The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice
Abstract
:1. Introduction
2. Results
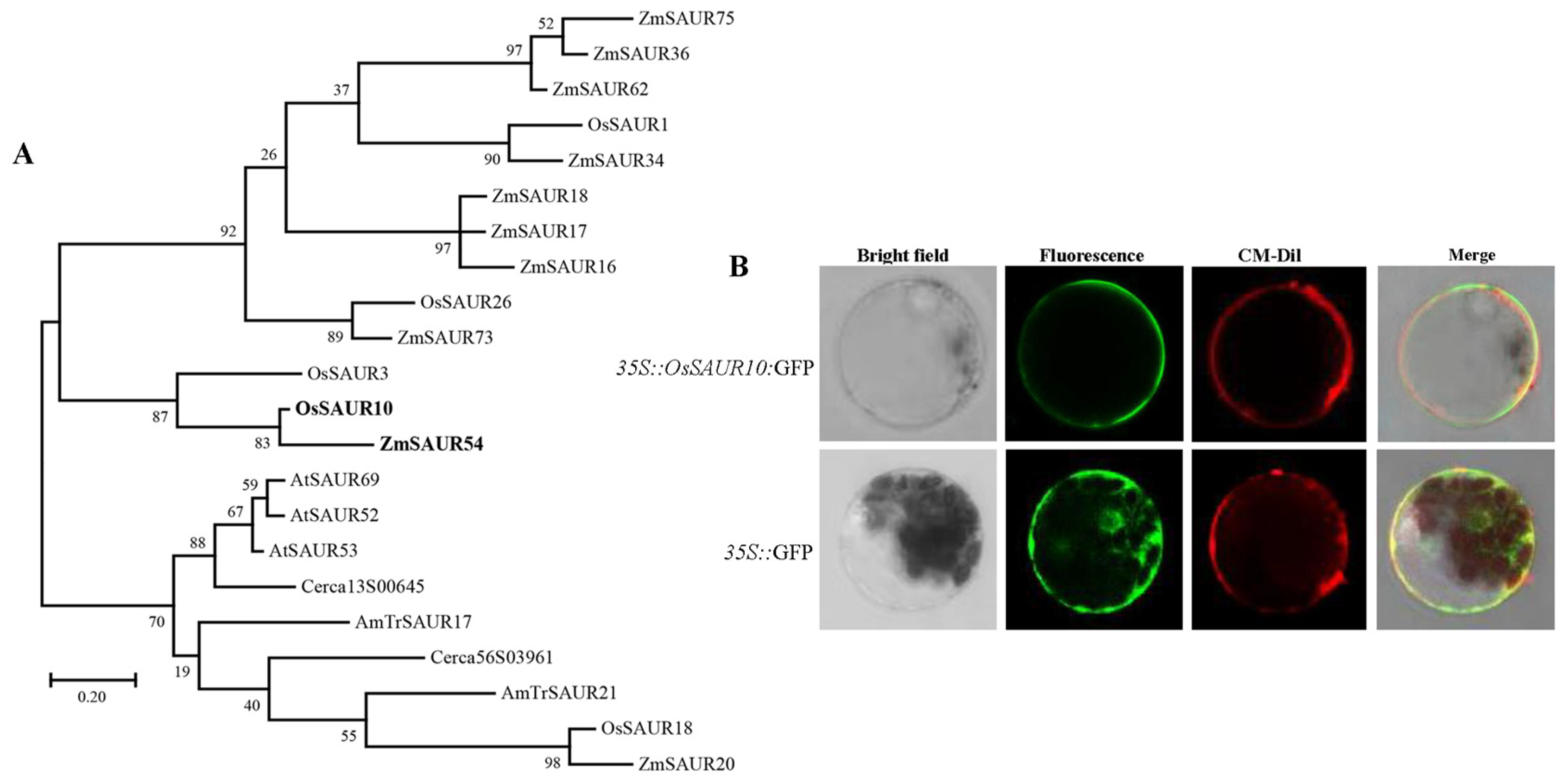
2.1. Structural and Phylogenetic Characterization of OsSAUR10
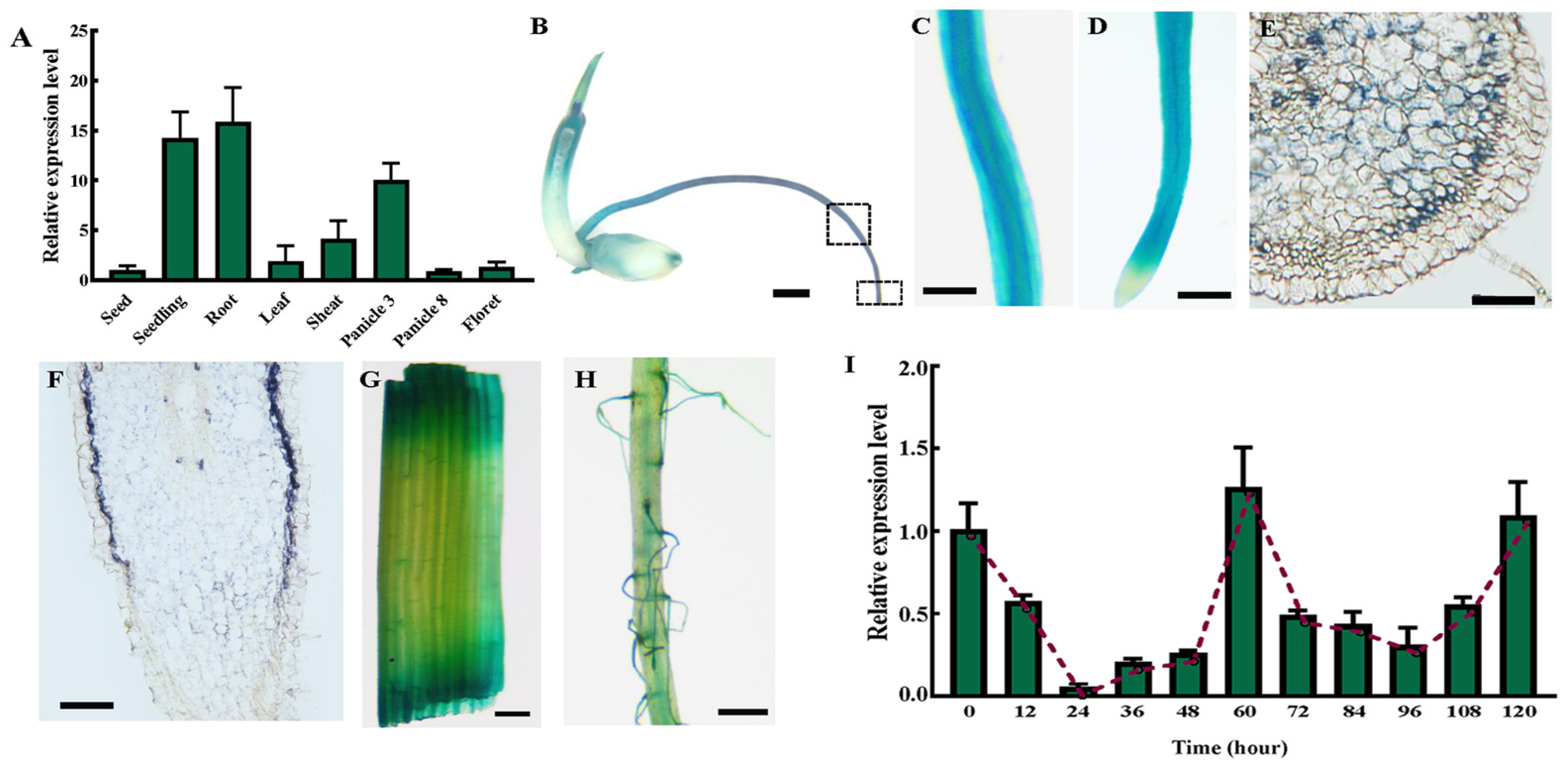
2.2. OsSAUR10 Had a Distinctive Expression Pattern and Was Expressed in a Circadian Manner
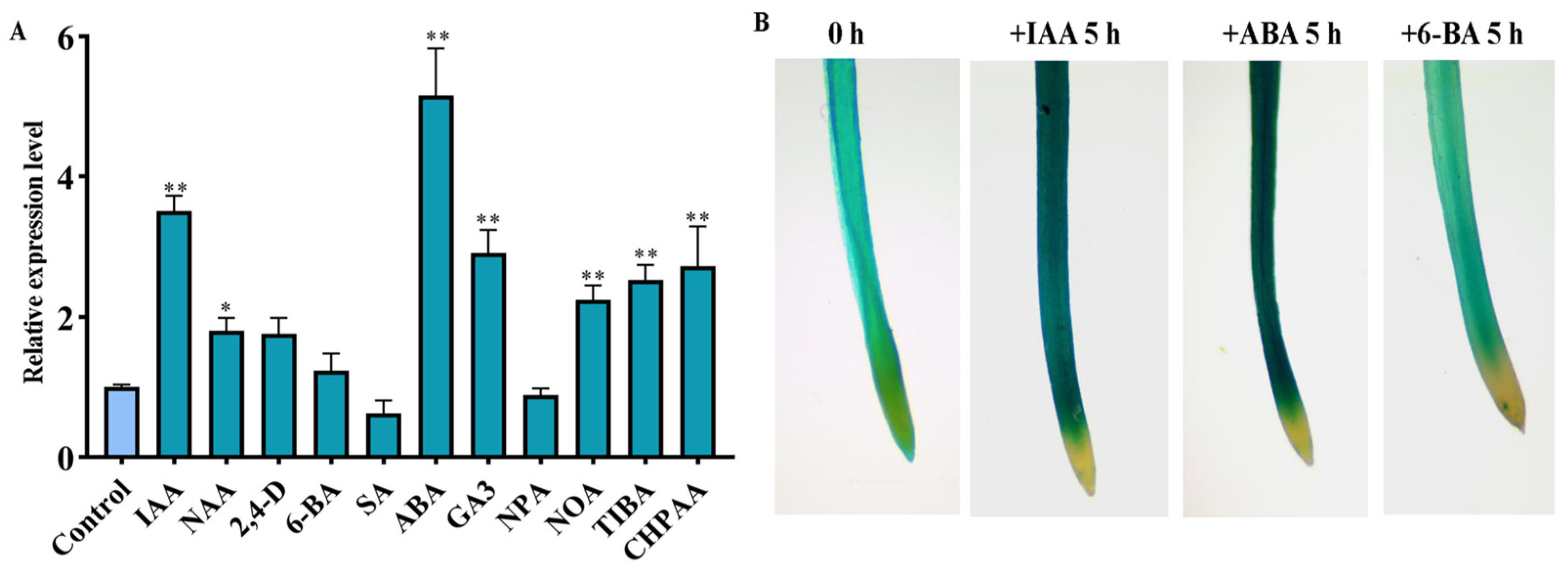
2.3. OsSAUR10 Positively Responded to Varied Phytohormone Signals
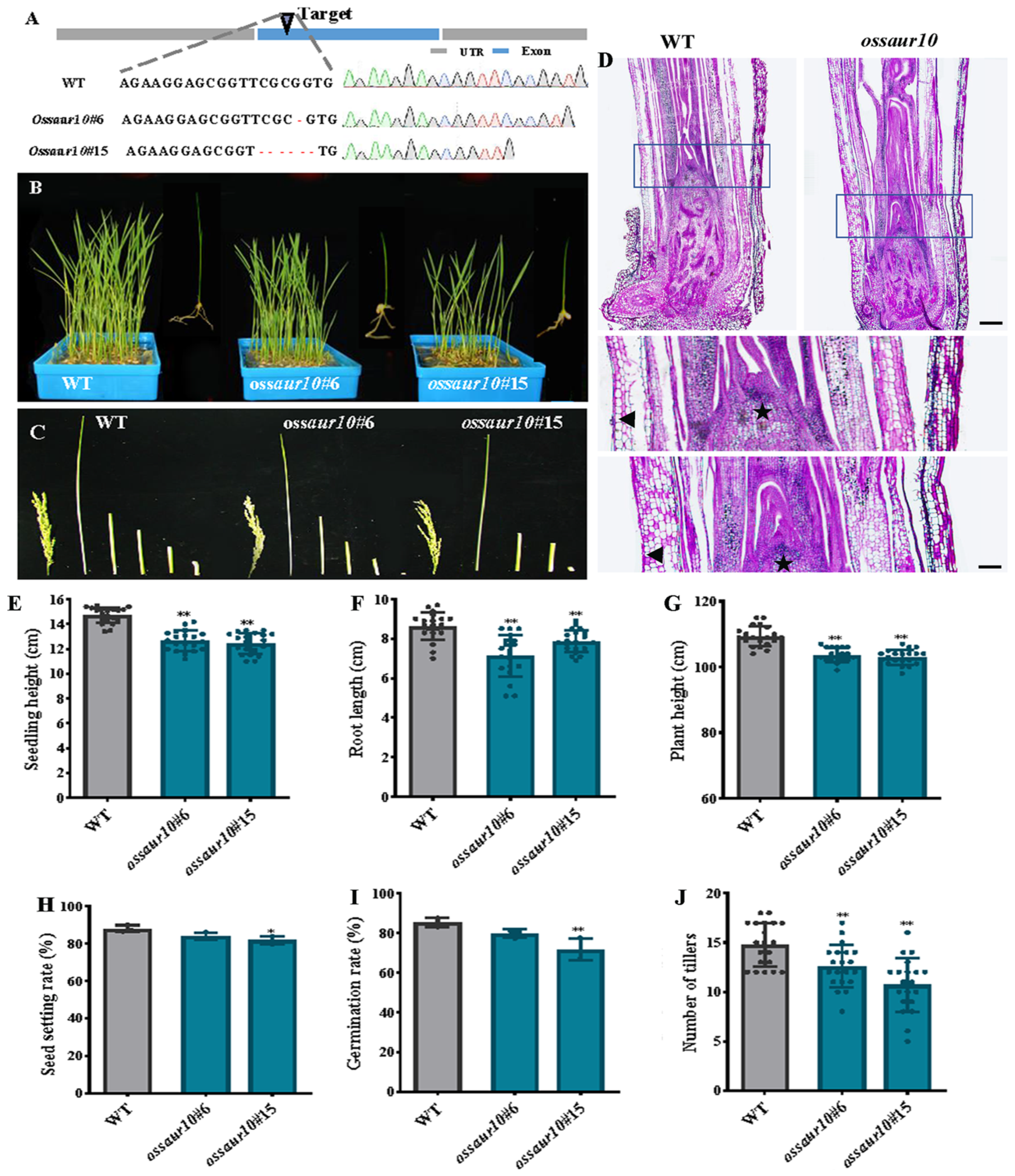
2.4. The OsSAUR10 Mutants Impeded Seedling Growth
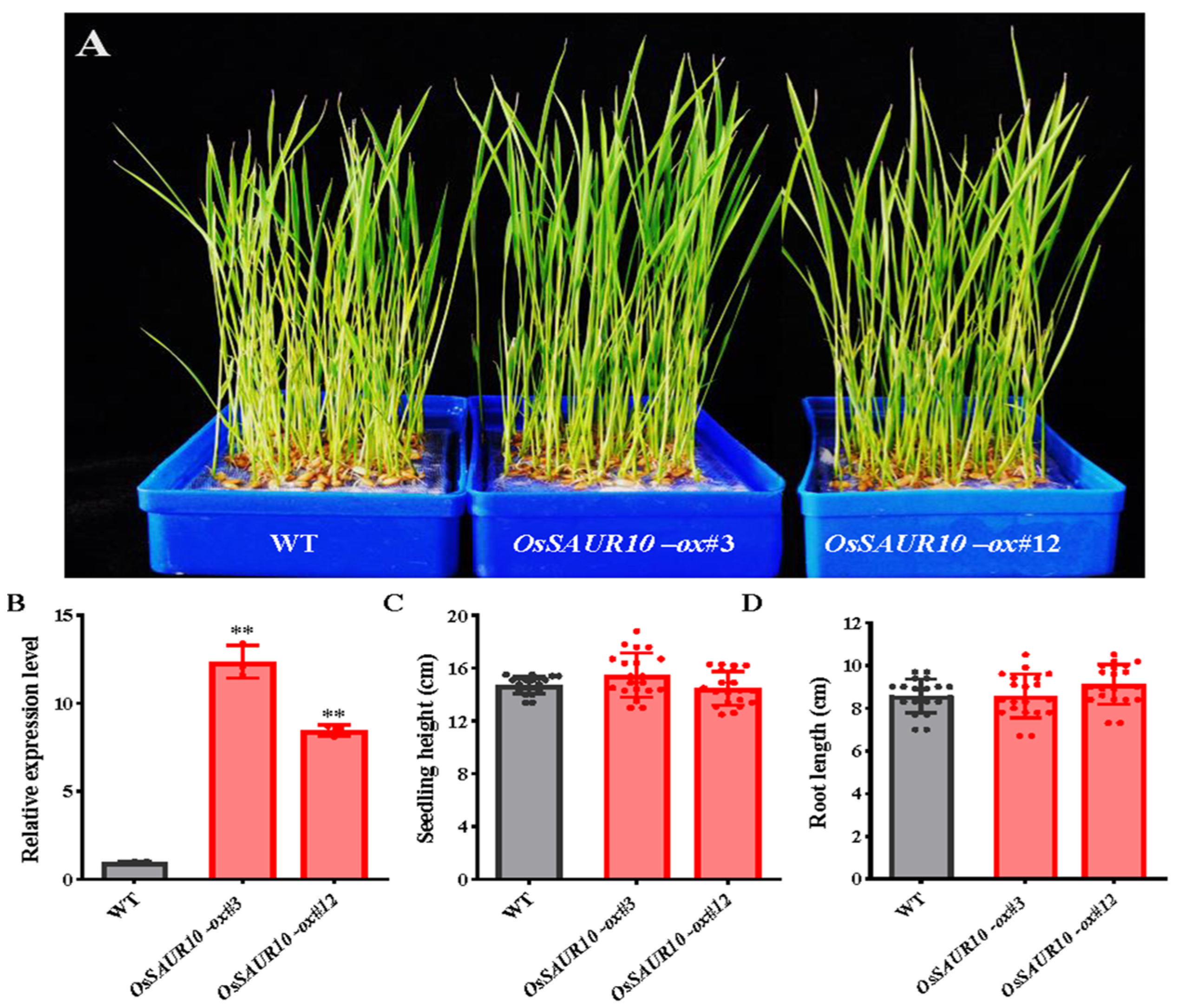
2.5. OsSAUR10-Overexpression Displayed No Apparent Effect on Seedling Growth
2.6. OsSAUR10 Influenced Auxin Synthesis and Transport
3. Discussion
3.1. OsSAUR10 Positively Regulates Seedling Growth in Rice
3.2. Tissue-Specific and Circadian-Regulated OsSAUR10 Is Inducible by Varied Phytohormones
3.3. OsSAUR10 Regulates Seedlings’ Growth by Impacting Auxin Production and Its Polar Transport
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogenetic and Promoter Analysis
4.3. Subcellular Localization Assay
4.4. GUS Staining
4.5. RNA Isolation and qRT-PCR Analysis
4.6. Vector Construction and Transgenic Rice Plants
4.7. Histological Observation
