Time-Dependent Proteomic Signatures Associated with Embryogenic Callus Induction in Carica papaya L.
Abstract
:1. Introduction
2. Results
2.1. Embryogenic Callus Induction
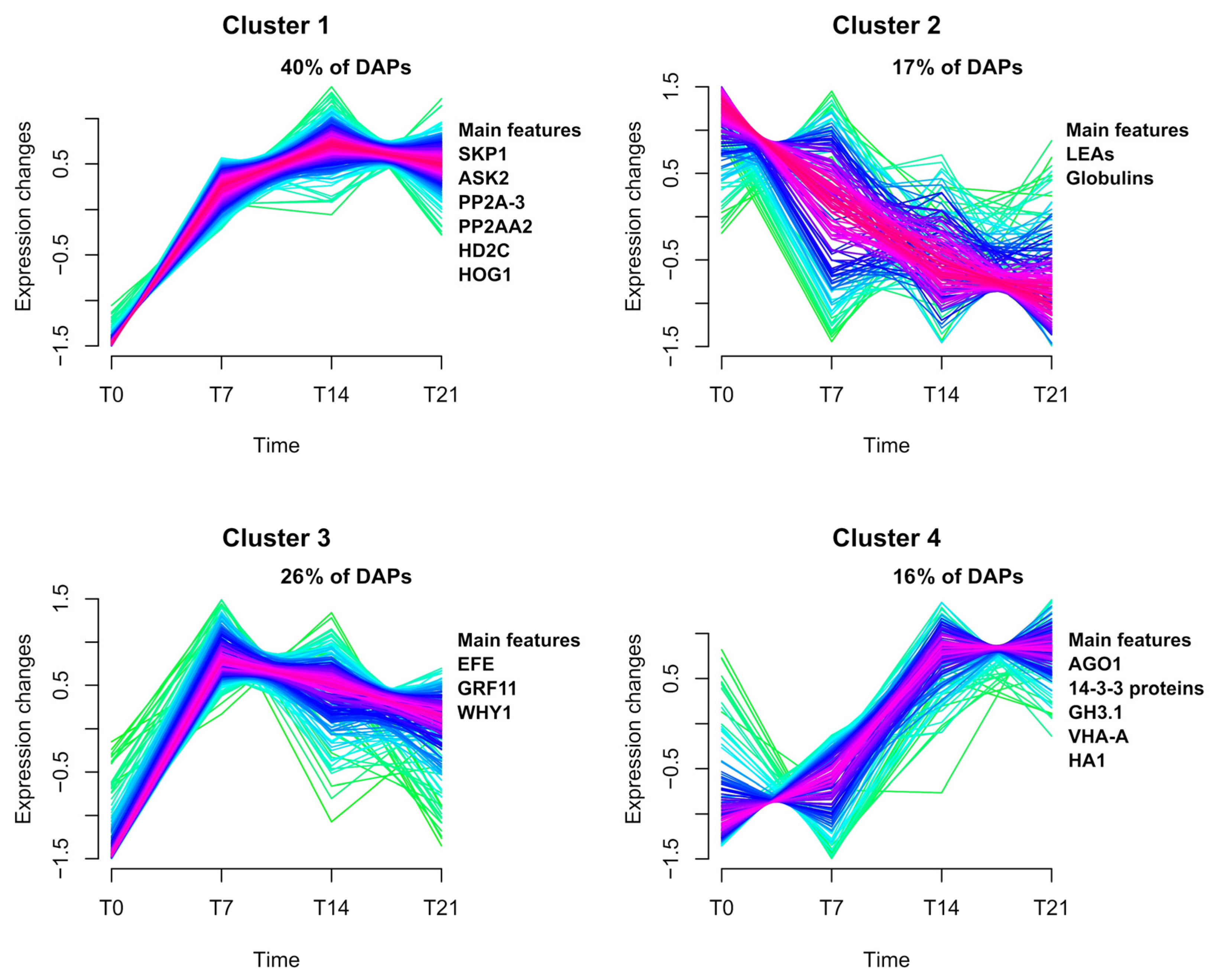
2.2. Proteomics Analysis during Embryogenic Callus Induction
2.3. Functional analysis of Proteomic Data
2.4. Identification of Regulatory Proteins Differentially Accumulated during Induction
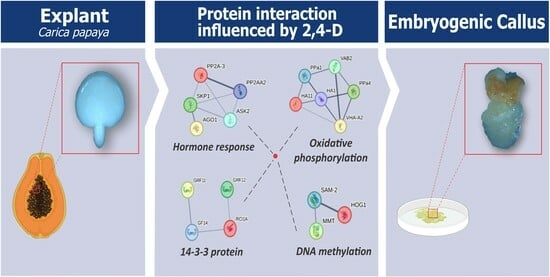
2.5. Prediction of Protein-Protein Interaction Networks
3. Discussion
3.1. The Explant Contains Storage Reserves That Are Consumed during the Induction of Embryogenic Callus through Changes in Energy Metabolism
3.2. Acquisition of Embryogenic Competence in Somatic Cells Involves Modulation of Regulatory Proteins and Epigenetic Mechanisms
3.3. Proteins Involved in Hormone Responses Are Induced Early after Callus Induction
4. Materials and Methods
4.1. Plant Material and Callus Induction
4.2. Protein Extraction and Digestion
4.3. Bottom-Up Proteomics Analysis
4.4. Proteomics Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Salinas, I.; Salehi, M.; Hueso, J.J.; Cuevas, J. Assessment of two sex-determining procedures in ‘BH-65’ papaya from an economical and developmental point of view. Fruits 2018, 73, 184–190. [Google Scholar] [CrossRef]
- Xavier, L.; Almeida, F.A.; Pinto, V.B.; Silveira, V. Large-scale regeneration of hermaphrodite emblings of Carica papaya L. Golden using early molecular sex determination during embryogenic callus multiplication. Plant Cell Tissue Organ Cult. 2021, 146, 643–649. [Google Scholar] [CrossRef]
- Gouvea, D.S.; Chagas, K.; Cipriano, J.L.D.; Lopes, J.C.; Schmildt, E.R.; Otoni, W.C.; Schmildt, O.; Araujo, C.P.D.; Alexandre, R.S. Somatic embryogenesis in the commercial papaya hybrid UENF/Caliman 01 relying on plantlet production from sexed adult hermaphrodite donor plants. An. Acad. Bras. Cienc. 2019, 91, e20180504. [Google Scholar] [CrossRef]
