Maize Terpene Synthase 8 (ZmTPS8) Contributes to a Complex Blend of Fungal-Elicited Antibiotics
Abstract
:1. Introduction
2. Results
2.1. Identification of Previously Undetected Maize Sesquiterpenoids in Stems following Elicitation
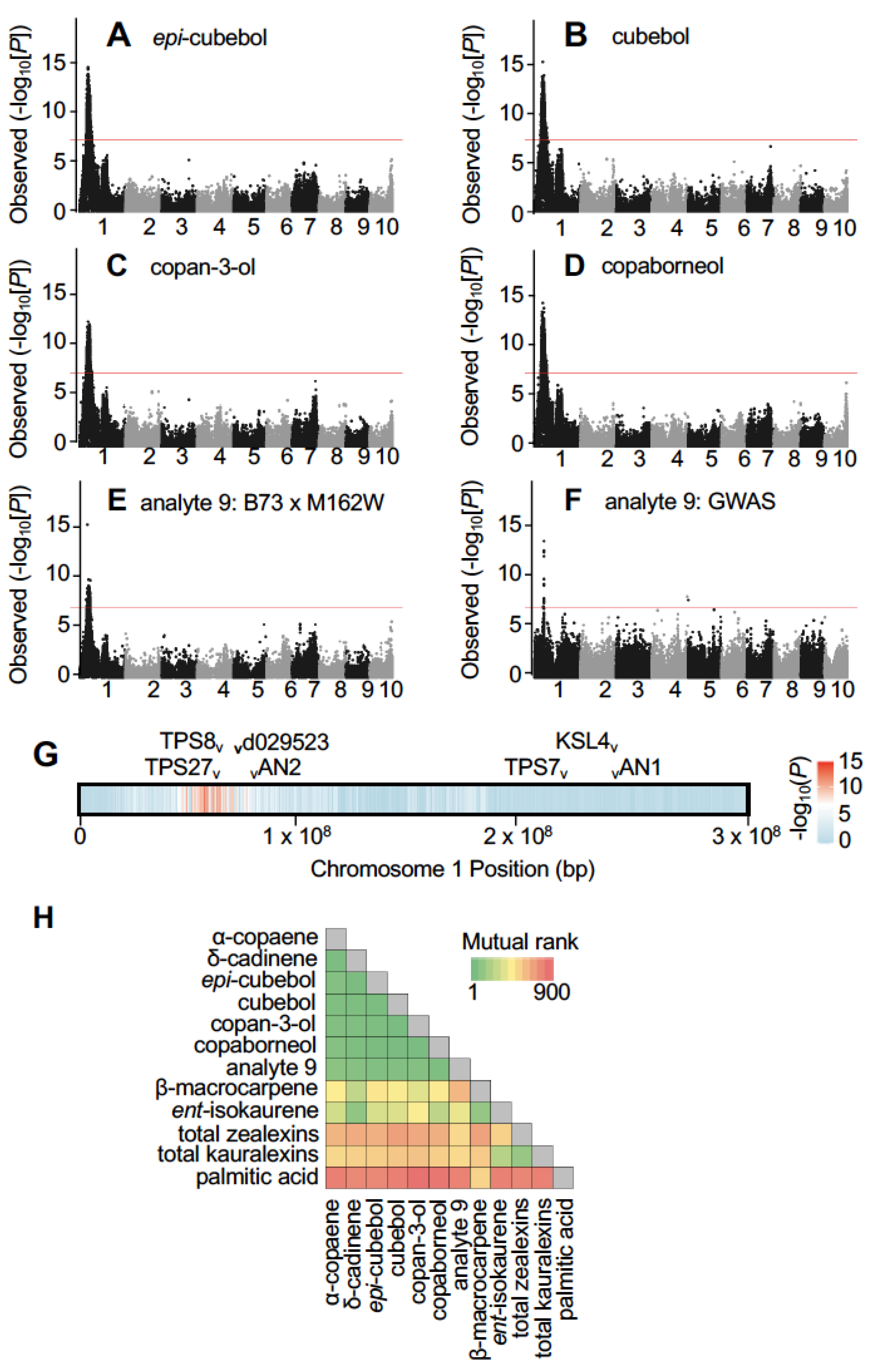
2.2. Association Mapping Using the B73 × M162W Recombinant Inbred Line (RIL) Population Identifies a Locus on Chromosome 1 Controlling Oxygenated Maize Sesquiterpenoids
2.3. Heterologous Expression of ZmTPS27 in N. benthamiana Supports Function as a Geraniol Synthase
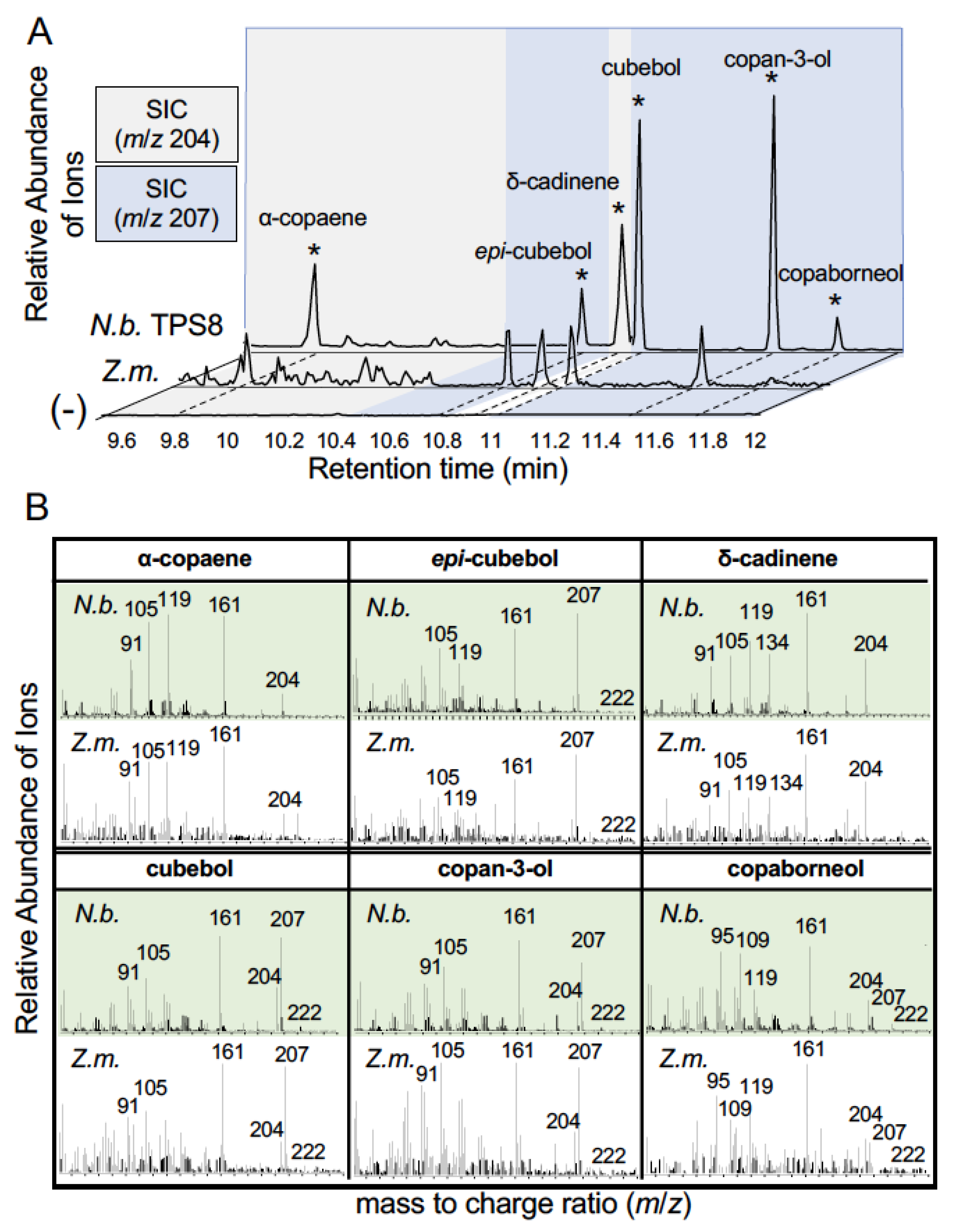
2.4. Previously Undetected Maize Sesquiterpenoids Are Products of ZmTPS8
2.5. ZmCYP71Z19 Oxidizes a ZmTPS8 Product to Produce an Unknown Sesquiterpene Acid Which Exists Endogenously in Fungal-Elicited Maize Tissues
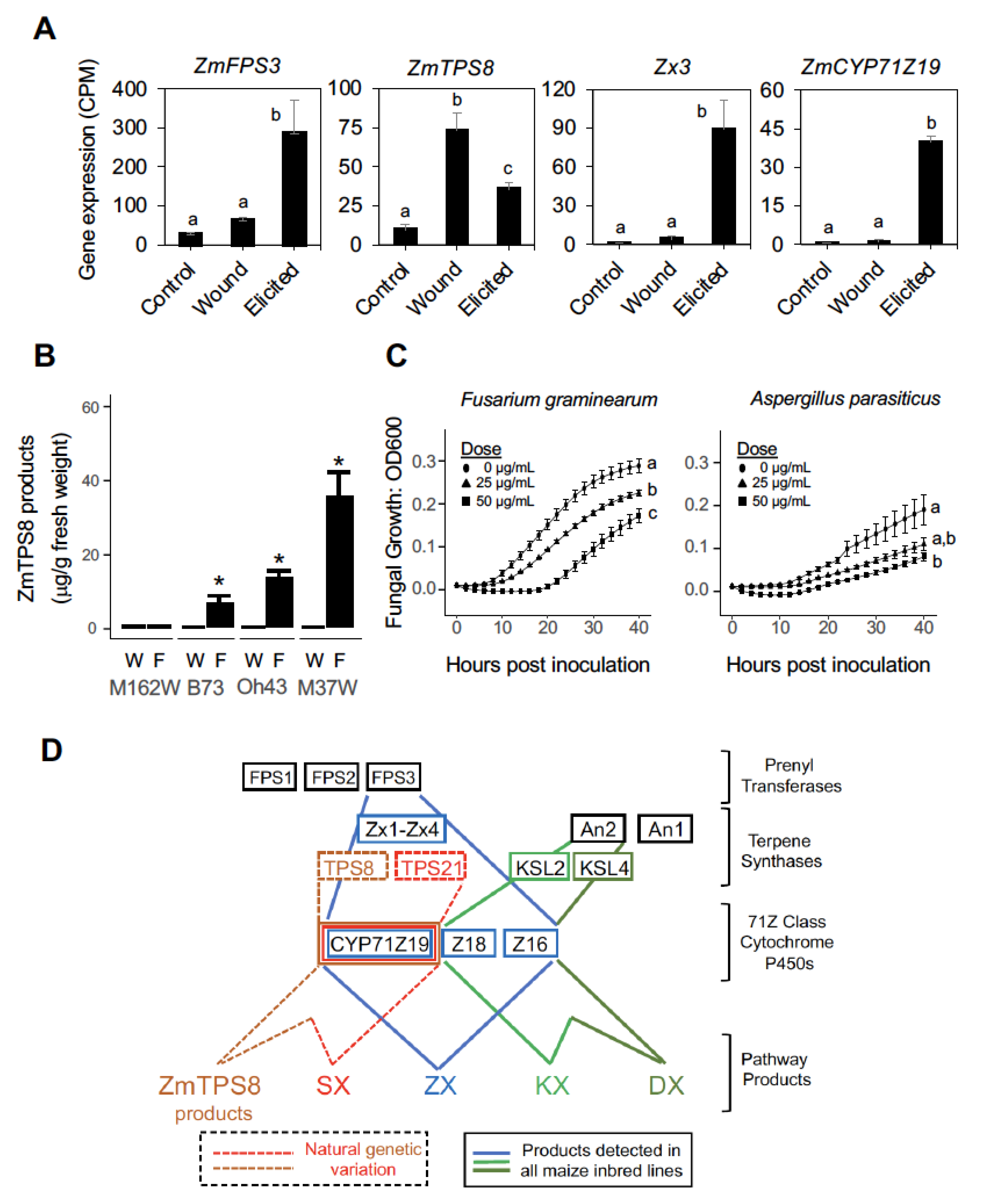
2.6. Antifungal ZmTPS8 Products Are Linked to Complex Pathway Regulation Involving Both Wounding and Fungal Elicitation at Different Biosynthetic Steps
3. Discussion
4. Materials and Methods
4.1. Plant and Fungal Materials
4.2. Maize Stem Tissues Used for Metabolite-Lead Association Analyses
4.3. Identification and Analyses of Plant Metabolites
4.4. Maize Forward Genetics Studies Using Metabolite Association Analyses
4.5. Transient Heterologous Co-Expression Assays in N. benthamiana
4.6. 3′-RNA-Seq Analyses of Control, Wounded and Fungal-Elicited B73 Stems
4.7. Construction of the ZmTPS8 Phylogenetic Tree among Diverse Inbred Lines
4.8. Purification of Cubebol from Piper Cubeba Essential Oil
4.9. In Vitro Antifungal Activity Assays with Cubebol
4.10. Statistics and Mutual Rank Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, Y.; Northen, T.R.; Khalil, A.; Huffaker, A.; Schmelz, E.A. Getting back to the grass roots: Harnessing specialized metabolites for improved crop stress resilience. Curr. Opin. Biotechnol. 2021, 70, 174–186. [Google Scholar] [CrossRef]
- Lacchini, E.; Goossens, A. Combinatorial Control of Plant Specialized Metabolism: Mechanisms, Functions, and Consequences. Annu. Rev. Cell Dev. Biol. 2020, 36, 291–313. [Google Scholar] [CrossRef]
- Ding, Y.Z.; Huffaker, A.; Kollner, T.G.; Weckwerth, P.; Robert, C.A.M.; Spencer, J.L.; Lipka, A.E.; Schmelz, E.A. Selinene Volatiles Are Essential Precursors for Maize Defense Promoting Fungal Pathogen Resistance. Plant Physiol. 2017, 175, 1455–1468. [Google Scholar] [CrossRef]