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.A.; Guilfoyle, T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 1987, 9, 611–623. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; Lou, Y.; Ming, F.; Jin, Y. Genome-wide identification and characterization of small auxin-up RNA (SAUR) gene family in plants: Evolution and expression profiles during normal growth and stress response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, X.; Cao, J. Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J. Integr. Plant Biol. 2014, 56, 133–150. [Google Scholar] [CrossRef]
- Lv, W.; He, X.; Guo, H.; Lan, H.; Jiao, Y.; Li, L.; Lian, Y.; Wang, Z.; Xin, Z.; Ren, Y.; et al. Genome-wide identification of tasaur gene family members in hexaploid wheat and functional characterization of TaSAUR66-5B in improving nitrogen use efficiency. Int. J. Mol. Sci. 2022, 23, 7574. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef]
- Sullivan, M.L.; Green, P.J. Mutational analysis of the DST element in tobacco cells and transgenic plants: Identification of residues critical for mRNA instability. RNA 1996, 2, 308–315. [Google Scholar]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef]
- Xu, Y.X.; Xiao, M.Z.; Liu, Y.; Fu, J.L.; He, Y.; Jiang, D.A. The small auxin-up RNA OsSAUR45 affects auxin synthesis and transport in rice. Plant Mol. Biol. 2017, 94, 97–107. [Google Scholar] [CrossRef]
- Knauss, S.; Rohrmeier, T.; Lehle, L. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J. Biol. Chem. 2003, 278, 23936–23943. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- He, S.L.; Hsieh, H.L.; Jauh, G.Y. SMALL AUXIN UP RNA62/75 are required for the translation of transcripts essential for pollen tube growth. Plant Physiol. 2018, 178, 626–640. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, Y.; Li, M.; Lamin-Samu, A.T.; Yang, D.; Yu, X.; Izhar, M.; Jan, I.; Ali, M.; Lu, G. The Arabidopsis SMALL AUXIN UP RNA32 protein regulates ABA-mediated responses to drought stress. Front. Plant Sci. 2021, 12, 625493. [Google Scholar] [CrossRef]
- Kathare, P.K.; Dharmasiri, S.; Dharmasiri, N. SAUR53 regulates organ elongation and apical hook development in Arabidopsis. Plant Signal. Behav. 2018, 13, e1514896. [Google Scholar] [CrossRef]
- Kong, Y.; Zhu, Y.; Gao, C.; She, W.; Lin, W.; Chen, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013, 54, 609–621. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inze, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef]
