Transcriptome Analysis of Macrophytes’ Myriophyllum spicatum Response to Ammonium Nitrogen Stress Using the Whole Plant Individual
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material Growth
2.2. Experimental Design
2.3. RNA Extraction, cDNA Library Construction, and Transcriptome Sequencing
2.4. De Novo Transcriptome Assembly and Annotation
2.5. Reads Mapping and Analysis of Differentially Expressed Genes
2.6. Identification and Construction of Co-Expressed Gene Networks
3. Results
3.1. Basic Information on the Transcriptome Sequence
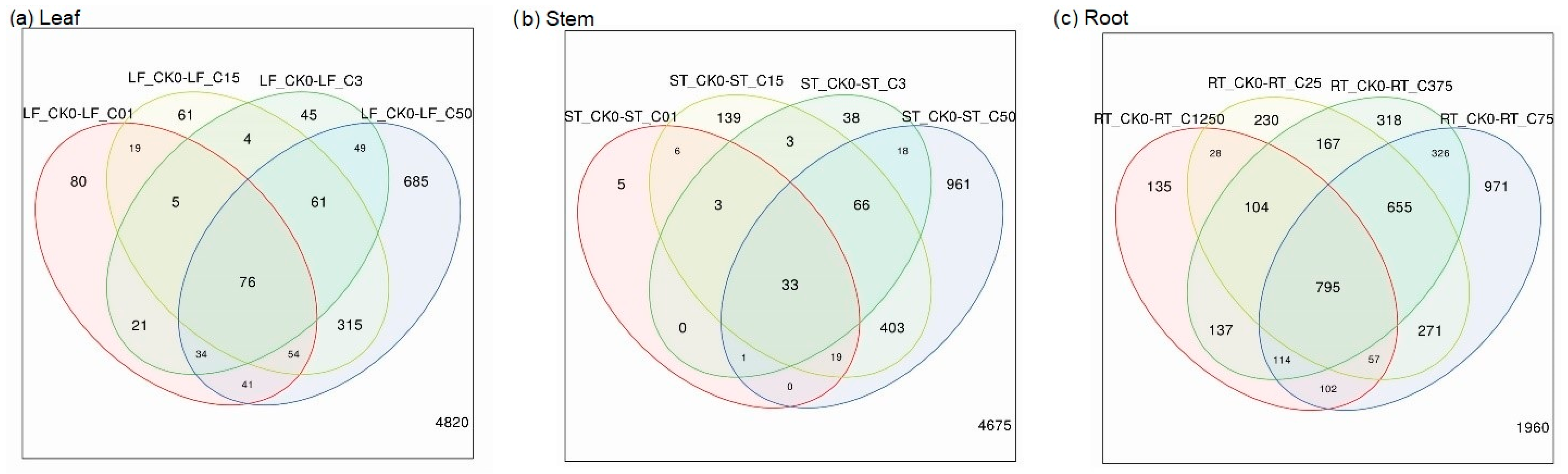
3.2. Overlaps of Differentially Expressed Genes in Leaf, Stem, and Roots after Exposure to Ammonium Treatments
3.3. K-Mean Clusters of Differentially Expressed Genes and GO Enrichment of Most Variable Genes
3.4. Principal Component Analysis (PCA) of the Genes
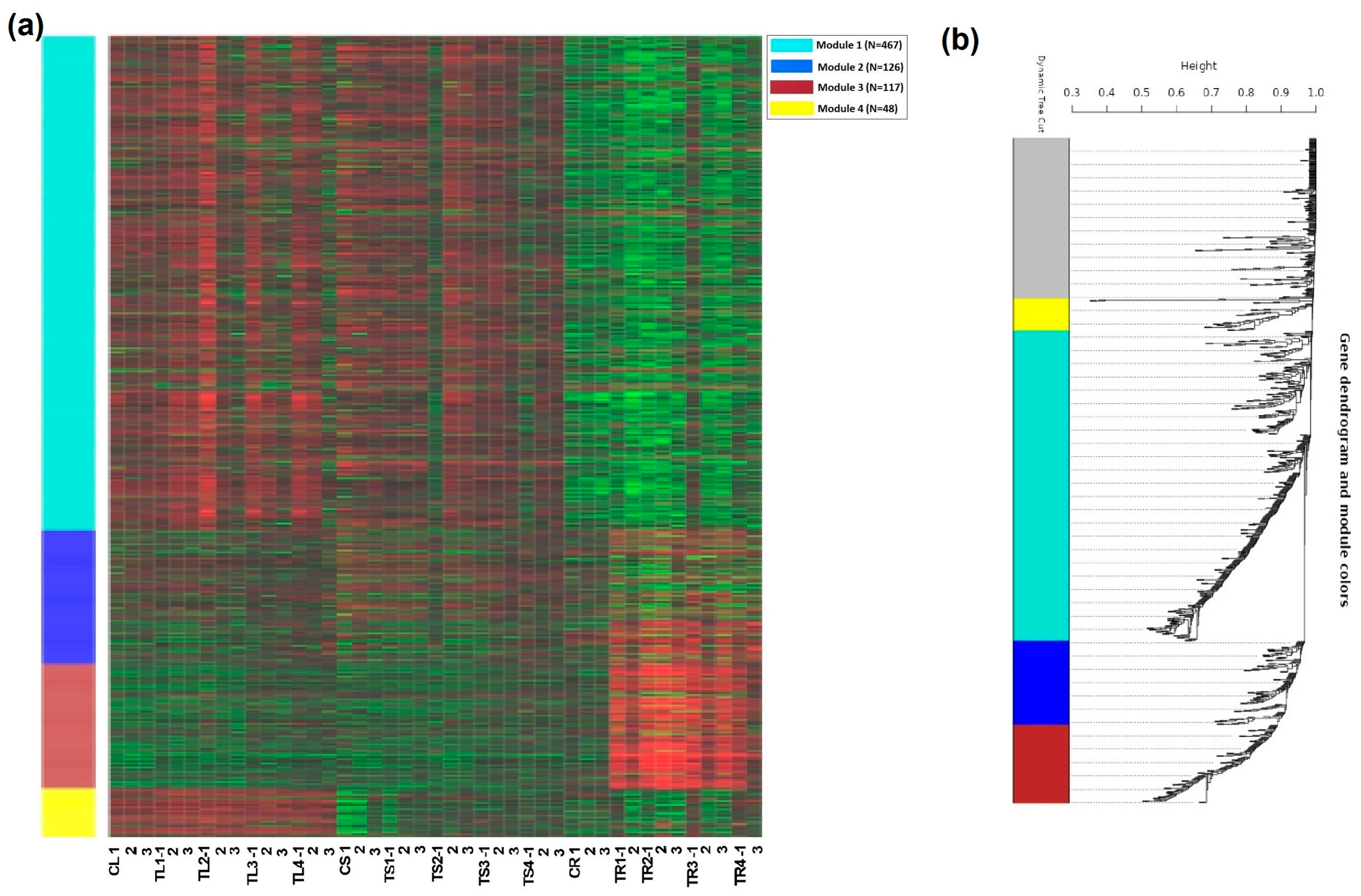
3.5. Expression Modules
3.6. Expression of the Top 10 Genes from the Selected Module
3.7. A Comparison of Gene Regulations between the Above-Ground and Below-Ground Parts
4. Discussion
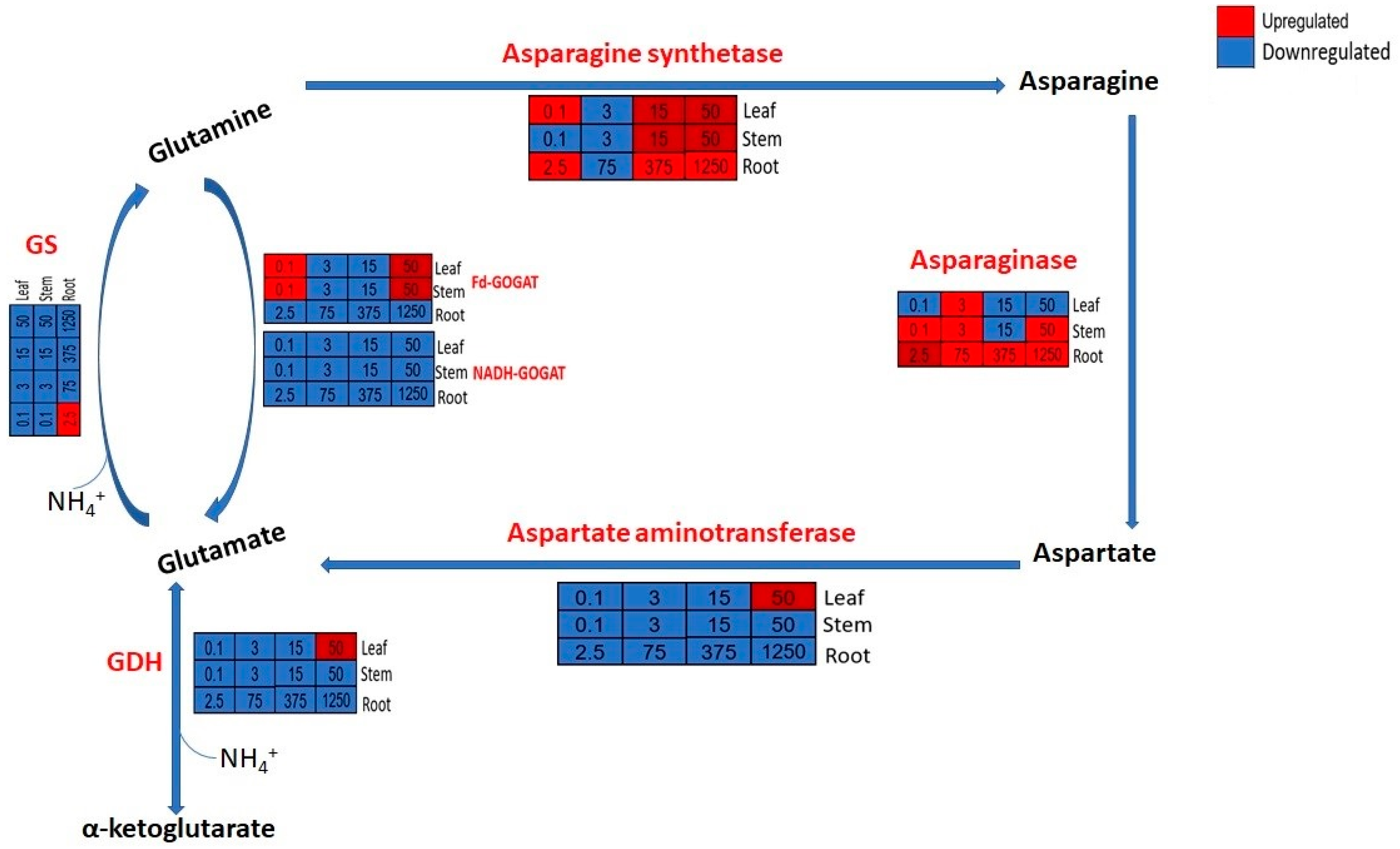
4.1. Response of Genes Related to Ammonium Assimilation
4.2. Response of Genes Related to Carbon Metabolism
4.3. Ammonium Transporter Genes and Glutathione S-Transferase Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frost, P.C.; Andrea, L.H. Human Shoreline Development and the nutrient stoichiometry of aquatic plant communities in Canadian Shield Lakes. Can. J. Fish. Aquat. Sci. 2012, 69, 1642–1650. [Google Scholar] [CrossRef]
- Jeppesen, E.; Söndergaard, M.; Söndergaard, M.; Christofferson, K. The structuring role of submerged macrophytes in lakes. Struct. Role Submerg. Macrophytes Lakes 1998, 131, 427–441. [Google Scholar] [CrossRef]
- Coban, O.; Kuschk, P.; Kappelmeyer, U.; Spott, O.; Martienssen, M.; Jetten, M.S.M.; Knoeller, K. Nitrogen transforming community in a horizontal subsurface-flow constructed wetland. Water Res. 2015, 74, 203–212. [Google Scholar] [CrossRef]