- Meihls, L.N.; Handrick, V.; Glauser, G.; Barbier, H.; Kaur, H.; Haribal, M.M.; Lipka, A.E.; Gershenzon, J.; Buckler, E.S.; Erb, M.; et al. Natural Variation in Maize Aphid Resistance Is Associated with 2,4-Dihydroxy-7-Methoxy-1,4-Benzoxazin-3-One Glucoside Methyltransferase Activity. Plant Cell 2013, 25, 2341–2355. [Google Scholar] [CrossRef] [Green Version]
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Nat. Chem. Biol. 2007, 3, 408–414. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Block, A.K.; Vaughan, M.M.; Schmelz, E.A.; Christensen, S.A. Biosynthesis and function of terpenoid defense compounds in maize (Zea mays). Planta 2019, 249, 21–30. [Google Scholar] [CrossRef]
- Bohlmann, J.; Keeling, C.I. Terpenoid biomaterials. Plant J. 2008, 54, 656–669. [Google Scholar] [CrossRef]
- Liang, J.; Liu, J.; Brown, R.; Jia, M.; Zhou, K.; Peters, R.J.; Wang, Q. Direct production of dihydroxylated sesquiterpenoids by a maize terpene synthase. Plant J. 2018, 94, 847–856. [Google Scholar] [CrossRef]
- Turlings, T.C.J.; Tumlinson, J.H.; Lewis, W.J. Exploitation of Herbivore-Induced Plant Odors by Host-Seeking Parasitic Wasps. Science 1990, 250, 1251–1253. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; Tholl, D. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-beta-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef] [Green Version]
- Rasmann, S.; Kollner, T.G.; Degenhardt, J.; Hiltpold, I.; Toepfer, S.; Kuhlmann, U.; Gershenzon, J.; Turlings, T.C.J. Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 2005, 434, 732–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degenhardt, J.; Hiltpold, I.; Kollner, T.G.; Frey, M.; Gierl, A.; Gershenzon, J.; Hibbard, B.E.; Ellersieck, M.R.; Turlings, T.C.J. Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Natl. Acad. Sci. USA 2009, 106, 13213–13218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, N.M.; Anderson, S.N.; Andorf, C.M.; Ahern, K.R.; Bai, F.; Barad, O.; Barbazuk, W.B.; Bass, H.W.; Baruch, K.; Ben-Zvi, G.; et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet. 2018, 50, 1282–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luck, K.; Chen, X.; Norris, A.M.; Chen, F.; Gershenzon, J.; Köllner, T.G. The reconstruction and biochemical characterization of ancestral genes furnish insights into the evolution of terpene synthase function in the Poaceae. Plant Mol. Biol. 2020, 104, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Weckwerth, P.R.; Poretsky, E.; Murphy, K.M.; Sims, J.; Saldivar, E.; Christensen, S.A.; Char, S.N.; Yang, B.; Tong, A.