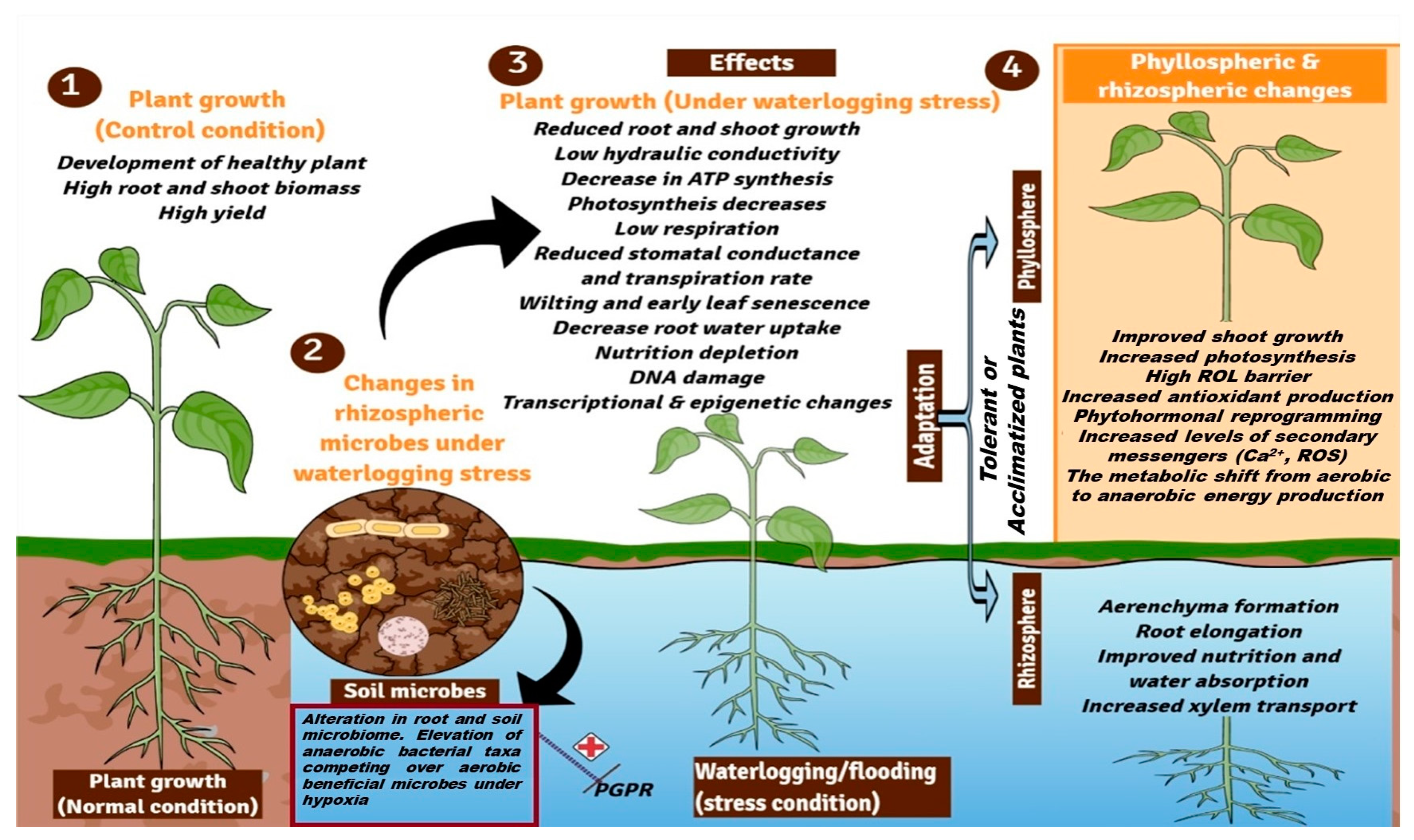
:1. Introduction
2. Flooding Affects Diverse Crop Traits
3. Adaptive Responses of Plants against Waterlogging Stress
4. Waterlogging-Mediated Signaling Mechanism in Plants
5. Strategies for Improving Waterlogging Tolerance in Plants: Past, Present, and Future
5.1. Past: Classical Breeding and Genetic Engineering Approaches Used for Waterlogging Tolerance in Plants
5.2. Present: Omics Approaches for Understanding Waterlogging Tolerance in Plants
5.3. Transcriptional, Metabolic, and Translational Profiling under Waterlogging Stress in Plants
5.4. Future: Integrated Omics and Panomics for Waterlogging Tolerance in Plants
6. Role of High-Throughput Phenotyping Tool in Waterlogging Stress
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voesenek, L.A.C.J.; Bailey-Serres, J. Flooding Tolerance: O2 Sensing and Survival Strategies. Curr. Opin. Plant Biol. 2013, 16, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Westra, S.; Fowler, H.J.; Evans, J.P.; Alexander, L.V.; Berg, P.; Johnson, F.; Kendon, E.J.; Lenderink, G.; Roberts, N. Future Changes to the Intensity and Frequency of Short-duration Extreme Rainfall. Rev. Geophys. 2014, 52, 522–555. [Google Scholar] [CrossRef] [Green Version]
- Masson-Delmotte, V.; Zhai, P.; Pörtner, H.-O.; Roberts, D.; Skea, J.; Shukla, P.R.; Pirani, A.; Moufouma-Okia, W.; Péan, C.; Pidcock, R. Global Warming of 1.5 °C; Cambridge University Press: Cambridge, UK, 2018; Volume 1. [Google Scholar]
- FAO. The Future of Food and Agriculture: Trends and Challenges; Food and Agriculture Organization of the United Nations Rome: Rome, Italy, 2017; Volume 4. [Google Scholar]
- Shabala, S. Physiological and Cellular Aspects of Phytotoxicity Tolerance in Plants: The Role of Membrane Transporters and Implications for Crop Breeding for Waterlogging Tolerance. New Phytol. 2011, 190, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Voesenek, L.A.C.J.; Sasidharan, R. Ethylene-and Oxygen Signalling-Drive Plant Survival during Flooding. Plant Biol. 2013, 15, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, J.; Salhab, M.; Jafino, B.A. Flood Exposure and Poverty in 188 Countries. Nat. Commun. 2022, 13, 3527. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making Sense of Low Oxygen Sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Peña-Castro, J.M. Submergence and Waterlogging Stress in Plants: A Review Highlighting Research Opportunities and Understudied Aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef]
- Yohan, Y. Influence of Waterlogging on Certain Biochemical and Yield Parameters of Pigeonpea (Cajanus cajan (L.) Millsp). Int. J. Pure Appl. Biosci. 2017, 5, 1862–1868. [Google Scholar] [CrossRef]
- Talukdar, D.; Sinjushin, A. Cytogenomics and Mutagenomics in Plant Functional Biology and Breeding. In PlantOmics: The Omics of Plant Science; Springer: New Delhi, India, 2015; pp. 113–156. [Google Scholar]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating Omic Approaches for Abiotic Stress Tolerance in Soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef]
- Shah, T.; Xu, J.; Zou, X.; Cheng, Y.; Nasir, M.; Zhang, X. Omics Approaches for Engineering Wheat Production under Abiotic Stresses. Int. J. Mol. Sci. 2018, 19, 2390. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Tyagi, A.; Bae, H. Ionomic Approaches for Discovery of Novel Stress-resilient Genes in Plants. Int. J. Mol. Sci. 2021, 22, 7182. [Google Scholar] [CrossRef]
- Yadav, C.B.; Pandey, G.; Muthamilarasan, M.; Prasad, M. Epigenetics and Epigenomics of Plants. Plant Genet. Mol. Biol. 2018, 164, 237–261. [Google Scholar]
- Jogaiah, S.; Govind, S.R.; Tran, L.S.P. Systems Biology-Based Approaches toward Understanding Drought Tolerance in Food Crops. Crit. Rev. Biotechnol. 2013, 33, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Luo, J. Metabolite-Based Genome-Wide Association Studies in Plants. Curr. Opin. Plant Biol. 2015, 24, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Nakabayashi, R.; Yang, Z.; Okazaki, Y.; Yonemaru, J.I.; Ebana, K.; Yano, M.; Saito, K. Metabolome-Genome-Wide Association Study Dissects Genetic Architecture for Generating Natural Variation in Rice Secondary Metabolism. Plant J. 2015, 81, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS Meets Germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525. [Google Scholar] [CrossRef] [Green Version]
- Wen, X. Bayesian Model Selection in Complex Linear Systems, as Illustrated in Genetic Association Studies. Biometrics 2014, 70, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Dhanapal, A.P.; Ray, J.D.; Singh, S.K.; Hoyos-Villegas, V.; Smith, J.R.; Purcell, L.C.; King, C.A.; Fritschi, F.B. Genome-Wide Association Analysis of Diverse Soybean Genotypes Reveals Novel Markers for Nitrogen Traits. Plant Genome 2015, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Weckwerth, W. Green Systems Biology-From Single Genomes, Proteomes and Metabolomes to Ecosystems Research and Biotechnology. J. Proteom. 2011, 75, 284–305. [Google Scholar] [CrossRef] [Green Version]
- Weckwerth, W. Toward a Unification of System-Theoretical Principles in Biology and Ecology—The Stochastic Lyapunov Matrix Equation and Its Inverse Application. Front. Appl. Math. Stat. 2019, 5, 29. [Google Scholar] [CrossRef]
- Xu, X.; Wang, H.; Qi, X.; Xu, Q.; Chen, X. Waterlogging-Induced Increase in Fermentation and Related Gene Expression in the Root of Cucumber (Cucumis sativus L.). Sci. Hortic. 2014, 179, 388–395. [Google Scholar] [CrossRef]
