Functional Modules in the Meristems: “Tinkering” in Action
Abstract
:1. Introduction
2. Meristems—The Functional Module at the Level of the Organism
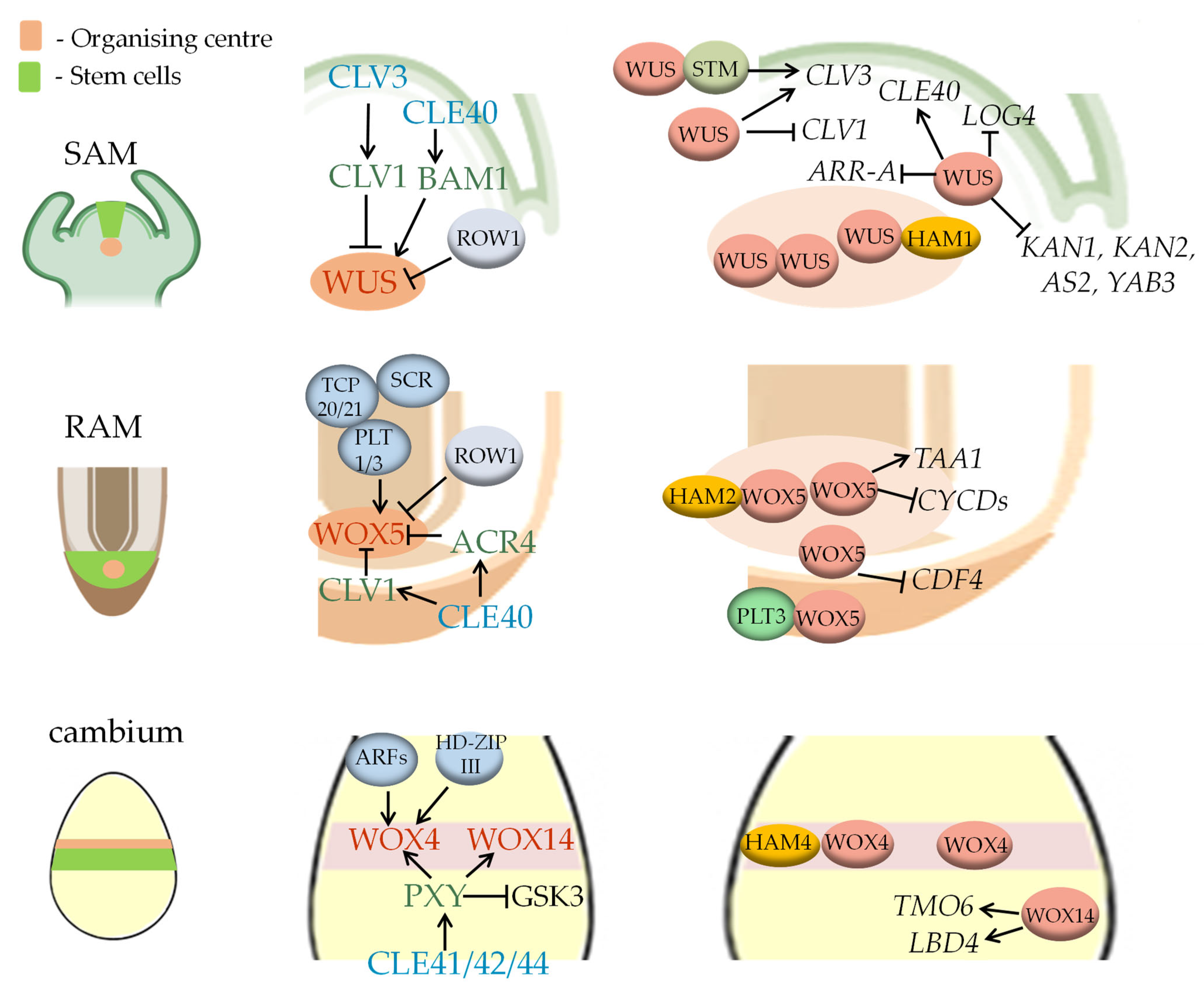
3. Molecular Module 1: The WOX-CLAVATA System
3.1. The WOX-CLAVATA System in the SAM
3.2. The WOX-CLAVATA System in the RAM
| Transcription Factor | Target Gene | Encoding Protein and Its Functions | Reference |
|---|---|---|---|
| WUS | CLV3 | Peptide phytohormone; negatively regulates WUS expression and size of SC pool in the SAM | [18,66] |
| CLE40 | Peptide phytohormone; positively regulates WUS expression and size of SC pool in the SAM | [47] | |
| CLV1 | Receptor of CLE peptides in the SAM and RAM | [38,66] | |
| ARR5 | Type-A ARR, repressors of CK signalling | [38,65] | |
| ARR5 | Type-A ARR, repressors of CK signalling | [38,65] | |
| ARR7 | Type-A ARR, repressors of CK signalling | [38,65] | |
| ARR15 | Type-A ARR, repressors of CK signalling | [38,65] | |
| LOG4 | Phosphoribohydrolase; catalyses the last step of CK biosynthesis | [70] | |
| ARF5/MP | Auxin-responsive TF; regulates the transport of IAA and the initiation of leaves and roots | [71] | |
| AS2 | TF; regulates the initiation of leaf primordium | [66] | |
| KAN1 | TF; regulates the development of abaxial side of leaf primordium | [66] | |
| KAN2 | TF; regulates the development of abaxial side of leaf primordium | [66] | |
| YAB3 | TF; regulates the development of abaxial side of leaf primordium | [66] | |
| WOX5 | CDF4 | TF; regulates columella SC differentiation | [19,105] |