-d.; et al. Genetic elucidation of interconnected antibiotic pathways mediating maize innate immunity. Nat. Plants 2020, 6, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Murphy, K.M.; Poretsky, E.; Mafu, S.; Yang, B.; Char, S.N.; Christensen, S.A.; Saldivar, E.; Wu, M.; Wang, Q.; et al. Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat. Plants 2019, 5, 1043–1056. [Google Scholar] [CrossRef]
- Schnee, C.; Kollner, T.G.; Held, M.; Turlings, T.C.J.; Gershenzon, J.; Degenhardt, J. The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. Acad. Sci. USA 2006, 103, 1129–1134. [Google Scholar] [CrossRef] [Green Version]
- Kollner, T.G.; Held, M.; Lenk, C.; Hiltpold, I.; Turlings, T.C.J.; Gershenzon, J.; Degenhardt, J. A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 2008, 20, 482–494. [Google Scholar] [CrossRef] [Green Version]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.C.J.; Sartor, R.; Shen, Z.; Briggs, S.P.; Vaughan, M.M.; Alborn, H.T.; et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef] [Green Version]
- Poretsky, E.; Dressano, K.; Weckwerth, P.; Ruiz, M.; Char, S.N.; Shi, D.; Abagyan, R.; Yang, B.; Huffaker, A. Differential activities of maize plant elicitor peptides as mediators of immune signaling and herbivore resistance. Plant J. 2020, 104, 1582–1602. [Google Scholar] [CrossRef]
- Poretsky, E.; Ruiz, M.; Ahmadian, N.; Steinbrenner, A.D.; Dressano, K.; Schmelz, E.A.; Huffaker, A. Comparative analyses of responses to exogenous and endogenous antiherbivore elicitors enable a forward genetics approach to identify maize gene candidates mediating sensitivity to herbivore-associated molecular patterns. Plant J. 2021, 108, 1295–1316. [Google Scholar] [CrossRef]
- Zhuang, X.F.; Kollner, T.G.; Zhao, N.; Li, G.L.; Jiang, Y.F.; Zhu, L.C.; Ma, J.X.; Degenhardt, J.; Chen, F. Dynamic evolution of herbivore-induced sesquiterpene biosynthesis in sorghum and related grass crops. Plant J. 2012, 69, 70–80. [Google Scholar] [CrossRef]