- Zhang, P.; Lyu, D.; Jia, L.; He, J.; Qin, S. Physiological and de Novo Transcriptome Analysis of the Fermentation Mechanism of Cerasus Sachalinensis Roots in Response to Short-Term Waterlogging. BMC Genom. 2017, 18, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camisón, Á.; Ángela Martín, M.; Dorado, F.J.; Moreno, G.; Solla, A. Changes in Carbohydrates Induced by Drought and Waterlogging in Castanea sativa. Trees-Struct. Funct. 2020, 34, 579–591. [Google Scholar] [CrossRef]
- Jaeger, C.; Gessler, A.; Biller, S.; Rennenberg, H.; Kreuzwieser, J. Differences in C Metabolism of Ash Species and Provenances as a Consequence of Root Oxygen Deprivation by Waterlogging. J. Exp. Bot. 2009, 60, 4335–4345. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; López, R.; Venturas, M.; Pita, P.; Gordaliza, G.G.; Gil, L.; Rodríguez-Calcerrada, J. Greater Resistance to Flooding of Seedlings of Ulmus Laevis than Ulmus Minor Is Related to the Maintenance of a More Positive Carbon Balance. Trees 2015, 29, 835–848. [Google Scholar] [CrossRef]
- Kozlowski, T.T. Responses of Woody Plants to Flooding and Salinity. Tree Physiol. 1997, 17, 490. [Google Scholar] [CrossRef]
- Shukla, V.; Lombardi, L.; Iacopino, S.; Pencik, A.; Novak, O.; Perata, P.; Giuntoli, B.; Licausi, F. Endogenous Hypoxia in Lateral Root Primordia Controls Root Architecture by Antagonizing Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 538–551. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.B. Long-Distance Signalling from Roots to Shoots Assessed: The Flooding Story. J. Exp. Bot. 2002, 53, 175–181. [Google Scholar] [CrossRef]
- Limami, A.M. Adaptations of Nitrogen Metabolism to Oxygen Deprivation in Plants. Plant Cell Monogr. 2014, 21, 209–221. [Google Scholar] [CrossRef]
- Emdadul Haque, M.; Kawaguchi, K.; Komatsu, S. Analysis of Proteins in Aerenchymatous Seminal Roots of Wheat Grown in Hypoxic Soils under Waterlogged Conditions. Protein Pept. Lett. 2011, 18, 912–924. [Google Scholar] [CrossRef]
- Dawood, T.; Yang, X.; Visser, E.J.W.; Te Beek, T.A.H.; Kensche, P.R.; Cristescu, S.M.; Lee, S.; Floková, K.; Nguyen, D.; Mariani, C.; et al. A Co-Opted Hormonal Cascade Activates Dormant Adventitious Root Primordia upon Flooding in Solanum Dulcamara. Plant Physiol. 2016, 170, 2351–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Qiao, X.; Tian, Z.; Zhang, X.; Zou, X.; Cheng, Y.; Lu, G.; Zeng, L.; Fu, G.; Ding, X.; et al. Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress. Plants 2018, 7, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamang, B.G.; Magliozzi, J.O.; Maroof, M.A.S.; Fukao, T. Physiological and Transcriptomic Characterization of Submergence and Reoxygenation Responses in Soybean Seedlings. Plant Cell Environ. 2014, 37, 2350–2365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.H.; Hwang, S.J.; Waqas, M.; Khan, A.L.; Lee, J.H.; Lee, J.D.; Nguyen, H.T.; Lee, I.J. Comparative Analysis of Endogenous Hormones Level in Two Soybean (Glycine max L.) Lines Differing in Waterlogging Tolerance. Front. Plant Sci. 2015, 6, 714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, B.; Zhang, J.; Li, X.; Fan, X.; Dong, S.; Liu, P.; Zhao, B. Effects of Waterlogging on the Yield and Growth of Summer Maize under Field Conditions. Can. J. Plant Sci. 2014, 94, 23–31. [Google Scholar] [CrossRef]
- Shen, C.; Yuan, J.; Qiao, H.; Wang, Z.; Liu, Y.; Ren, X.; Wang, F.; Liu, X.; Zhang, Y.; Chen, X.; et al. Transcriptomic and Anatomic Profiling Reveal the Germination Process of Different Wheat Varieties in Response to Waterlogging Stress. BMC Genet. 2020, 21, 93. [Google Scholar] [CrossRef]
- Luan, H.; Li, H.; Li, Y.; Chen, C.; Li, S.; Wang, Y.; Yang, J.; Xu, M.; Shen, H.; Qiao, H. Transcriptome Analysis of Barley (Hordeum vulgare L.) under Waterlogging Stress, and Overexpression of the HvADH4 Gene Confers Waterlogging Tolerance in Transgenic Arabidopsis. BMC Plant Biol. 2023, 23, 62. [Google Scholar] [CrossRef]
- Fry, E.L.; Zhu, F.; Greenwood, B. Adapting to Environmental Change. In Microbiomes of Soils, Plants and Animals; Cambridge University Press: Cambridge, UK, 2020; pp. 154–181. [Google Scholar]
- Ali, S.; Tyagi, A.; Mushtaq, M.; Al-Mahmoudi, H.; Bae, H. Harnessing Plant Microbiome for Mitigating Arsenic Toxicity in Sustainable Agriculture. Environ. Pollut. 2022, 300, 118940. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Park, S.; Mir, R.A.; Mushtaq, M.; Bhat, B.; Al-Mahmoudi, H.; Bae, H. Deciphering the Plant Microbiome to Improve Drought Tolerance: Mechanisms and Perspectives. Environ. Exp. Bot. 2022, 201, 104933. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Bae, H. Plant Microbiome: An Ocean of Possibilities for Improving Disease Resistance in Plants. Microorganisms 2023, 11, 392. [Google Scholar] [CrossRef]
- Francioli, D.; Cid, G.; Kanukollu, S.; Ulrich, A.; Hajirezaei, M.R.; Kolb, S. Flooding Causes Dramatic Compositional Shifts and Depletion of Putative Beneficial Bacteria on the Spring Wheat Microbiota. Front. Microbiol. 2021, 12, 773116. [Google Scholar] [CrossRef]
- Hamonts, K.; Clough, T.J.; Stewart, A.; Clinton, P.W.; Richardson, A.E.; Wakelin, S.A.; O’Callaghan, M.; Condron, L.M. Effect of Nitrogen and Waterlogging on Denitrifier Gene Abundance, Community Structure and Activity in the Rhizosphere of Wheat. FEMS Microbiol. Ecol. 2013, 83, 568–584. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhu, D.; Lin, X. Effects of Water Management and Organic Fertilization with SRI Crop Practices on Hybrid Rice Performance and Rhizosphere Dynamics. Paddy Water Environ. 2011, 9, 33–39. [Google Scholar] [CrossRef]
- Li, H.Y.; Zhao, C.A.; Liu, C.J.; Xu, X.F. Endophytic Fungi Diversity of Aquatic/Riparian Plants and Their Antifungal Activity in Vitro. J. Microbiol. 2010, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.Y.; Cao, Y.; Zhang, K.Q. Metagenomic Insights into Communities, Functions of Endophytes, and Their Associates with Infection by Root-Knot Nematode, Meloidogyne Incognita, in Tomato Roots. Sci. Rep. 2015, 5, 17087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vishwanathan, K.; Zienkiewicz, K.; Liu, Y.; Janz, D.; Feussner, I.; Polle, A.; Haney, C.H. Ectomycorrhizal Fungi Induce Systemic Resistance against Insects on a Nonmycorrhizal Plant in a CERK1-Dependent Manner. New Phytol. 2020, 228, 728–740. [Google Scholar] [CrossRef]