- Gastaldi, V.; Lucero, L.E.; Ferrero, L.V.; Ariel, F.D.; Gonzalez, D.H. Class-I TCP Transcription factors activate the SAUR63 Gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 2020, 182, 2096–2110. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, Y.S.; Yoon, H.K.; Park, C.M. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007, 172, 150–157. [Google Scholar] [CrossRef]
- Chae, K.; Isaacs, C.G.; Reeves, P.H.; Maloney, G.S.; Muday, G.K.; Nagpal, P.; Reed, J.W. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. Cell Mol. Biol. 2012, 71, 684–697. [Google Scholar] [CrossRef] [PubMed]
- van Mourik, H.; van Dijk, A.D.J.; Stortenbeker, N.; Angenent, G.C.; Bemer, M. Divergent regulation of Arabidopsis SAUR genes: A focus on the SAUR10-clade. BMC Plant Biol. 2017, 17, 245. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Chen, Y.; Li, M.; Kong, Y.; Zhu, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily of small auxin up RNA genes: Potential implications. Plant Signal. Behav. 2013, 8, e25283. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef]
- Bai, M.Y.; Shang, J.X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.P.; Wang, Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef]
- Favero, D.S.; Le, K.N.; Neff, M.M. Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J. 2017, 89, 1133–1145. [Google Scholar] [CrossRef]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Qiu, T.; Qi, M.; Ding, X.; Zheng, Y.; Zhou, T.; Chen, Y.; Han, N.; Zhu, M.; Bian, H.; Wang, J. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2020, 125, 805–819. [Google Scholar] [CrossRef]
- Strader, L.C.; Zhao, Y. Auxin perception and downstream events. Curr. Opin. Plant Biol. 2016, 33, 8–14. [Google Scholar] [CrossRef]
- Ishida, J.K.; Wakatake, T.; Yoshida, S.; Takebayashi, Y.; Kasahara, H.; Wafula, E.; de Pamphilis, C.W.; Namba, S.; Shirasu, K. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell 2016, 28, 1795–1814. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.; Xing, J.; Yan, L.; Wang, R.; Zhao, Y. The YUCCA-Auxin-WOX11 module controls crown root development in Rice. Front. Plant Sci. 2018, 9, 523. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Skupa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klima, P.; Carna, M.; Rolcik, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Dal Bosco, C.; Dovzhenko, A.; Palme, K. Intracellular auxin transport in pollen: PIN8, PIN5 and PILS5. Plant Signal. Behav. 2012, 7, 1504–1505. [Google Scholar] [CrossRef]