- Christensen, S.A.; Sims, J.; Vaughan, M.M.; Hunter, C.; Block, A.; Willett, D.; Alborn, H.T.; Huffaker, A.; Schmelz, E.A. Commercial hybrids and mutant genotypes reveal complex protective roles for inducible terpenoid defenses in maize. J. Exp. Bot. 2018, 69, 1693–1705. [Google Scholar] [CrossRef] [Green Version]
- Mafu, S.; Ding, Y.Z.; Murphy, K.M.; Yaacoobi, O.; Addison, J.B.; Wang, Q.; Shen, Z.X.; Briggs, S.P.; Bohlmann, J.; Castro-Falcon, G.; et al. Discovery, Biosynthesis and Stress-Related Accumulation of Dolabradiene-Derived Defenses in Maize. Plant Physiol. 2018, 176, 2677–2690. [Google Scholar] [CrossRef]
- Murphy, K.M.; Edwards, J.; Louie, K.B.; Bowen, B.P.; Sundaresan, V.; Northen, T.R.; Zerbe, P. Bioactive diterpenoids impact the composition of the root-associated microbiome in maize (Zea mays). Sci. Rep. 2021, 11, 333. [Google Scholar] [CrossRef]
- Fontana, A.; Held, M.; Fantaye, C.A.; Turlings, T.C.; Degenhardt, J.; Gershenzon, J. Attractiveness of Constitutive and Herbivore-Induced Sesquiterpene Blends of Maize to the Parasitic Wasp Cotesia marginiventris (Cresson). J. Chem. Ecol. 2011, 37, 582–591. [Google Scholar] [CrossRef] [Green Version]
- McMullen, M.D.; Kresovich, S.; Villeda, H.S.; Bradbury, P.; Li, H.; Sun, Q.; Flint-Garcia, S.; Thornsberry, J.; Acharya, C.; Bottoms, C.; et al. Genetic Properties of the Maize Nested Association Mapping Population. Science 2009, 325, 737–740. [Google Scholar] [CrossRef] [Green Version]
- Garms, S.; Köllner, T.G.; Boland, W. A multiproduct terpene synthase from Medicago truncatula generates cadalane sesquiterpenes via two different mechanisms. J. Org. Chem. 2010, 75, 5590–5600. [Google Scholar] [CrossRef]
- Poretsky, E.; Huffaker, A. MutRank: An R shiny web-application for exploratory targeted mutual rank-based coexpression analyses integrated with user-provided supporting information. PeerJ 2020, 8, e10264. [Google Scholar] [CrossRef]
- Dong, L.; Miettinen, K.; Goedbloed, M.; Verstappen, F.W.; Voster, A.; Jongsma, M.A.; Memelink, J.; van der Krol, S.; Bouwmeester, H.J. Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: Similar activity but difference in subcellular localization. Metab. Eng. 2013, 20, 198–211. [Google Scholar] [CrossRef]
- Cornwell, C.P.; Reddy, N.; Leach, D.N.; Wyllie, S.G. Origin of (+)-δ-cadinene and the cubenols in the essential oils of the Myrtaceae. Flavour Fragr. J. 2000, 15, 352–361. [Google Scholar] [CrossRef]