- Xu, X.; Ji, J.; Ma, X.; Xu, Q.; Qi, X.; Chen, X. Comparative Proteomic Analysis Provides Insight into the Key Proteins Involved in Cucumber (Cucumis sativus L.) Adventitious Root Emergence under Waterlogging Stress. Front. Plant Sci. 2016, 7, 1515. [Google Scholar] [CrossRef] [Green Version]
- Eysholdt-Derzsó, E.; Sauter, M. Hypoxia and the Group VII Ethylene Response Transcription Factor HRE2 Promote Adventitious Root Elongation in Arabidopsis. Plant Biol. 2019, 21, 103–108. [Google Scholar] [CrossRef] [Green Version]
- Visser, E.J.W.; Voesenek, L.A.C.J. Acclimation to Soil Flooding-Sensing and Signal-Transduction. Plant Soil 2005, 274, 197–214. [Google Scholar] [CrossRef] [Green Version]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-Gibberellin Signaling Underlies Adaptation of Rice to Periodic Flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Caruso, P.; Baldoni, E.; Mattana, M.; Pietro Paolo, D.; Genga, A.; Coraggio, I.; Russo, G.; Picchi, V.; Reforgiato Recupero, G.; Locatelli, F. Ectopic Expression of a Rice Transcription Factor, Mybleu, Enhances Tolerance of Transgenic Plants of Carrizo Citrange to Low Oxygen Stress. Plant Cell Tissue Organ Cult. 2012, 109, 327–339. [Google Scholar] [CrossRef]
- Borella, J.; Becker, R.; Lima, M.C.; de Oliveira, D.D.S.C.; Braga, E.J.B.; de Oliveira, A.C.B.; Do Amarante, L. Nitrogen Source Influences the Antioxidative System of Soybean Plants under Hypoxia and Re-Oxygenation. Sci. Agric. 2019, 76, 51–62. [Google Scholar] [CrossRef]
- Doupis, G.; Kavroulakis, N.; Psarras, G.; Papadakis, I.E. Growth, Photosynthetic Performance and Antioxidative Response of ‘Hass’ and ‘Fuerte’ Avocado (Persea americana Mill.) Plants Grown under High Soil Moisture. Photosynthetica 2017, 55, 655–663. [Google Scholar] [CrossRef]
- Tyagi, A.; Sharma, S.; Ali, S.; Gaikwad, K. Crosstalk between H2S and NO: An emerging signalling pathway during waterlogging stress in legume crops. Plant Biol. 2022, 24(4), 576–586. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Sharma, S.; Srivastava, H.; Singh, N.K.; Gaikwad, K. In silico characterization and homology modeling of cytosolic APX gene predicts novel glycine residue modulating waterlogging stress response in pigeon pea. PeerJ 2021, 9, e10888. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Shen, H.; Pan, Y.; Guo, B.; Lv, C.; Xu, R. Elucidating the Hypoxic Stress Response in Barley (Hordeum vulgare L.) during Waterlogging: A Proteomics Approach. Sci. Rep. 2018, 8, 9655. [Google Scholar] [CrossRef]
- Alpuerto, J.B.; Hussain, R.M.F.; Fukao, T. The Key Regulator of Submergence Tolerance, SUB1A, Promotes Photosynthetic and Metabolic Recovery from Submergence Damage in Rice Leaves. Plant Cell Environ. 2016, 39, 672–684. [Google Scholar] [CrossRef] [Green Version]
- Hartman, S.; Sasidharan, R.; Voesenek, L.A.C.J. The Role of Ethylene in Metabolic Acclimations to Low Oxygen. New Phytol. 2021, 229, 64–70. [Google Scholar] [CrossRef]
- Hess, N.; Klode, M.; Anders, M.; Sauter, M. The Hypoxia Responsive Transcription Factor Genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis Are Differentially Regulated by Ethylene. Physiol. Plant. 2011, 143, 41–49. [Google Scholar] [CrossRef]
- Gasch, P.; Fundinger, M.; Müller, J.T.; Lee, T.; Bailey-Serres, J.; Mustropha, A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved Cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef] [Green Version]
- Giuntoli, B.; Perata, P. Group VII Ethylene Response Factors in Arabidopsis: Regulation and Physiological Roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A Group VII Ethylene Response Factor Gene, ZmEREB180, Coordinates Waterlogging Tolerance in Maize Seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimamura, S.; Nishimura, T.; Koshiba, T.; Yamamoto, R.; Hiraga, S.; Nakamura, T.; Komatsu, S. Effects of Anti-Auxins on Secondary Aerenchyma Formation in Flooded Soybean Hypocotyls. Plant Prod. Sci. 2016, 19, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Sun, F.; Gao, R.; Dong, H. RAP2.6L Overexpression Delays Waterlogging Induced Premature Senescence by Increasing Stomatal Closure More than Antioxidant Enzyme Activity. Plant Mol. Biol. 2012, 79, 609–622. [Google Scholar] [CrossRef]
- McDonald, M.P.; Visser, E.J.W. A Study of the Interaction between Auxin and Ethylene in Wild Type and Transgenic Ethylene-Insensitive Tobacco during Adventitious Root Formation Induced by Stagnant Root Zone Conditions. Plant Biol. 2003, 5, 550–556. [Google Scholar] [CrossRef]
- Qi, X.; Li, Q.; Ma, X.; Qian, C.; Wang, H.; Ren, N.; Shen, C.; Huang, S.; Xu, X.; Xu, Q.; et al. Waterlogging-Induced Adventitious Root Formation in Cucumber Is Regulated by Ethylene and Auxin through Reactive Oxygen Species Signalling. Plant Cell Environ. 2019, 42, 1458–1470. [Google Scholar] [CrossRef]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal Interplay during Adventitious Root Formation in Flooded Tomato Plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef]
- Wang, G.; Fan, W.; Peng, F. Physiological Responses of the Young Peach Tree to Water-Logging and Spraying SA at Different Timing. J. Fruit Sci. 2015, 32, 872–878. [Google Scholar]
- Kamal, A.H.M.; Komatsu, S. Jasmonic Acid Induced Protein Response to Biophoton Emissions and Flooding Stress in Soybean. J. Proteom. 2016, 133, 33–47. [Google Scholar] [CrossRef]