- Scheffer, M.; Hosper, S.H.; Meijer, M.-L.; Moss, B.; Jeppesen, E. Alternative equilibria in shallow lakes. Trends Ecol. Evol. 1993, 8, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Tyagi, A.K. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol. J. 2007, 5, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Zurbriggen, M.D.; Hajirezaei, M.R.; Carrillo, N. Engineering the future. Development of transgenic plants with enhanced tolerance to adverse environments. Biotechnol. Genet. Eng. Rev. 2010, 27, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, W.; Li, J.; Yang, Z. Growth, oxidative stress responses, and gene transcription of juvenile bighead carp (Hypophthalmichthys nobilis) under chronic-term exposure to ammonia. Environ. Toxicol. Chem. 2014, 33, 1726–1731. [Google Scholar] [CrossRef]
- Shengqi, S.; Zhou, Y.; Qin, J.G.; Wang, W.; Yao, W.; Song, L. Physiological Responses of Egeria Densa to High Ammonium Concentration and Nitrogen Deficiency. Chemosphere 2012, 86, 538–545. [Google Scholar] [CrossRef]
- Cao, T.; Xie, P.; Ni, L.; Wu, A.; Zhang, M.; Wu, S.; Smolders, A.J.P. The role of NH4 + toxicity in the decline of the submersed macrophyte Vallisneria natans in lakes of the Yangtze River basin, China. Mar. Freshw. Res. 2007, 58, 581–587. [Google Scholar] [CrossRef]
- Bittsánszky, A.; Pilinszky, K.; Gyulai, G.; Komives, T. Overcoming ammonium toxicity. Plant Sci. 2015, 231, 184–190. [Google Scholar] [CrossRef]
- Zhou, Q.; Gao, J.; Zhang, R.; Zhang, R. Ammonia Stress on Nitrogen Metabolism in Tolerant Aquatic Plant—Myriophyllum aquaticum. Ecotoxicol. Environ. Saf. 2017, 143, 102–110. [Google Scholar] [CrossRef]
- Lam, H.M.; Wong, P.; Chan, H.K.; Yam, K.M.; Chen, L.; Chow, C.M.; Coruzzi, G.M. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 2003, 132, 926–935. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 2006, 140, 444–456. [Google Scholar] [CrossRef]
- Qu, C.; Hao, B.; Xu, X.; Wang, Y.; Yang, C.; Xu, Z.; Liu, G. Functional Research on Three Presumed Asparagine Synthetase Family Members in Poplar. Genes 2019, 10, 326. [Google Scholar] [CrossRef]
- Wong, H.K.; Chan, H.K.; Coruzzi, G.M.; Lam, H.M. Correlation of ASN2 Gene Expression with Ammonium Metabolism in Arabidopsis. Plant Physiol. 2004, 134, 332–338. [Google Scholar] [CrossRef]