- Almeida, F.A.; Vale, E.M.; Reis, R.S.; Santa-Catarina, C.; Silveira, V. LED lamps enhance somatic embryo maturation in association with the differential accumulation of proteins in the Carica papaya L. ‘Golden’ embryogenic callus. Plant Physiol. Biochem. 2019, 143, 109–118. [Google Scholar] [CrossRef]
- Al-Shara, B.; Taha, R.M.; Mohamad, J.; Elias, H.; Khan, A. Somatic embryogenesis and plantlet regeneration in the Carica papaya L. Cv. Eksotika. Plants 2020, 9, 360. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Wójcik, A.M.; Gaj, M.D. Epigenetic regulation of auxin-induced somatic embryogenesis in plants. Int. J. Mol. Sci. 2020, 21, 2307. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Jaskóła, K.; Gąsiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef]
- Aguilar-Hernández, V.; Loyola-Vargas, V.M. Advanced proteomic approaches to elucidate somatic embryogenesis. Front. Plant Sci. 2018, 9, 1658. [Google Scholar] [CrossRef]
- Botini, N.; Almeida, F.A.; Cruz, K.; Reis, R.S.; Vale, E.M.; Garcia, A.B.; Santa-Catarina, C.; Silveira, V. Stage-specific protein regulation during somatic embryo development of Carica papaya L. ‘Golden’. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140561. [Google Scholar] [CrossRef]
- Vale, E.M.; Santana, D.B.; Reis, R.S.; Sousa, K.R.; de Souza Filho, G.A.; Oliveira, J.G.D.; Santa-Catarina, C.; Silveira, V. Mitochondrial proteomics reveals new insights into embryogenic competence acquisition in Carica papaya L. callus. J. Proteom. 2022, 252, 104434. [Google Scholar] [CrossRef]
- Kumar, V.; Jha, P.; Van Staden, J. LEAFY COTYLEDONs (LECs): Master regulators in plant embryo development. Plant Cell Tissue Organ Cult. 2020, 140, 475–487. [Google Scholar] [CrossRef]
- Gliwicka, M.; Nowak, K.; Cieśla, E.; Gaj, M.D. Expression of seed storage product genes (CRA1 and OLEO4) in embryogenic cultures of somatic tissues of Arabidopsis. Plant Cell Tissue Organ Cult. 2012, 109, 235–245. [Google Scholar] [CrossRef]
- Ghelis, T.; Bolbach, G.r.; Clodic, G.; Habricot, Y.; Miginiac, E.; Sotta, B.; Jeannette, E. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in arabidopsis seeds and suspension cells. Plant Physiol. 2008, 148, 1668–1680. [Google Scholar] [CrossRef]