- Hudgins, J.W.; Franceschi, V.R. Methyl Jasmonate-Induced Ethylene Production Is Responsible for Conifer Phloem Defense Responses and Reprogramming of Stem Cambial Zone for Traumatic Resin Duct Formation. Plant Physiol. 2004, 135, 2134–2149. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Liu, D.; Li, L.; Wu, J.; Wang, S.; Li, X. Effects of Spraying Plant Growth Regulators on Peanut Growth and Yield & Quality under Waterlogging Stress. J. Hum. Agric. 2018, 44, 129. [Google Scholar]
- Kang, Y.Y.; Guo, S.R.; Li, J.; Duan, J.J. Effect of Root Applied 24-Epibrassinolide on Carbohydrate Status and Fermentative Enzyme Activities in Cucumber (Cucumis sativus L.) Seedlings under Hypoxia. Plant Growth Regul. 2009, 57, 259–269. [Google Scholar] [CrossRef]
- Ma, Y.H.; Guo, S.R. 24-Epibrassinolide Improves Cucumber Photosynthesis under Hypoxia by Increasing CO2 Assimilation and Photosystem II Efficiency. Photosynthetica 2014, 52, 96–104. [Google Scholar] [CrossRef]
- Li, H.; Vaillancourt, R.; Mendham, N.; Zhou, M. Comparative Mapping of Quantitative Trait Loci Associated with Waterlogging Tolerance in Barley (Hordeum vulgare L.). BMC Genom. 2008, 9, 401. [Google Scholar] [CrossRef] [Green Version]
- Mano, Y.; Muraki, M.; Komatsu, T.; Fujimori, M.; Akiyama, F.; Takamizo, T. Varietal Difference in Pre-Germination Flooding Tolerance and Waterlogging Tolerance at the Seedling Stage in Maize Inbred Lines. Jpn. J. Crop Sci. 2002, 71, 361–367. [Google Scholar] [CrossRef] [Green Version]
- Boru, G.; Van Ginkel, M.; Kronstad, W.E.; Boersma, L. Expression and Inheritance of Tolerance to Waterlogging Stress in Wheat. Euphytica 2001, 117, 91–98. [Google Scholar] [CrossRef]
- Cai, S.; Cao, Y.; Fang, X. Studies on the Variability and Combining Ability of Waterlogging Tolerance in Common Wheat. Jiangsu J. Agric. Sci. 1996, 12, 1–5. [Google Scholar]
- Yang, C.; Shibin, C.; Zhaosu, W. Studies on Genetic Features of Waterlogging Tolerance in Wheat. Jiangsu J. Agric. Sci. 1995, 11, 11–15. [Google Scholar]
- Ikeda, T. Studies on the Wet-Injury Resistance of Wheat and Barley Varieties. (II) Varietal Difference of Wet-Injury Resistance of Wheat and Barley. Bull. Div. Plant Breed. Cultiv. Tokai-Kinki Natl. Agric. Exp. Stn. 1955, 2, 11–16. [Google Scholar]
- Reyna, N.; Cornelious, B.; Shannon, J.G.; Sneller, C.H. Evaluation of a QTL for Waterlogging Tolerance in Southern Soybean Germplasm. Crop. Sci. 2003, 43, 2077–2082. [Google Scholar] [CrossRef]
- Hamaci, Y.; Yoshino, M.; Furusho, M.; Yoshida, T. Index of Screening for Wet Endurance in Malting Barley. Jpn. J. Breed. 1990, 40, 361–366. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.W.; Cao, Y.; Cai, S.B.; Xiong, E.H.; Zhu, W. Genetic Evaluation of Waterlogging Tolerance in Triticum Macha. Jiangsu J. Agric. Sci. 1997, 13, 73–75. [Google Scholar]
- Sachs, M.M. Molecular Genetic Basis of Metabolic Adaptation to Anoxia in Maize and Its Possible Utility for Improving Tolerance of Crops to Soil Waterlogging. In Interacting Stresses on Plants in a Changing Climate; Springer: Berlin/Heidelberg, Germany, 1993; pp. 375–393. [Google Scholar]
- Hamachi, Y.; Furusho, M.; Yoshida, T. Heritability of Wet Endurance in Malting Barley. Jpn. J. Breed. 1989, 39, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Mazur, B.J.; Tingey, S.V. Genetic Mapping and Introgression of Genes of Agronomic Importance. Curr. Opin. Biotechnol. 1995, 6, 175–182. [Google Scholar] [CrossRef]
- Campbell, M.T.; Proctor, C.A.; Dou, Y.; Schmitz, A.J.; Phansak, P.; Kruger, G.R.; Zhang, C.; Walia, H. Genetic and Molecular Characterization of Submergence Response Identifies Subtol6 as a Major Submergence Tolerance Locus in Maize. PLoS ONE 2015, 10, e0120385. [Google Scholar] [CrossRef]
- Mano, Y.; Omori, F.; Muraki, M.; Takamizo, T. QTL Mapping of Adventitious Root Formation under Flooding Conditions in Tropical Maize (Zea mays L.) Seedlings. Breed. Sci. 2005, 55, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Mano, Y.; Muraki, M.; Fujimori, M.; Takamizo, T.; Kindiger, B. Identification of QTL Controlling Adventitious Root Formation during Flooding Conditions in Teosinte (Zea mays Ssp. Huehuetenangensis) Seedlings. Euphytica 2005, 142, 33–42. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, B.; Yu, F.; Li, L.; Wang, M.; Xue, Y.; Zhang, Z.; Yan, J.; Yue, B.; Zheng, Y.; et al. Identification of Major QTL for Waterlogging Tolerance Using Genome-Wide Association and Linkage Mapping of Maize Seedlings. Plant Mol. Biol. Report. 2013, 31, 594–606. [Google Scholar] [CrossRef]
- Qiu, F.; Zheng, Y.; Zhang, Z.; Xu, S. Mapping of QTL Associated with Waterlogging Tolerance during the Seedling Stage in Maize. Ann. Bot. 2007, 99, 1067–1081. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Miura, K.; Asano, K.; Yamamoto, E.; Mori, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. A Major QTL Confers Rapid Internode Elongation in Response to Water Rise in Deepwater Rice. Breed. Sci. 2007, 57, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Ballesteros, D.C.; Mason, R.E.; Addison, C.K.; Andrea Acuña, M.; Nelly Arguello, M.; Subramanian, N.; Miller, R.G.; Sater, H.; Gbur, E.E.; Miller, D.; et al. Tolerance of Wheat to Vegetative Stage Soil Waterlogging Is Conditioned by Both Constitutive and Adaptive QTL. Euphytica 2015, 201, 329–343. [Google Scholar] [CrossRef]
- Ma, Y.U.; Mao, S.L.; Chen, G.Y.; Liu, Y.X.; Wei, L.I.; Wei, Y.M.; Liu, C.J.; Zheng, Y.L. QTLs for Waterlogging Tolerance at Germination and Seedling Stages in Population of Recombinant Inbred Lines Derived from a Cross Between Synthetic and Cultivated Wheat Genotypes. J. Integr. Agric. 2014, 13, 31–39. [Google Scholar] [CrossRef]