- Tian, H.; Fu, J.; Drijber, R.A.; Gao, Y. Expression patterns of five genes involved in nitrogen metabolism in two winter wheat (Triticum aestivum L.) genotypes with high and low nitrogen utilization efficiencies. J. Cereal Sci. 2015, 61, 48–54. [Google Scholar] [CrossRef]
- Rennenberg, H.; Wildhagen, H.; Ehlting, B. Nitrogen nutrition of poplar trees. Plant Biol. 2010, 12, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Di, D.W.; Li, G.J.; Kronzucker, H.J.; Shi, W.M. Spatio-temporal dynamics in global rice gene expression (Oryza sativa L.) in response to high ammonium stress. J. Plant Physiol. 2017, 212, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, X.; Chen, J.; Shang, S.; Zhu, M.; Liang, S.; Zang, Y. Comparative studies on the response of Zostera marina leaves and roots to ammonium stress and effects on nitrogen metabolism. Aquat. Toxicol. 2021, 240, 105965. [Google Scholar] [CrossRef]
- Wang, R.; Xu, S.; Sun, H.; Feng, S.; Jiang, C.; Zhou, S.; Wu, S.; Zhuang, G.; Chen, B.; Bai, Z.; et al. The complex regulatory network allows Myriophyllum aquaticum to thrive under high-concentration ammonia toxicity. Sci. Rep. 2019, 9, 4801. [Google Scholar] [CrossRef]
- Wang, W.; Li, R.; Zhu, Q.; Tang, X.; Zhao, Q. Transcriptomic and physiological analysis of common duckweed Lemna minor responses to NH4+ toxicity. BMC Plant Biol. 2016, 16, 92. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Hao, D.L.; Song, Z.Z.; Yang, G.Z.; Wang, L.; Su, Y.H. RNA-Seq analysis of differentially expressed genes in rice under varied nitrogen supplies. Gene 2015, 555, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Hao, D.L.; Jin, M.; Li, Y.; Liu, Z.T.; Huang, Y.N.; Chen, T.X.; Su, Y.H. Internal ammonium excess induces ROS-mediated reactions and causes carbon scarcity in rice. BMC Plant Biol. 2020, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.; Liu, F.; Luo, P.; Wang, Y.; Liu, D.; Wu, X.; Zhang, Z.; Wu, J. Transcriptomic and physiological analysis revealed the ammonium tolerance mechanisms of Myriophyllum aquaticum. Environ. Exp. Bot. 2021, 187, 104462. [Google Scholar] [CrossRef]
- Xian, L.; Zhang, Y.; Cao, Y.; Wan, T.; Gong, Y.; Dai, C.; Ochieng, W.A.; Nasimiyu, A.T.; Li, W.; Liu, F. GDH plays an important role in the ammonium detoxification of submerged macrophytes. Sci. Total Environ. 2020, 722, 137859. [Google Scholar] [CrossRef]