- Tang, L.P.; Zhang, X.S.; Su, Y.H. Regulation of cell reprogramming by auxin during somatic embryogenesis. aBIOTECH 2020, 1, 185–193. [Google Scholar] [CrossRef]
- Amara, I.; Zaidi, I.; Masmoudi, K.; Ludevid, M.D.; Pagés, M.; Goday, A.; Brini, F. Insights into Late Embryogenesis Abundant (LEA) proteins in plants: From structure to the functions. Am. J. Plant Sci. 2014, 5, 3440–3455. [Google Scholar] [CrossRef]
- Morończyk, J.; Brąszewska, A.; Wójcikowska, B.; Chwiałkowska, K.; Nowak, K.; Wójcik, A.M.; Kwaśniewski, M.; Gaj, M.D. Insights into the histone acetylation-mediated regulation of the transcription factor genes that control the embryogenic transition in the somatic cells of Arabidopsis. Cells 2022, 11, 863. [Google Scholar] [CrossRef]
- Wickramasuriya, A.M.; Dunwell, J.M. Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genom. 2015, 16, 301. [Google Scholar] [CrossRef]
- Yakovlev, I.A.; Carneros, E.; Lee, Y.; Olsen, J.E.; Fossdal, C.G. Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta 2016, 243, 1237–1249. [Google Scholar] [CrossRef]
- Xavier, L.R.; Almeida, F.A.; Pinto, V.B.; Passamani, L.Z.; Santa-Catarina, C.; de Souza Filho, G.A.; Mooney, B.P.; Thelen, J.J.; Silveira, V. Integrative proteomics and phosphoproteomics reveals phosphorylation networks involved in the maintenance and expression of embryogenic competence in sugarcane callus. J. Plant Physiol. 2022, 268, 153587. [Google Scholar] [CrossRef]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What these terms mean in the era of molecular plant biology? Front. Plant Sci. 2019, 10, 536. [Google Scholar] [CrossRef]
- Osorio-Montalvo, P.; Sáenz-Carbonell, L.; De-la-Peña, C. 5-Azacytidine: A promoter of epigenetic changes in the quest to improve plant somatic embryogenesis. Int. J. Mol. Sci. 2018, 19, 3182. [Google Scholar] [CrossRef]
- Almeida, F.A.; Passamani, L.Z.; Santa-Catarina, C.; Mooney, B.P.; Thelen, J.J.; Silveira, V. Label-free quantitative phosphoproteomics reveals signaling dynamics involved in embryogenic competence acquisition in sugarcane. J. Proteome Res. 2020, 19, 4145–4157. [Google Scholar] [CrossRef]