- Tang, H.V.; Berryman, D.L.; Mendoza, J.; Yactayo-Chang, J.P.; Li, Q.B.; Christensen, S.A.; Hunter, C.T.; Best, N.; Soubeyrand, E.; Akhtar, T.A.; et al. Dedicated farnesyl diphosphate synthases circumvent isoprenoid-derived growth-defense tradeoffs in Zea mays. Plant J. 2022, 112, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Karban, R.; Baldwin, I.T. Induced Responses to Herbivory; University of Chicago Press: Chicago, IL, USA, 1997; 319p. [Google Scholar]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef]
- Barry, D.; Alfaro, D.; Darrah, L.L. Relation of European Corn Borer (Lepidoptera: Pyralidae) Leaf-Feeding Resistance and Dimboa Content in Maize. Environ. Entomol. 1994, 23, 177–182. [Google Scholar] [CrossRef]
- Wouters, F.C.; Blanchette, B.; Gershenzon, J.; Vassao, D.G. Plant defense and herbivore counter-defense: Benzoxazinoids and insect herbivores. Phytochem. Rev. 2016, 15, 1127–1151. [Google Scholar] [CrossRef] [Green Version]
- Turlings, T.C.J.; Erb, M. Tritrophic Interactions Mediated by Herbivore-Induced Plant Volatiles: Mechanisms, Ecological Relevance, and Application Potential. Annu. Rev. Entomol. 2018, 63, 433–452. [Google Scholar] [CrossRef]
- Hirsch, C.N.; Foerster, J.M.; Johnson, J.M.; Sekhon, R.S.; Muttoni, G.; Vaillancourt, B.; Penagaricano, F.; Lindquist, E.; Pedraza, M.A.; Barry, K.; et al. Insights into the Maize Pan-Genome and Pan-Transcriptome. Plant Cell 2014, 26, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Schmelz, E.A.; Kaplan, F.; Huffaker, A.; Dafoe, N.J.; Vaughan, M.M.; Ni, X.; Rocca, J.R.; Alborn, H.T.; Teal, P.E. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 5455–5460. [Google Scholar] [CrossRef] [Green Version]
- Huffaker, A.; Kaplan, F.; Vaughan, M.M.; Dafoe, N.J.; Ni, X.; Rocca, J.R.; Alborn, H.T.; Teal, P.E.A.; Schmelz, E.A. Novel Acidic Sesquiterpenoids Constitute a Dominant Class of Pathogen-Induced Phytoalexins in Maize. Plant Physiol. 2011, 156, 2082–2097. [Google Scholar] [CrossRef] [Green Version]
- Schnee, C.; Kollner, T.G.; Gershenzon, J.; Degenhardt, J. The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-beta-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol. 2002, 130, 2049–2060. [Google Scholar] [CrossRef] [Green Version]