- Apudo, A.A.; Cao, Y.; Wakibia, J.; Li, W.; Liu, F. Physiological plastic responses to acute NH4+-N toxicity in Myriophyllum spicatum L. cultured in high and low nutrient Conditions. Environ. Exp. Bot. 2016, 130, 79–85. [Google Scholar] [CrossRef]
- Ochieng, W.A.; Xian, L.; Nasimiyu, A.T.; Muthui, S.W.; Ndirangu, L.N.; Otieno, D.O.; Wan, T.; Liu, F. Exploring the ammonium detoxification mechanism of young and mature leaves of the macrophyte Potamogeton lucens. Aquat. Toxicol. 2021, 237, 105879. [Google Scholar] [CrossRef]
- Xian, L.; Ochieng, W.A.; Muthui, S.W.; Otieno, D.O.; Yu, S.; Li, W.; Yan, X.; Yu, Q.; Liu, F. The Above-Ground Part of Submerged Macrophytes Plays an Important Role in Ammonium Utilization. Front. Plant Sci. 2022, 13, 865578. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 15, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization, and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Zhang, X.; Jonassen, I. RASflow: An RNA-Seq analysis workflow with Snakemake. BMC Bioinform. 2020, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Wickert, E.; Marcondes, J.; Lemos, M.V.; Lemos, E.G.M. Nitrogen assimilation in citrus based on CitEST data mining. Genet. Mol. Biol. 2007, 30, 810–818. [Google Scholar] [CrossRef]
- Ruiz, J.M.; Rivero, R.M.; Romero, L. Comparative effect of Al, Se, and Mo toxicity on NO3- assimilation in sunflower (Helianthus annuus L.). Plants J. Environ. Manag. 2007, 83, 207–212. [Google Scholar] [CrossRef]
- Suá Rez, M.F.; Gallardo, F.; Cantó, F.R.; García-Gutié, R.A.; Claros, M.G.; Cá Novas, F.M. Molecular and enzymatic analysis of ammonium assimilation in woody plants. J. Exp. Bot. 2002, 53, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Skopelitis, D.S.; Paranychianakis, N.V.; Paschalidis, K.A.; Pliakonis, E.D.; Delis, I.D.; Yakoumakis, D.I.; Kouvarakis, A.; Papadakis, A.K.; Stephanou, E.G.; Roubelakis-Angelakis, K.A. Abiotic stress generates ROS that signals the expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 2006, 18, 2767–2781. [Google Scholar] [CrossRef] [PubMed]