- Yue, K.; Sandal, P.; Williams, E.L.; Murphy, E.; Stes, E.; Nikonorova, N.; Ramakrishna, P.; Czyzewicz, N.; Montero-Morales, L.; Kumpf, R.; et al. PP2A-3 interacts with ACR4 and regulates formative cell division in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2016, 113, 1447–1452. [Google Scholar] [CrossRef]
- Karampelias, M.; Neyt, P.; De Groeve, S.; Aesaert, S.; Coussens, G.; Rolčík, J.; Bruno, L.; De Winne, N.; Van Minnebruggen, A.; Van Montagu, M.; et al. ROTUNDA3 function in plant development by phosphatase 2A-mediated regulation of auxin transporter recycling. Proc. Natl. Acad. Sci. USA 2016, 113, 2768–2773. [Google Scholar] [CrossRef]
- Chiu, L.-W.; Heckert, M.J.; You, Y.; Albanese, N.; Fenwick, T.; Siehl, D.L.; Castle, L.A.; Tao, Y. Members of the GH3 family of proteins conjugate 2,4-D and dicamba with aspartate and glutamate. Plant Cell Physiol. 2018, 59, 2366–2380. [Google Scholar] [CrossRef]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2006, 18, 626–638. [Google Scholar] [CrossRef]
- Yoon, G.M.; Kieber, J.J. 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 2013, 25, 1016–1028. [Google Scholar] [CrossRef]
- Catalá, R.; López-Cobollo, R.; Mar Castellano, M.; Angosto, T.; Alonso, J.M.; Ecker, J.R.; Salinas, J. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 2014, 26, 3326–3342. [Google Scholar] [CrossRef]
- Bai, B.; Su, Y.H.; Yuan, J.; Zhang, X.S. Induction of somatic embryos in arabidopsis requires local YUCCA expression mediated by the down-regulation of ethylene biosynthesis. Mol. Plant 2013, 6, 1247–1260. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Nanjo, Y.; Skultety, L.; Uváčková, L.u.; Klubicová, K.; Hajduch, M.; Komatsu, S. Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteome Res. 2012, 11, 372–385. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 2014, 11, 167–170. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2018, 47, D442–D450. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing, R version 4.3.1 (2023-06-16 ucrt)—“Beagle Scouts”; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef] [PubMed]