- Wang, J.R.; Hu, H.; Wang, G.H.; Li, J.; Chen, J.Y.; Wu, P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant 2009, 2, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef]
- Wu, L.; Luo, Z.; Shi, Y.; Jiang, Y.; Li, R.; Miao, X.; Yang, F.; Li, Q.; Zhao, H.; Xue, J.; et al. A cost-effective tsCUT&Tag method for profiling transcription factor binding landscape. J. Integr. Plant Biol. 2022, 64, 2033–2038. [Google Scholar] [CrossRef]
- Huang, X.L.; Lu, Z.H.; Wang, X.; Ouyang, Y.D.; Chen, W.; Xie, K.L.; Wang, D.Y.; Luo, M.; Luo, J.; Yao, J.L. Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 2016, 87, 305–317. [Google Scholar] [CrossRef]
- He, Y.; Zhang, T.; Sun, H.; Zhan, H.; Zhao, Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 2020, 7, 152. [Google Scholar] [CrossRef]
- Xu, Y.; Zong, W.; Hou, X.; Yao, J.; Liu, H.; Li, X.; Zhao, Y.; Xiong, L. OsARID3, an AT-rich interaction domain-containing protein, is required for shoot meristem development in rice. Plant J. 2015, 83, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, S.S.; Liu, Y.Q.; Ling, S.; Chen, C.S.; Yao, J.L. HOTHEAD-Like HTH1 is involved in anther cutin biosynthesis and is required for pollen fertility in rice. Plant Cell Physiol. 2017, 58, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhou, T.; Hu, G.; Wu, H.; Han, Z.; Xiao, J.; Li, X.; Xing, Y. An ethyl methanesulfonate-induced neutral mutant-bridging method efficiently identifies spontaneously mutated genes in rice. Plant J. Cell Mol. Biol. 2020, 104, 1129–1141. [Google Scholar] [CrossRef]
- Wu, C.; Li, X.; Yuan, W.; Chen, G.; Kilian, A.; Li, J.; Xu, C.; Li, X.; Zhou, D.X.; Wang, S.; et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003, 35, 418–427. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Lu, Z.; Zhai, L.; Li, N.; Yan, H. The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice. Plants 2023, 12, 3880. https://doi.org/10.3390/plants12223880
Huang X, Lu Z, Zhai L, Li N, Yan H. The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice. Plants. 2023; 12(22):3880. https://doi.org/10.3390/plants12223880
Chicago/Turabian StyleHuang, Xiaolong, Zhanhua Lu, Lisheng Zhai, Na Li, and Huiqing Yan. 2023. "The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice" Plants 12, no. 22: 3880. https://doi.org/10.3390/plants12223880