| CYCD1;1 | D-class cyclin; promotes the G1/S transition in the plant cell cycle | [105,106] | |
| CYCD3;1 | D-class cyclin; promotes the G1/S transition in the plant cell cycle | [105,106] | |
| TAA1 | Tryptophan aminotransferase; catalyses the first step of auxin biosynthesis via indol-3-pyruvate | [104] |
3.3. The WOX-CLAVATA System in the Cambium
3.4. WOX-CLAVATA Systems in Other Meristems
4. Module 2: Florigen-Activating Complex (FAC)
4.1. FACs in Flowering Control
4.2. FACs in the Regulation of Bud Dormancy
4.3. FACs in the Regulation of Storage Organ Formation
5. Changes in Regulatory Modules: Nature and Man Worked Hand in Hand
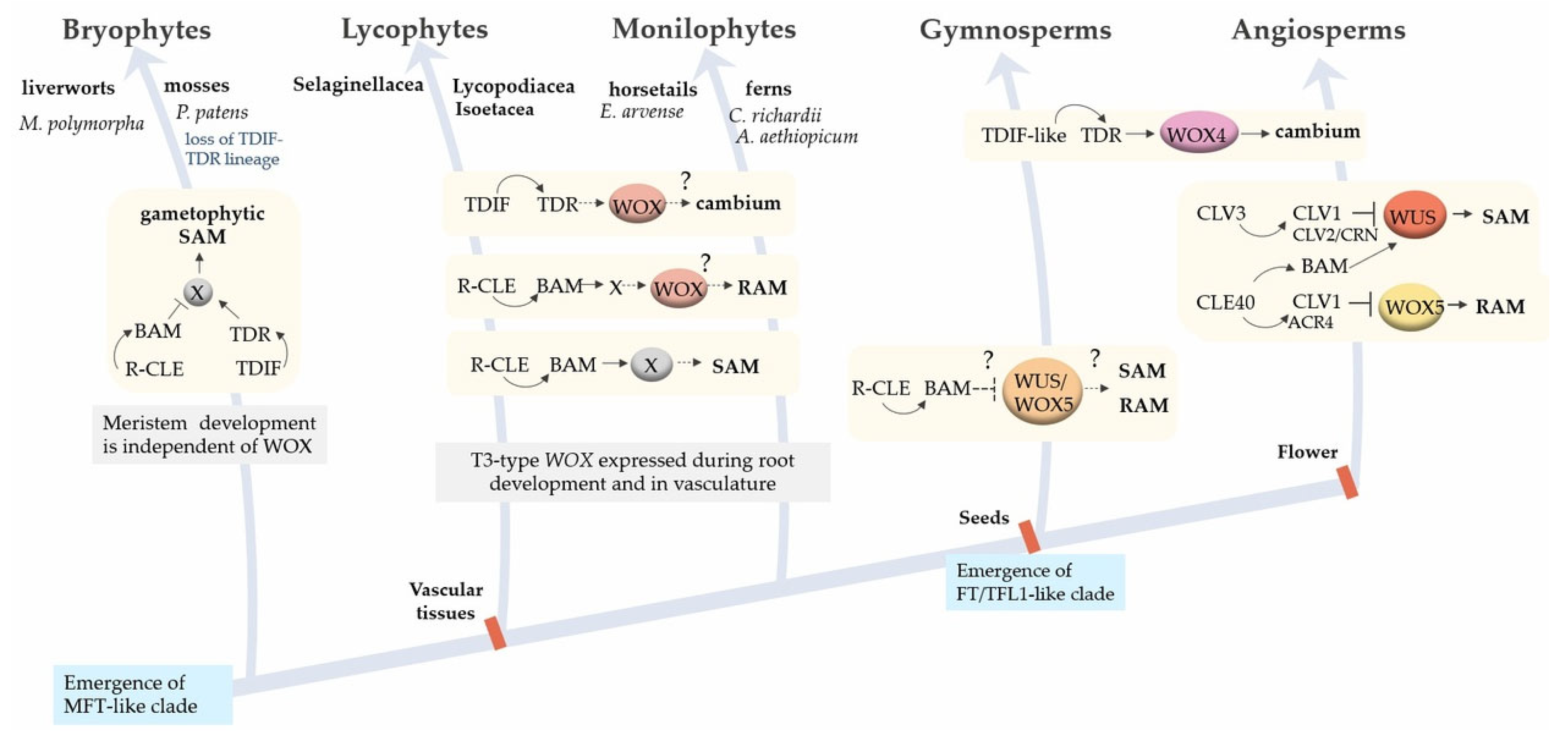
5.1. Changes in Regulatory Modules during Evolution
5.1.1. Evolution of Meristems as Morphological Modules
5.1.2. Evolution of the WOX-CLAVATA Module
WOX TFs
CLE Peptides
LRR-RLKs
5.1.3. Evolution of the FAC Module
5.2. Changes in Regulatory Modules during in the Course of Plant Breeding
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Notov, A.A. On Functional Organization and Individual Development of Modular Objects. Wulfenia 2005, 12, 65–85. [Google Scholar]
- Klingenberg, C.P. Morphological Integration and Developmental Modularity. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 115–132. [Google Scholar] [CrossRef]
- Afonkin, S.Y. Biokombinatorika Ili Blochnyi Printsyp Organizasii Zhizni [Bio-Combination Theory or Block Principle of the Organisation of a Life]. Biology 2003, 35. [Google Scholar]
- Jacob, F. Evolution and Tinkering. Science 1977, 196, 1161–1166. [Google Scholar] [CrossRef]