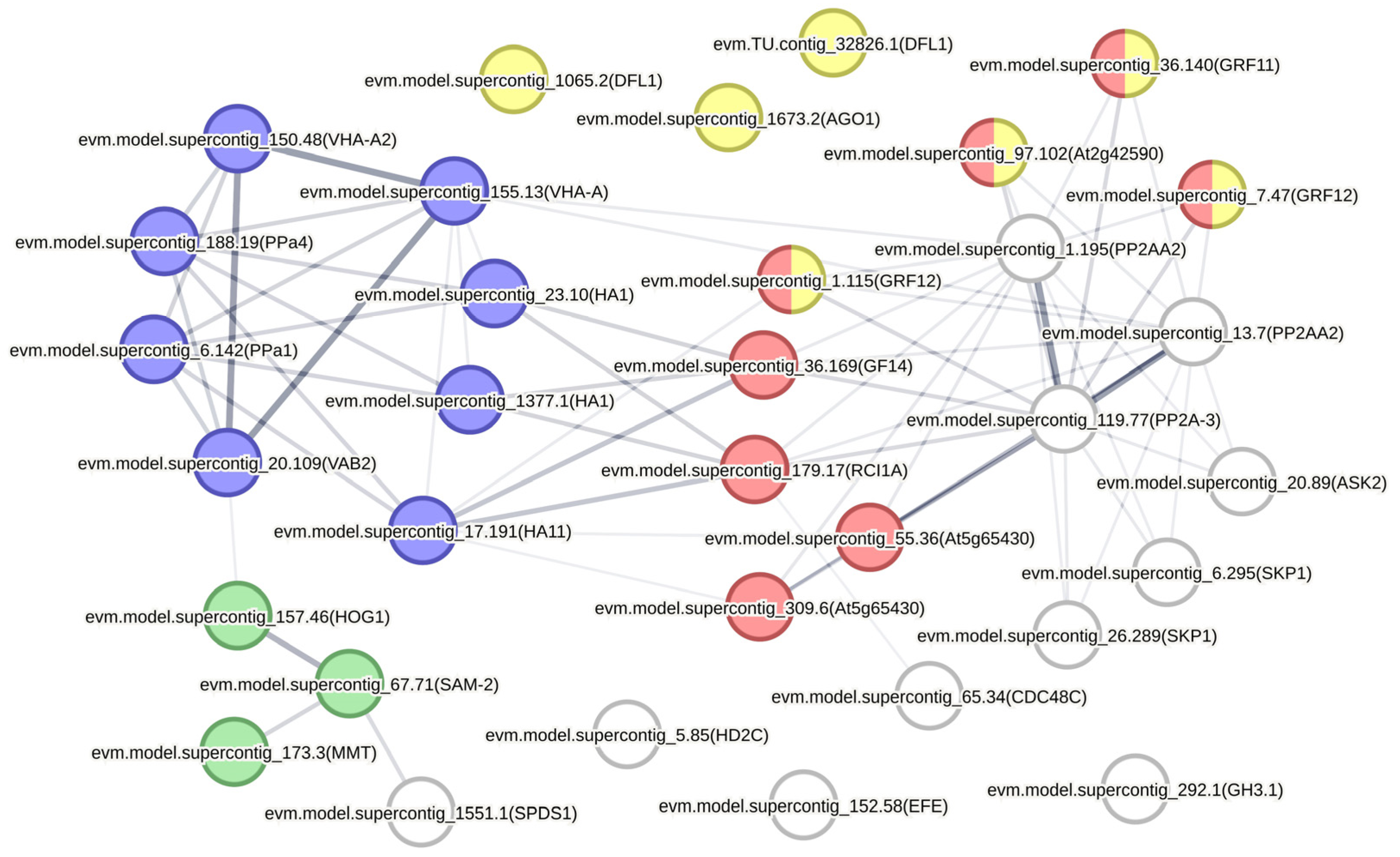
| Protein ID | BLAST Top Hit | Arabidopsis Orthologs | Cluster |
|---|---|---|---|
| evm.model.supercontig_1673.2 | protein argonaute 1 | AGO1 | 4 |
| evm.model.supercontig_20.89 | SKP1-like protein 1B | ASK2 | 1 |
| evm.model.supercontig_97.102 | 14-3-3-like protein D isoform X2 | At2g42590 | 1 |
| evm.model.supercontig_309.6 | 14-3-3-like protein B | At5g65430 | 4 |
| evm.model.supercontig_55.36 | 14-3-3-like protein GF14 kappa isoform X2 | At5g65430 | 4 |
| evm.model.supercontig_65.34 | cell division cycle protein 48 homolog | CDC48C | Unique_T7+ |
| evm.model.supercontig_1065.2 | indole-3-acetic acid-amido synthetase GH3.6 | DFL1 | Unique_T7+ |
| evm.TU.contig_32826.1 | indole-3-acetic acid-amido synthetase GH3.6 | DFL1 | Unique_T7+ |
| evm.model.supercontig_152.58 | 1-aminocyclopropane-1-carboxylate oxidase | EFE | 3 |
| evm.model.supercontig_36.169 | 14-3-3-like protein | GF14 | 1 |
| evm.model.supercontig_292.1 | probable indole-3-acetic acid-amido synthetase GH3.1 | GH3.1 | 4 |
| evm.model.supercontig_36.140 | 14-3-3-like protein | GRF11 | 3 |
| evm.model.supercontig_7.47 | putative 14-3-3 protein | GRF12 | 4 |
| evm.model.supercontig_1.115 | 14-3-3-like protein GF14 iota | GRF12 | 4 |
| evm.model.supercontig_23.10 | plasma membrane ATPase 4 | HA1 | 4 |
| evm.model.supercontig_1377.1 | plasma membrane ATPase 4 | HA1 | Unique_T7+ |
| evm.model.supercontig_17.191 | ATPase 11, plasma membrane-type | HA11 | 1 |
| evm.model.supercontig_5.85 | histone deacetylase HDT1 | HD2C | 1 |
| evm.model.supercontig_157.46 | adenosylhomocysteinase | HOG1 | 1 |
| evm.model.supercontig_173.3 | methionine S-methyltransferase | MMT | Unique_T7+ |
| evm.model.supercontig_119.77 | serine/threonine-protein phosphatase PP2A-2 catalytic subunit | PP2A-3 | 1 |
| evm.model.supercontig_13.7 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | PP2AA2 | 1 |