- Xue, D.W.; Zhou, M.X.; Zhang, X.Q.; Chen, S.; Wei, K.; Zeng, F.R.; Mao, Y.; Wu, F.B.; Zhang, G.P. Identification of QTLs for Yield and Yield Components of Barley under Different Growth Conditions. J. Zhejiang Univ. Sci. B 2010, 11, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Osman, K.A.; Tang, B.; Wang, Y.; Chen, J.; Yu, F.; Li, L.; Han, X.; Zhang, Z.; Yan, J.; Zheng, Y.; et al. Dynamic QTL Analysis and Candidate Gene Mapping for Waterlogging Tolerance at Maize Seedling Stage. PLoS ONE 2013, 8, e79305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhou, G.; Shabala, S.; Koutoulis, A.; Shabala, L.; Johnson, P.; Li, C.; Zhou, M. Identification of Aerenchyma Formation-Related QTL in Barley That Can Be Effective in Breeding for Waterlogging Tolerance. Theor. Appl. Genet. 2016, 129, 1167–1177. [Google Scholar] [CrossRef]
- Mano, Y.; Omori, F.; Takamizo, T.; Kindiger, B.; Bird, R.M.; Loaisiga, C.H.; Takahashi, H. QTL Mapping of Root Aerenchyma Formation in Seedlings of a Maize × Rare Teosinte “Zea nicaraguensis” cross. Plant Soil 2007, 295, 103–113. [Google Scholar] [CrossRef]
- Zhou, M.; Johnson, P.; Zhou, G.; Li, C.; Lance, R. Quantitative Trait Loci for Waterlogging Tolerance in a Barley Cross of Franklin × YuYaoXiangTian Erleng and the Relationship between Waterlogging and Salinity Tolerance. Crop. Sci. 2012, 52, 2082–2088. [Google Scholar] [CrossRef]
- Cavanagh, C.; Morell, M.; Mackay, I.; Powell, W. From Mutations to MAGIC: Resources for Gene Discovery, Validation and Delivery in Crop Plants. Curr. Opin. Plant Biol. 2008, 11, 215–221. [Google Scholar] [CrossRef]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.L.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-Parent Advanced Generation Inter-Cross (MAGIC) Populations in Rice: Progress and Potential for Genetics Research and Breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Dell’Acqua, M.; Gatti, D.M.; Pea, G.; Cattonaro, F.; Coppens, F.; Magris, G.; Hlaing, A.L.; Aung, H.H.; Nelissen, H.; Baute, J.; et al. Genetic Properties of the MAGIC Maize Population: A New Platform for High Definition QTL Mapping in Zea mays. Genome Biol. 2015, 16, 167. [Google Scholar] [CrossRef] [Green Version]
- Sannemann, W.; Huang, B.E.; Mathew, B.; Léon, J. Multi-Parent Advanced Generation Inter-Cross in Barley: High-Resolution Quantitative Trait Locus Mapping for Flowering Time as a Proof of Concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Du, H.; Shen, X.; Huang, Y.; Huang, M.; Zhang, Z. Overexpression of Vitreoscilla Hemoglobin Increases Waterlogging Tolerance in Arabidopsis and Maize. BMC Plant Biol. 2016, 16, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Peng, R.H.; Fan, H.Q.; Xiong, A.S.; Yao, Q.H.; Cheng, Z.M.; Li, Y. Vitreoscilla Hemoglobin Overexpression Increases Submergence Tolerance in Cabbage. Plant Cell Rep. 2005, 23, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.C.; Hu, Y.L.; Zhong, J.; Wang, L.X.; Guo, J.Y.; Lin, Z.P. Improvement of the Hydroponic Growth and Waterlogging Tolerance of Petunias by the Introduction of Vhb Gene. Acta Bot. Sin. 2003, 45, 205–210. [Google Scholar]
- Raineri, J.; Caraballo, L.; Rigalli, N.; Portapila, M.; Otegui, M.E.; Chan, R.L. HaHB11 Transformed Maize Has Improved Yield under Waterlogging and Defoliation in Control and Field Conditions. bioRxiv 2021, 2021-10. [Google Scholar]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. NAC Transcription Factor speedy hyponastic growth Regulates Flooding-Induced Leaf Movement in Arabidopsis. Plant Cell 2013, 25, 4941–4955. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Ji, J.; Xu, Q.; Qi, X.; Weng, Y.; Chen, X. The major-effect quantitative trait locus csarn6.1 encodes an aaa atpase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. Plant J. 2018, 93, 917–930. [Google Scholar] [CrossRef] [Green Version]
- Yin, D.; Sun, D.; Han, Z.; Ni, D.; Norris, A.; Jiang, C.Z. PhERF2, an Ethylene-Responsive Element Binding Factor, Plays an Essential Role in Waterlogging Tolerance of Petunia. Hortic. Res. 2019, 6, 83. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive Expression of a Stabilized Transcription Factor Group VII Ethylene Response Factor Enhances Waterlogging Tolerance in Wheat without Penalizing Grain Yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef]
- Luan, H.; Guo, B.; Shen, H.; Pan, Y.; Hong, Y.; Lv, C.; Xu, R. Overexpression of Barley Transcription Factor HvERF2.11 in Arabidopsis Enhances Plant Waterlogging Tolerance. Int. J. Mol. Sci. 2020, 21, 1982. [Google Scholar] [CrossRef] [Green Version]
- Cabello, J.V.; Giacomelli, J.I.; Piattoni, C.V.; Iglesias, A.A.; Chan, R.L. The Sunflower Transcription Factor HaHB11 Improves Yield, Biomass and Tolerance to Flooding in Transgenic Arabidopsis Plants. J. Biotechnol. 2016, 222, 73–83. [Google Scholar] [CrossRef]
- Xuan, L.; Hua, J.; Zhang, F.; Wang, Z.; Pei, X.; Yang, Y.; Yin, Y.; Creech, D.L. Identification and Functional Analysis of Thadh1 and Thadh4 Genes Involved in Tolerance to Waterlogging Stress in Taxodium Hybrid ‘Zhongshanshan 406’. Genes 2021, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Quimio, C.A.; Torrizo, L.B.; Setter, T.L.; Ellis, M.; Grover, A.; Abrigo, E.M.; Oliva, N.P.; Ella, E.S.; Carpena, A.L.; Ito, O.; et al. Enhancement of Submergence Tolerance in Transgenic Rice Overproducing Pyruvate Decarboxylase. J. Plant Physiol. 2000, 156, 516–521. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A Is an Ethylene-Response-Factor-like Gene That Confers Submergence Tolerance to Rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukao, T.; Bailey-Serres, J. Submergence Tolerance Conferred by Sub1A Is Mediated by SLR1 and SLRL1 Restriction of Gibberellin Responses in Rice. Proc. Natl. Acad. Sci. USA 2008, 105, 16814–16819. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The Ethylene Response Factors SNORKEL1 and SNORKEL2 Allow Rice to Adapt to Deep Water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Ismond, K.P.; Dolferus, R.; De Pauw, M.; Dennis, E.S.; Good, A.G. Enhanced Low Oxygen Survival in Arabidopsis through Increased Metabolic Flux in the Fermentative Pathway. Plant Physiol. 2003, 132, 1292–1302. [Google Scholar] [CrossRef] [Green Version]