- Tercé-Laforgue, T.; Dubois, F.; Ferrario-Méry, S.; de Crecenzo, M.A.P.; Sangwan, R.; Hirel, B. Glutamate dehydrogenase of tobacco is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiol. 2004, 136, 4308–4317. [Google Scholar] [CrossRef] [PubMed]
- Zhonghua, T.; Yanju, L.; Xiaorui, G.; Yuangang, Z. The combined effects of salinity and nitrogen forms on Catharanthus roseus: The role of internal ammonium and free amino acids during salt stress. J. Plant Nutr. Soil Sci. 2011, 174, 135–144. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of a glutamate dehydrogenase gene, MgGDH, from Magnaporthe grisea confers tolerance to dehydration stress in transgenic rice. Planta 2015, 241, 727–740. [Google Scholar] [CrossRef]
- Kanamori, T.; Matsumoto, H. Asparagine biosynthesis by Oryza sativa seedlings. Phytochemistry 1974, 13, 1407–1412. [Google Scholar] [CrossRef]
- Stewart, C.R. The effect of ammonium, glutamine, methionine sulfoxamine, and azaserine on asparagine synthetase in soybean leaves. Plant Sci. Lett. 1979, 14, 269. [Google Scholar] [CrossRef]
- de La Torre, F.; Cañas, R.A.; Pascual, M.B.; Avila, C.; Cánovas, F.M. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 2014, 65, 5527–5534. [Google Scholar] [CrossRef]
- Dornfeld, K.; Madden, M.; Skildum, A.; Wallace, K.B. Aspartate facilitates mitochondrial function, growth arrest, and survival during doxorubicin exposure. Cell Cycle 2015, 14, 3282–3291. [Google Scholar] [CrossRef]
- Wadsworth, G.J. The plant aspartate aminotransferase gene family. Physiol. Plant. 1997, 100, 998–1006. [Google Scholar] [CrossRef]
- Rudrabhatla, P.; Reddy, M.M.; Rajasekharan, R. Genome-wide analysis and experimentation of plant serine/threonine/ tyrosine-specific protein kinases. Plant Mol. Biol. 2006, 60, 293–319. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhang, F.; Beckles, D.M. A Cytosolic Protein Kinase STY46 in Arabidopsis thaliana Is Involved in Plant Growth and Abiotic Stress Response. Plants 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Atkins, C.A.; Patterson, B.D.; Graham, D. Plant carbonic anhydrases I. Distribution of types among species. Plant Physiol. 1972, 50, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.M.; Brown, R.H.; Black, C.C. CO2 compensation concentration, rate of photosynthesis, and carbonic anhydrase activity of plants. Weed Sci. 1970, 18, 399–403. [Google Scholar] [CrossRef]