| evm.model.supercontig_1.195 | serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | PP2AA2 | 1 |
| evm.model.supercontig_6.142 | soluble inorganic pyrophosphatase-like | PPa1 | Unique_T7+ |
| evm.model.supercontig_188.19 | soluble inorganic pyrophosphatase 4 | PPa4 | Unique_T7+ |
| evm.model.supercontig_179.17 | 14-3-3-like protein A | RCI1A | 1 |
| evm.model.supercontig_67.71 | S-adenosylmethionine synthase 2 | SAM-2 | 2 |
| evm.model.supercontig_26.289 | SKP1-like protein 1B | SKP1 | 1 |
| evm.model.supercontig_6.295 | SKP1-like protein 1B | SKP1 | Unique_T7+ |
| evm.model.supercontig_1551.1 | spermidine synthase-like | SPDS1 | Unique_T7+ |
| evm.model.supercontig_20.109 | V-type proton ATPase subunit B 2 | VAB2 | 1 |
| evm.model.supercontig_155.13 | V-type proton ATPase catalytic subunit A | VHA-A | 4 |
| evm.model.supercontig_150.48 | V-type proton ATPase subunit a3 | VHA-A2 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xavier, L.R.; Corrêa, C.C.G.; da Paschoa, R.P.; Vieira, K.d.S.; Pacheco, D.D.R.; Gomes, L.d.E.S.; Duncan, B.C.; da Conceição, L.d.S.; Pinto, V.B.; Santa-Catarina, C.; et al. Time-Dependent Proteomic Signatures Associated with Embryogenic Callus Induction in Carica papaya L. Plants 2023, 12, 3891. https://doi.org/10.3390/plants12223891
Xavier LR, Corrêa CCG, da Paschoa RP, Vieira KdS, Pacheco DDR, Gomes LdES, Duncan BC, da Conceição LdS, Pinto VB, Santa-Catarina C, et al. Time-Dependent Proteomic Signatures Associated with Embryogenic Callus Induction in Carica papaya L. Plants. 2023; 12(22):3891. https://doi.org/10.3390/plants12223891
Chicago/Turabian StyleXavier, Lucas Rodrigues, Caio Cezar Guedes Corrêa, Roberta Pena da Paschoa, Karina da Silva Vieira, Daniel Dastan Rezabala Pacheco, Lucas do Espirito Santo Gomes, Bárbara Cardoso Duncan, Laís dos Santos da Conceição, Vitor Batista Pinto, Claudete Santa-Catarina, and et al. 2023. "Time-Dependent Proteomic Signatures Associated with Embryogenic Callus Induction in Carica papaya L." Plants 12, no. 22: 3891. https://doi.org/10.3390/plants12223891