- Dolferus, R.; Wolansky, M.; Carroll, R.; Miyashita, Y.; Ismond, K.; Good, A. Functional Analysis of Lactate Dehydrogenase during Hypoxic Stress in Arabidopsis. Funct. Plant Biol. 2008, 35, 131–140. [Google Scholar] [CrossRef]
- Hunt, P.W.; Klok, E.J.; Trevaskis, B.; Watts, R.A.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. Increased Level of Hemoglobin 1 Enhances Survival of Hypoxic Stress and Promotes Early Growth in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 17197–17202. [Google Scholar] [CrossRef] [Green Version]
- Grichko, V.P.; Glick, B.R. Flooding Tolerance of Transgenic Tomato Plants Expressing the Bacterial Enzyme ACC Deaminase Controlled by the 35S, RolD or PRB-1b Promoter. Plant Physiol. Biochem. 2001, 39, 19–25. [Google Scholar] [CrossRef]
- Zhang, J.; Van Toai, T.; Huynh, L.; Preiszner, J. Development of Flooding-Tolerant Arabidopsis Thaliana by Autoregulated Cytokinin Production. Mol. Breed. 2000, 6, 135–144. [Google Scholar] [CrossRef]
- Huynh, L.N.; VanToai, T.; Streeter, J.; Banowetz, G. Regulation of Flooding Tolerance of SAG12:Ipt Arabidopsis Plants by Cytokinin. J. Exp. Bot. 2005, 56, 1397–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.L.; Wang, G.; Wang, T.; Jia, Z.H.; Guo, Z.R.; Zhang, J.Y. Adrap2.3, a Novel Ethylene Response Factor VII from Actinidia Deliciosa, Enhanceswaterlogging Resistance in Transgenic Tobacco through Improving Expression Levels of Pdc and Adh Genes. Int. J. Mol. Sci. 2019, 20, 1189. [Google Scholar] [CrossRef] [Green Version]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of Abiotic Stress on Plants: A Systems Biology Perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; Van Dongen, J.T.; Giuntoli, B.; Novi, G.; Santaniello, A.; Geigenberger, P.; Perata, P. HRE1 and HRE2, Two Hypoxia-Inducible Ethylene Response Factors, Affect Anaerobic Responses in Arabidopsis thaliana. Plant J. 2010, 62, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Hoeren, F.U.; Dolferus, R.; Wu, Y.; Peacock, W.J.; Dennis, E.S. Evidence for a Role for AtMYB2 in the Induction of the Arabidopsis Alcohol Dehydrogenase Gene (ADH1) by Low Oxygen. Genetics 1998, 149, 479–490. [Google Scholar] [CrossRef]
- Bond, D.M.; Wilson, I.W.; Dennis, E.S.; Pogson, B.J.; Jean Finnegan, E. Vernalization Insensitive 3 (VIN3) Is Required for the Response of Arabidopsis thaliana Seedlings Exposed to Low Oxygen Conditions. Plant J. 2009, 59, 576–587. [Google Scholar] [CrossRef]
- Christianson, J.A.; Wilson, I.W.; Llewellyn, D.J.; Dennis, E.S. The Low-Oxygen-Induced NAC Domain Transcription Factor ANAC102 Affects Viability of Arabidopsis Seeds Following Low-Oxygen Treatment. Plant Physiol. 2009, 149, 1724–1738. [Google Scholar] [CrossRef] [Green Version]
- Tournaire-Roux, C.; Sutka, M.; Javot, H.; Gout, E.; Gerbeau, P.; Luu, D.T.; Bligny, R.; Maurel, C. Cytosolic PH Regulates Root Water Transport during Anoxic Stress through Gating of Aquaporins. Nature 2003, 425, 393–397. [Google Scholar] [CrossRef]
- Aroca, R.; Porcel, R.; Ruiz-Lozano, J.M. Regulation of Root Water Uptake under Abiotic Stress Conditions. J. Exp. Bot. 2012, 63, 43–57. [Google Scholar] [CrossRef]
- Steffens, B.; Geske, T.; Sauter, M. Aerenchyma Formation in the Rice Stem and Its Promotion by H2O2. New Phytol. 2011, 190, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Zheng, L.; Li, J.; Mao, Y.; Zhang, R.; Niu, X.; Geng, M.; Zhang, X.; Huang, W.; Luo, K.; et al. Transcriptomic Profiling Suggests Candidate Molecular Responses to Waterlogging in Cassava. PLoS ONE 2022, 17, e0261086. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Sharma, S.; Srivastava, H.; Singh, A.; Kaila, T.; Ali, S.; Gaikwad, A.B.; Singh, N.K.; Gaikwad, D. Transcriptome Profiling of Two Contrasting Pigeon Pea (Cajanus cajan) Genotypes in Response to Waterloging Stress. Front. Genet. 2023, 13, 3757. [Google Scholar] [CrossRef] [PubMed]
- Butsayawarapat, P.; Juntawong, P.; Khamsuk, O.; Somta, P. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Tolerant Zombi Pea (Vigna vexillata) Reveals Energy Conservation and Root Plasticity Controlling Waterlogging Tolerance. Plants 2019, 8, 264. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Liu, X.; Zhang, Z.; Liu, N.; Li, D.; Hu, L. Melatonin Improved Waterlogging Tolerance in Alfalfa (Medicago sativa) by Reprogramming Polyamine and Ethylene Metabolism. Front. Plant Sci. 2019, 10, 4. [Google Scholar] [CrossRef]
- Casarotto, G.; Kaspary, T.E.; Cutti, L.; Thomas, A.L.; Neto, J.F.B. Expression of Genes Related to Soil Flooding Tolerance in Soybeans. Acta Sci. Agron. 2019, 41, e42709. [Google Scholar] [CrossRef]