- Atkins, C.A. Occurrence and some properties of carbonic anhydrate from legume root nodules. Phytochemistry 1974, 13, 93–98. [Google Scholar] [CrossRef]
- Villarejo, A.; Burén, S.; Larsson, S.; Déjardin, A.; Monné, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005, 17, 1224–1231. [Google Scholar] [CrossRef]
- Thompson, M.; Gamage, D.; Hirotsu, N.; Martin, A.; Seneweera, S. Effects of Elevated Carbon Dioxide on Photosynthesis and Carbon Partitioning: A Perspective on Root Sugar Sensing and Hormonal Crosstalk. Front. Physiol. 2017, 8, 253220. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Kronzucker, H.J. The nitrogen-potassium intersection: Membranes, metabolism, and mechanism. Plant Cell Environ. 2017, 40, 2029–2041. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Lejay, L.; Gojon, A.; Ninnemann, O.; Frommer, W.B.; Von Wirén, N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 1999, 11, 937–948. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Wood, C.C.; Roeb, G.W.; Udvardi, M.K. Characterization of Arabidopsis Atamt2, a High-Affinity Ammonium Transporter of the Plasma Membrane. Plant Physiol. 2002, 130, 1788–1796. [Google Scholar] [CrossRef]
- Suenaga, A.; Moriya, K.; Sonoda, Y.; Ikeda, A.; Von Wirén, N.; Hayakawa, T.; Yamaguchi, J.; Yamaya, T. Constitutive Expression of a Novel-Type Ammonium Transporter OsAMT2 in Rice Plants. Plant Cell Physiol. 2003, 44, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Betti, M.; Pérez-Delgado, C.; García-Calderón, M.; Díaz, P.; Monza, J.; Márquez, A.J. Cellular Stress Following Water Deprivation in the Model Legume Lotus japonicus. Cells 2012, 1, 1089–1106. [Google Scholar] [CrossRef] [PubMed]



| Plant Part | Ammonium Concentrations (mg/L) | ||||
|---|---|---|---|---|---|
| Above-ground | CK0 | 0.1 | 3 | 15 | 50 |
| Below-ground | CK0 | 2.5 | 75 | 375 | 1250 |
| Symbol | Control (C) | T1 | T2 | T3 | T4 |
| Sample | Clean Reads | Base Num_Clean | GC% | >Q30 | >Q20 | Clean Rate |
|---|---|---|---|---|---|---|
| Leaf_CK0_1 | 9483174 | 1365253325 | 44.96% | 85.4% | 94.78% | 90.29% |