- Gouinguené, S.P.; Turlings, T.C.J. The Effects of Abiotic Factors on Induced Volatile Emissions in Corn Plants. Plant Physiol. 2002, 129, 1296–1307. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chen, H.; Yuan, J.S.; Köllner, T.G.; Chen, Y.; Guo, Y.; Zhuang, X.; Chen, X.; Zhang, Y.J.; Fu, J.; et al. The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 2018, 16, 1778–1787. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Stoopen, G.; Yalpani, N.; Vervoort, J.; de Vos, R.; Voster, A.; Verstappen, F.W.; Bouwmeester, H.J.; Jongsma, M.A. Metabolic engineering of geranic acid in maize to achieve fungal resistance is compromised by novel glycosylation patterns. Metab. Eng. 2011, 13, 414–425. [Google Scholar] [CrossRef]
- Guo, J.; Qi, J.; He, K.; Wu, J.; Bai, S.; Zhang, T.; Zhao, J.; Wang, Z. The Asian corn borer Ostrinia furnacalis feeding increases the direct and indirect defence of mid-whorl stage commercial maize in the field. Plant Biotechnol. J. 2019, 17, 88–102. [Google Scholar] [CrossRef] [Green Version]
- Boland, W.; Garms, S. Induced volatiles of Medicago truncatula: Molecular diversity and mechanistic aspects of a multiproduct sesquiterpene synthase from M. truncatula. Flavour Fragr. J. 2010, 25, 114–116. [Google Scholar] [CrossRef]
- Mao, H.; Liu, J.; Ren, F.; Peters, R.J.; Wang, Q. Characterization of CYP71Z18 indicates a role in maize zealexin biosynthesis. Phytochemistry 2016, 121, 4–10. [Google Scholar] [CrossRef]
- Köllner, T.G.; Schnee, C.; Gershenzon, J.; Degenhardt, J. The sesquiterpene hydrocarbons of maize (Zea mays) form five groups with distinct developmental and organ-specific distributions. Phytochemistry 2004, 65, 1895–1902. [Google Scholar] [CrossRef]
- Zerbe, P.; Bohlmann, J. Plant diterpene synthases: Exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 2015, 33, 419–428. [Google Scholar] [CrossRef]
- Liang, Z.; Qiu, Y.; Schnable, J.C. Genome-Phenome Wide Association in Maize and Arabidopsis Identifies a Common Molecular and Evolutionary Signature. Mol. Plant 2020, 13, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.M.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef]
- Schmelz, E.A.; Engelberth, J.; Tumlinson, J.H.; Block, A.; Alborn, H.T. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 2004, 39, 790–808. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry; Allured publishing corporation: Carol Stream, IL, USA, 2007; Volume 456. [Google Scholar]