- Oh, M.W.; Nanjo, Y.; Komatsu, S. Gel-Free Proteomic Analysis of Soybean Root Proteins Affected by Calcium under Flooding Stress. Front. Plant Sci. 2014, 5, 559. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Deng, Z.; Zhang, W.; Meng, Z.; Chang, X.; Lv, M. Effect of Waterlogging Duration at Different Growth Stages on the Growth, Yield and Quality of Cotton. PLoS ONE 2017, 12, e0169029. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.J.; Roy, S.K.; Cho, S.W.; Kwon, S.J.; Kun, C.; Chun, H.C.; Woo, S.H. Proteome Analysis of Sesame Leaves in Response to Waterlogging Stress at Vegetative and Flowering Stages. Biol. Plant. 2019, 63, 733–749. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, D.G.; Lee, S.H.; Kang, K.Y.; Bahk, J.D.; Choi, M.S.; Lee, I.J.; Renaut, J.; Lee, B.H. A Comparative Proteomic Analysis of Tomato Leaves in Response to Waterlogging Stress. Physiol. Plant. 2007, 131, 555–570. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A.C.J. Flooding Stress: Acclimations and Genetic Diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, K.J.; Zabalza, A.; Van Dongen, J.T. Regulation of Respiration When the Oxygen Availability Changes. Physiol. Plant. 2009, 137, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.T.; Schurr, U.; Pfister, M.; Geigenberger, P. Phloem Metabolism and Function Have to Cope with Low Internal Oxygen. Plant Physiol. 2003, 131, 1529–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benamar, A.; Rolletschek, H.; Borisjuk, L.; Avelange-Macherel, M.H.; Curien, G.; Mostefai, H.A.; Andriantsitohaina, R.; Macherel, D. Nitrite-Nitric Oxide Control of Mitochondrial Respiration at the Frontier of Anoxia. Biochim. Biophys. Acta-Bioenerg. 2008, 1777, 1268–1275. [Google Scholar] [CrossRef]
- Lothier, J.; Diab, H.; Cukier, C.; Limami, A.M.; Tcherkez, G. Metabolic Responses to Waterlogging Differ between Roots and Shoots and Reflect Phloem Transport Alteration in Medicago truncatula. Plants 2020, 9, 1373. [Google Scholar] [CrossRef]
- Komatsu, S.; Nakamura, T.; Sugimoto, Y.; Sakamoto, K. Proteomic and Metabolomic Analyses of Soybean Root Tips Under Flooding Stress. Protein Pept. Lett. 2014, 21, 865–884. [Google Scholar] [CrossRef]
- Cui, J.; Davanture, M.; Zivy, M.; Lamade, E.; Tcherkez, G. Metabolic Responses to Potassium Availability and Waterlogging Reshape Respiration and Carbon Use Efficiency in Oil Palm. New Phytol. 2019, 223, 310–322. [Google Scholar] [CrossRef]
- Zhao, N.; Li, C.; Yan, Y.; Cao, W.; Song, A.; Wang, H.; Chen, S.; Jiang, J.; Chen, F. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Waterlogging-Tolerant Chrysanthemum morifolium Cultivars under Waterlogging Stress and Reoxygenation Conditions. Int. J. Mol. Sci. 2018, 19, 1455. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Bai, D.; Zhong, Y.; Lin, M.; Sun, L.; Qi, X.; Hu, C.; Fang, J. Full-Length Transcriptome and RNA-Seq Analyses Reveal the Mechanisms Underlying Waterlogging Tolerance in Kiwifruit (Actinidia valvata). Int. J. Mol. Sci. 2022, 23, 3237. [Google Scholar] [CrossRef]
- Cui, J.; Abadie, C.; Carroll, A.; Lamade, E.; Tcherkez, G. Responses to K Deficiency and Waterlogging Interact via Respiratory and Nitrogen Metabolism. Plant Cell Env. 2019, 42, 647–658. [Google Scholar] [CrossRef]
- Cid, G.A.; Francioli, D.; Kolb, S.; Tandron-Moya, Y.A.; von Wiren, N.; Hajirezaei, M.-R. Elucidating the Systemic Response of Wheat Plants under Waterlogging Based on Transcriptomic and Metabolic Approaches. bioRxiv 2022, 2022-08. [Google Scholar] [CrossRef]
- Pazhamala, L.T.; Kudapa, H.; Weckwerth, W.; Millar, A.H.; Varshney, R.K. Systems Biology for Crop Improvement. Plant Genome 2021, 14, e20098. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of Integrating Data to Uncover Genotype-Phenotype Interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Chen, L.; Hao, L.; Parry, M.A.J.; Phillips, A.L.; Hu, Y.G. Progress in TILLING as a Tool for Functional Genomics and Improvement of Crops. J. Integr. Plant Biol. 2014, 56, 425–443. [Google Scholar] [CrossRef] [Green Version]
- Negrão, S.; Julkowska, M.M. Plant Phenotyping. eLS 2020, 1–14. [Google Scholar] [CrossRef]


| Species | WS Condition | Affected Traits | References |
|---|---|---|---|
| Triticum aestivum L. | 1 wk | Dry weight of stem and root ↓ Length of root ↓ Ratio of root/shoot ↓ Root aerenchyma ↑ | [33] |
| Solanum dulcamara | 1 wk | Stem region ↓ and adventitious root ↑ (ET↑, ABA↓) | [34] |
| Brassica napus L. | 3 d | Length of root and shoot ↓ Fresh weight ↓ | [35] |
| Glycine max (L.) Merr. cv. “Williams 82” | 10 d | Length of root ↓ Development of lateral root and root hairs ↓ | [36] |
| Glycine max L. (S99-2281) | 10 d | Length of shoot ↓ Fresh weight of shoot and root ↓ Root aerenchyma ↑ Adventitious root ↑ | [37] |
| Zea mays L. (DH605, ZD958) | 3 and 6 d | Height of plant and ear ↓ Leaf area index ↓ Yield ↓ Bald tip ↑ | [38] |
| Triticum aestivum L. (ZM22) | 72 h | Germination ↓ Coleoptile height ↓ Amyloplast ↑ | [39] |
| Hordeum vulgare L. (Franklin) | 21 d | Leaf area ↓ Dry and fresh weight of shoot ↓ Plant height ↓ Total length and number of adventitious root ↑ Leaf aerenchyma ↑ Chlorosis and age of leaf ↑ | [40] |
| Species | Mapping Population Type | QTL | Traits | References |
|---|---|---|---|---|
| Maize | BC3F4, RILs, F2, F2:3 | Subtol6, Qarf7.04–7.05, Qarf8.05, sdw9-4, tdw9-2, tdw9-3 | Leaf chlorosis, mean leaf senescence score, adventitious root formation, shoot dry weight, total dry weight | [90,91,92,93,94] |
| Rice | F2 | qTIL1 C9285, qTIL1 T65, Sub1, qTIL12 C9285, qTIL12 W0120, qNEI12 C9285, qNEI12 W0120, qLEI12 C9285, qLEI12 W0120 | Number and total internode length, green leaf recovery, number of elongated internodes | [95] |
| Wheat | RILs | QRfbio.ua-1B-WGH, QSfbio.ua-1B-WGH, QSpadpost.ua-1B-WF, QSpad.ua-1D.5, GRI-7A | Shoot and root fresh biomass, chlorophyll content, shoot and root dry biomass, seed germination rate | [96,97] |
| Barley | DH lines | KWw2.1, GSw1.1/2.1, tfy1.1-1, QWI.YyFr.2H, tfy1.2-1/2.1-1, tfy1.1-2, QWL.YeFr.4H, QTL-AER, QTL-WL-4H, yfy2.2-3, GYw1.2 | Kernel weight, grains per spike, leaf chlorosis, plant healthiness, yellow leaf percentage, survival rate, aerenchyma formation, waterlogging tolerance, root porosity, grain yield | [52,78,98,99,100,101,102] |
| Gene | Transgenic Plant | Gene Source | Waterlogging Tolerance | References |
|---|---|---|---|---|
| Pdc1 (pyruvate decarboxylase isozyme 1) | O. sativa | O. sativa | Enhanced waterlogging tolerance | [118] |