| Leaf_CKO_2 | 44039636 | 6346954385 | 45.44% | 86.86% | 95.14% | 90.07% |
| Leaf_CKO_3 | 46669192 | 6737538060 | 45.13% | 87.14% | 95.27% | 90.93% |
| Leaf_C0.1_1 | 48367430 | 7071729005 | 45.47% | 87.27% | 94.84% | 94.7% |
| Leaf_C0.1_2 | 43056516 | 6279483747 | 45.97% | 86.18% | 94.36% | 94.41% |
| Leaf_C0.1_3 | 48723216 | 7105614339 | 45.82% | 86.66% | 94.58% | 94.58% |
| Leaf_C3_1 | 46622400 | 6752015620 | 46.42% | 88.19% | 95.64% | 90.82% |
| Leaf_C3_2 | 43454682 | 6223190987 | 45.49% | 85.5% | 94.62% | 89.01% |
| Leaf_C3_3 | 7974958 | 1147249076 | 45.01% | 85.18% | 94.7% | 90.25% |
| Leaf_C15_1 | 51618630 | 7533928462 | 46.27% | 86.7% | 94.58% | 94.64% |
| Leaf_C15_2 | 50278296 | 7349072444 | 46.94% | 87.34% | 94.82% | 94.63% |
| Leaf_C15_3 | 43884022 | 6414982330 | 47.11% | 87.48% | 94.89% | 94.88% |
| Leaf_C50_1 | 45608796 | 6660489072 | 47.48% | 86.78% | 94.58% | 93.82% |
| Leaf_C50_2 | 56796048 | 8257059066 | 46.01% | 86.85% | 95.06% | 92.48% |
| Leaf_C50_3 | 46767812 | 6811573000 | 47.13% | 86.57% | 94.7% | 93.62% |
| Sample | Clean Reads | Base Num_Clean | GC% | >Q30 | >Q20 | Clean Rate |
|---|---|---|---|---|---|---|
| Stem_CK0_1 | 43380046 | 6273729213 | 45.62% | 88.19% | 95.6% | 89.73% |
| Stem_CKO_2 | 38909370 | 5581236204 | 46.15% | 86.4% | 94.85% | 86.43% |
| Stem_CKO_3 | 48699588 | 7045460726 | 45.25% | 87.84% | 95.54% | 90.58% |
| Stem_C0.1_1 | 44618368 | 6528565752 | 45.34% | 87.59% | 94.98% | 94.75% |
| Stem_C0.1_2 | 52092526 | 7626597011 | 45.36% | 87.74% | 95.04% | 94.92% |
| Stem_C0.1_3 | 40865556 | 5973232795 | 45.19% | 87% | 94.74% | 94.95% |
| Stem_C3_1 | 44015712 | 6349464797 | 44.84% | 87.09% | 95.25% | 90.95% |
| Stem_C3_2 | 43578078 | 6229285621 | 45.45% | 85.24% | 94.49% | 89.44% |
| Stem_C3_3 | 43623748 | 6311383774 | 45.48% | 87.86% | 95.54% | 91.16% |
| Stem_C15_1 | 52479126 | 7679519421 | 45.71% | 87.48% | 94.92% | 94.97% |
| Stem_C15_2 | 50737878 | 7416480012 | 45.89% | 87.26% | 94.84% | 94.84% |
| Stem_C15_3 | 50729090 | 7415387552 | 45.87% | 87.22% | 94.8% | 94.59% |
| Stem_C50_1 | 48614830 | 7103220369 | 45.94% | 87.8% | 95.24% | 94.34% |
| Stem_C50_2 | 47811050 | 6985103878 | 46.19% | 87.07% | 94.74% | 94.54% |
| Stem_C50_3 | 45180526 | 6602860020 | 45.71% | 87.24% | 94.83% | 95% |