- Bos, R.; Woerdenbag, H.J.; Kayser, O.; Quax, W.J.; Ruslan, K. Elfami, Essential Oil Constituents of Piper cubeba L. fils. from Indonesia. J. Essent. Oil Res. 2007, 19, 14–17. [Google Scholar] [CrossRef]
- Samayoa, L.F.; Malvar, R.A.; Olukolu, B.A.; Holland, J.B.; Butron, A. Genome-wide association study reveals a set of genes associated with resistance to the Mediterranean corn borer (Sesamia nonagrioides L.) in a maize diversity panel. BMC Plant Biol. 2015, 15, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Sadre, R.; Kuo, P.; Chen, J.; Yang, Y.; Banerjee, A.; Benning, C.; Hamberger, B. Cytosolic lipid droplets as engineered organelles for production and accumulation of terpenoid biomaterials in leaves. Nat. Commun. 2019, 10, 853. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Portwood, J.L., 2nd; Woodhouse, M.R.; Cannon, E.K.; Gardiner, J.M.; Harper, L.C.; Schaeffer, M.L.; Walsh, J.R.; Sen, T.Z.; Cho, K.T.; Schott, D.A.; et al. MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Res. 2019, 47, D1146–D1154. [Google Scholar] [CrossRef] [Green Version]
- Deorowicz, S.; Debudaj-Grabysz, A.; Gudyś, A. FAMSA: Fast and accurate multiple sequence alignment of huge protein families. Sci. Rep. 2016, 6, 33964. [Google Scholar] [CrossRef] [PubMed]
- Steenwyk, J.L.; Buida III, T.J.; Li, Y.; Shen, X.-X.; Rokas, A. ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 2020, 18, e3001007. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Fu, J.; Ren, F.; Lu, X.; Mao, H.; Xu, M.; Degenhardt, J.; Peters, R.J.; Wang, Q. A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol. 2016, 170, 742–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, A.; Schaff, C.; Zhang, Z.; Lipka, A.E.; Tian, F.; Köllner, T.G.; Schnee, C.; Preiß, S.; Irmisch, S.; Jander, G.; et al. Characterization of biosynthetic pathways for the production of the volatile homoterpenes DMNT and TMTT in Zea mays. Plant Cell 2016, 28, 2651–2665. [Google Scholar] [CrossRef] [Green Version]