| OsSub1A (ethylene-response-factor-like submergence tolerance gene) | O. sativa | O. sativa | Enhanced waterlogging tolerance in rice plants by increasing the expression of ADH1 | [119,120,121] |
| Pdc1 (pyruvate decarboxylase isozyme 1) | A. thaliana | A. thaliana | Confers waterlogging tolerance | [122] |
| Pdc2 (pyruvate decarboxylase isozyme 2) | A. thaliana | A. thaliana | Enhanced waterlogging tolerance | [122] |
| AtACO5 (1-aminocyclopropane-1-carboxylic acid oxidase) and AtACS (acetyl-CoA synthetase) | A. thaliana | A. thaliana | Increased ET levels and waterlogging tolerance | [111] |
| AtLDH (lactate dehydrogenase) | A. thaliana | A. thaliana | Confers hypoxia tolerance by increasing PDC enzyme activity | [123] |
| AtRAP2.6L (member of ERF subfamily) | A. thaliana | A. thaliana | Enhanced the activity of antioxidant enzymes and transcript levels of ABA biosynthesis genes, stomatal closure | [68] |
| GLB1 class I hemoglobin (Hb) | A. thaliana | Parasponia andersonii | Enhanced resistance to hypoxia | [124] |
| Hb (hemoglobin) | Brassica oleracea | Vitreoscilla filiformis | Confers waterlogging tolerance | [109] |
| ACC (1-aminocyclopropane-1-carboxylic acid) deaminase | Solanum lycopersicum | Enterobacter | Confers waterlogging tolerance | [125] |
| ipt (isopentenyl transferase in cytokinin biosynthesis) | A. thaliana | A. thaliana | Confers waterlogging tolerance | [126,127] |
| ZmEREB180 (a group VII ethylene response factor gene) | Zea mays | Zea mays | Confers waterlogging tolerance by stimulating AR formation | [66] |
| AdRAP2.3 (member of ERF subfamily) | Actinidia deliciosa | Nicotiana tabacum | Enhanced ADH and PDC enzyme activities | [128] |
| HvERF2.11 (ethylene responsive factor 2) | Hordeum vulgare | A. thaliana | Stimulates the expression level of ET genes and also increases antioxidant enzyme activity | [115] |
| HaHB11 (homeodomain-leucine zipper I subfamily) | Helianthus annus | A. thaliana | Confers waterlogging tolerance | [116] |
| ThADH1 (alcohol dehydrogenase 1) and ThADH4 (alcohol dehydrogenase 4) | Populus alba | Taxodium mucronatum Tenore × Taxodium distichum (L.). Rich | Confers waterlogging and hypoxia tolerance | [117] |
| HvADH4 (alcohol dehydrogenase 4) | Hordeum vulgare | A. thaliana | Confers waterlogging tolerance by enhancing the antioxidant enzyme activity | [40] |
| Omics Study | Species | WS Condition | Key Genes/Metabolites/Proteins | References |
|---|---|---|---|---|
| Transcriptomics | Chrysanthemum morifolium (Nannongxuefeng) | 12 h | N-end rule pathway (RAP2.3, HRE2, ATE, PCO1, PCO2) ↑ ROS signaling (POD, AOX1a) ↑ Anaerobic respiration and carbohydrate metabolism (ADH, PDC, SUS1, PDC1) ↑ Hsp 83-like, Chaperone protein ClpB1-like, Snakin-2-like isoform X1 ↑ | [153] |
| Actinidia valvata (KR5) | 12, 24, 72 h | ROS scavenging pathway (POD, CAT) ↑, NADH-GOGAT/AlaAT, ERF77 ↑ | [154] | |
| Manihot esculenta Grantz | 6 d | Photosynthesis, RNA transport, RNA degradation, amino metabolism ↑ | [137] | |
| T. aestivum L. (ZM22) | 72 h | Oxidoreductase activity, biological response to ABA and SA ↑ | [39] | |
| Hordeum vulgare L. (Franklin) | 24 h | Metabolic process (biosynthesis of secondary metabolites and phenylpropanoid), transferase activity, catalytic activity ↑ | [40] | |
| 72 h | Oxidation–reduction process, protein binding, catalytic activity ↑ | |||
| Proteomics | B. napus L. (ZS9, tolerant cultivar) | 4, 8, 12 h | Oxidation–reduction process (BnaA09g29780D), response to ethylene (BnaA09g07120D) ↑ Abiotic stress response (BnaC08g02330D), (BnaC02g24210D), response to jasmonic acid (BnaC02g24210D) ↓ | [35] |
| B. napus L. (GH01, sensitive cultivar) | 4, 8, 12 h | Abiotic stress response (BnaC08g02330D), response to ethylene (BnaA09g07120D) ↑ Oxidation–reduction process (BnaA09g29780D), response to jasmonic acid (BnaC02g24210D) ↓ | ||
| G. max L. cultivar Enrei | 2 d | Fermentation and glycolysis-related proteins ↑ Degradation/synthesis/posttranslational modification of proteins, hormone/cell wall metabolisms, and DNA synthesis ↓ | [142] | |
| Sesamum indicum L., cv. Miryang 44 | 2, 3 d | Photosynthesis (OEE1), stress defense (HSPs, Chaperones), energy metabolism (ATPs, GS) ↑ | [144] | |
| Lycopersicon esculentum L. cv. Koma | 24, 48, 72 h | Stress and defense related (Hsp cognate 70, plastidic cysteine synthase1) ↑ Photosynthesis (rubisco large/small subunits, rubisco activase), biosynthesis and metabolism of protein (Cytochrome P450, glycinamide ribonucleotide synthetase) ↓ | [145] | |
| Metabolomics | M. truncatula | 7 and 21 d | Sugars, organic acid, aromatics, glycine, alanine, glutamine, lysine ↑ Nitrogenous compounds, threitol ↓ | [150] |
| H. annuus | 2, 7, 14 d | Alanine, sugars, polyols, aconitate, citrate, phosphate ↑ Aspartate, fumarate ↓ | [155] | |
| Elaeis guineensis | 1, 2, 3, 7 wks | Polyol (myoinositol) ↑ Aconitate, citrate, serine, asparine ↓ | [152] | |
| G. max L. cultivar Enrei | 2 d | Alanine, AMP, cysteine, DHAP, GABA, glycine ↑ 2-oxoglutarate, acetyl-CoA, allantonin, aspartic acid, fumarate, cinnamate, glutamine ↓ | [151] | |
| T. aestivum (Chinese spring) | 12 d | Glycine, alanine, GABA ↑ Asparagine, pyruvate ↓ | [156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyagi, A.; Ali, S.; Park, S.; Bae, H. Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants. Plants 2023, 12, 1544. https://doi.org/10.3390/plants12071544
Tyagi A, Ali S, Park S, Bae H. Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants. Plants. 2023; 12(7):1544. https://doi.org/10.3390/plants12071544
Chicago/Turabian StyleTyagi, Anshika, Sajad Ali, Suvin Park, and Hanhong Bae. 2023. "Exploring the Potential of Multiomics and Other Integrative Approaches for Improving Waterlogging Tolerance in Plants" Plants 12, no. 7: 1544. https://doi.org/10.3390/plants12071544