| Sample | Clean Reads | Base Num_Clean | GC% | >Q30 | >Q20 | Clean Rate |
|---|---|---|---|---|---|---|
| Root_CK0_1 | 9832836 | 1419104428 | 45.4% | 86.14% | 95.04% | 90.24% |
| Root_CKO_2 | 40122732 | 5727077094 | 45.28% | 85.02% | 94.36% | 87.59% |
| Root_CKO_3 | 39167564 | 5595243942 | 44.87% | 85.16% | 94.42% | 87.9% |
| Root_C2.5_1 | 61172098 | 8957002394 | 47.34% | 87.02% | 94.64% | 94.24% |
| Root_C_2.5_2 | 55210796 | 8064235149 | 48.88% | 86.3% | 94.26% | 94.02% |
| Root_C75_1 | 46046854 | 6791496790 | 49.27% | 90.36% | 96.76% | 94.43% |
| Root_C75_2 | 44461536 | 6558048748 | 49.56% | 90.2% | 96.69% | 93.76% |
| Root_C75_3 | 8530548 | 1229173572 | 47.53% | 85.92% | 94.91% | 90.14% |
| Root_C375_1 | 47354218 | 6900194618 | 50.47% | 85.42% | 93.8% | 93.56% |
| Root_C375_2 | 51698210 | 7554479584 | 49.22% | 86.34% | 94.26% | 94.28% |
| Root_C375_3 | 48025588 | 7017901998 | 46.73% | 86.42% | 94.4% | 94.8% |
| Root_C1250_1 | 53495218 | 7844730098 | 47.57% | 87.54% | 94.88% | 94.86% |
| Root_C1250_2 | 20264096 | 2954814344 | 52.79% | 89.72% | 96.4% | 63.52% |
| Root_C1250_3 | 53383328 | 7810306178 | 51.9% | 86.84% | 94.48% | 93.34% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochieng, W.A.; Wei, L.; Wagutu, G.K.; Xian, L.; Muthui, S.W.; Ogada, S.; Otieno, D.O.; Linda, E.L.; Liu, F. Transcriptome Analysis of Macrophytes’ Myriophyllum spicatum Response to Ammonium Nitrogen Stress Using the Whole Plant Individual. Plants 2023, 12, 3875. https://doi.org/10.3390/plants12223875
Ochieng WA, Wei L, Wagutu GK, Xian L, Muthui SW, Ogada S, Otieno DO, Linda EL, Liu F. Transcriptome Analysis of Macrophytes’ Myriophyllum spicatum Response to Ammonium Nitrogen Stress Using the Whole Plant Individual. Plants. 2023; 12(22):3875. https://doi.org/10.3390/plants12223875
Chicago/Turabian StyleOchieng, Wyckliffe Ayoma, Li Wei, Godfrey Kinyori Wagutu, Ling Xian, Samuel Wamburu Muthui, Stephen Ogada, Duncan Ochieng Otieno, Elive Limunga Linda, and Fan Liu. 2023. "Transcriptome Analysis of Macrophytes’ Myriophyllum spicatum Response to Ammonium Nitrogen Stress Using the Whole Plant Individual" Plants 12, no. 22: 3875. https://doi.org/10.3390/plants12223875