- Kollner, T.G.; Schnee, C.; Li, S.; Svatos, A.; Schneider, B.; Gershenzon, J. and Degenhardt, J. Protonation of a neutral (S)-β-bisabolene intermediate is involved in (S)-β-macrocarpene formation by the maize sesquiterpene synthases TPS6 and TPS11. J. Biol. Chem. 2008, 283, 20779–20788. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.M.; Ma, L.T.; Ding, Y.; Schmelz, E.A.; Zerbe, P. Funce products of a single maize sesquiterpene synthase form a volatile defense signathways in maize (Zea mays). Front. Plant Sci. 2018, 9, 1542. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.J.; Saparno, A.; Johnston, A.; Prisic, S.; Xu, M.; Allard, S.; Kathiresan, A.; Ouellet, T.; Peters, R.J. The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol. Biol. 2005, 59, 881–894. [Google Scholar] [CrossRef]
- Ren, F.; Mao, H.; Liang, J.; Liu, J.; Shu, K.; Wang, Q. Functional characterization of ZmTPS7 reveals a maize τ-cadinol synthase involved in stress response. Planta 2016, 244, 1065–1074. [Google Scholar] [CrossRef]
- Bensen, R.J.; Johal, G.S.; Crane, V.C.; Tossberg, J.T.; Schnable, P.S.; Meeley, R.B.; Briggs, S.P. Cloning and characterization of the maize An1 gene. Plant Cell 1995, 7, 75–84. [Google Scholar]
- Lin, C.; Shen, B.; Xu, Z.; Köllner, T.G.; Degenhardt, J.; Dooner, H.K. Characterization of the monoterpene synthase gene tps26, the ortholog of a gene induced by insect herbivory in maize. Plant Physiol. 2008, 146, 940–951. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Zheng, Z.; Dooner, H.K. A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: Characterization of wild-type and mutant alleles. Proc. Natl. Acad. Sci. USA 2000, 97, 14807–14812. [Google Scholar] [CrossRef] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saldivar, E.V.; Ding, Y.; Poretsky, E.; Bird, S.; Block, A.K.; Huffaker, A.; Schmelz, E.A. Maize Terpene Synthase 8 (ZmTPS8) Contributes to a Complex Blend of Fungal-Elicited Antibiotics. Plants 2023, 12, 1111. https://doi.org/10.3390/plants12051111
Saldivar EV, Ding Y, Poretsky E, Bird S, Block AK, Huffaker A, Schmelz EA. Maize Terpene Synthase 8 (ZmTPS8) Contributes to a Complex Blend of Fungal-Elicited Antibiotics. Plants. 2023; 12(5):1111. https://doi.org/10.3390/plants12051111
Chicago/Turabian StyleSaldivar, Evan V., Yezhang Ding, Elly Poretsky, Skylar Bird, Anna K. Block, Alisa Huffaker, and Eric A. Schmelz. 2023. "Maize Terpene Synthase 8 (ZmTPS8) Contributes to a Complex Blend of Fungal-Elicited Antibiotics" Plants 12, no. 5: 1111. https://doi.org/10.3390/plants12051111