- Inge-Vechtomov, S.G. Block Principle in the Theory of Evolution. Prospects and Paradoxes. In Fundamental’nye Zoologicheskie Issledovaniya (Fundamental Zoological Studies); KMK: St. Petersburg, Russia, 2004; pp. 74–88. [Google Scholar]
- Ivanov, V.B. The problem of stem cells in plants. Ontogenez 2003, 34, 253–261. [Google Scholar] [PubMed]
- Dodueva, I.E.; Tvorogova, V.E.; Azarakhsh, M.; Lebedeva, M.A.; Lutova, L.A. Plant Stem Cells: Unity and Diversity. Russ. J. Genet. Appl. Res. 2017, 7, 385–403. [Google Scholar] [CrossRef]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in Regulating Stem Cell Fate in the Arabidopsis Shoot Meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A Signaling Module Controlling the Stem Cell Niche in Arabidopsis Root Meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef]
- Gaillochet, C.; Lohmann, J.U. The Never-Ending Story: From Pluripotency to Plant Developmental Plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef]
- Ivanov, V.B. Stem cells in the root and the problem of stem cells in plants. Ontogenez 2007, 38, 406–419. [Google Scholar] [CrossRef]
- Coudert, Y.; Novák, O.; Harrison, C.J. A KNOX-Cytokinin Regulatory Module Predates the Origin of Indeterminate Vascular Plants. Curr. Biol. 2019, 29, 2743–2750.e5. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of Plant Stem Cell Function by Conserved Interacting Transcriptional Regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef]
- Poliushkevich, L.O.; Gancheva, M.S.; Dodueva, I.E.; Lutova, L.A. Receptors of CLE Peptides in Plants. Russ. J. Plant Physiol. 2020, 67, 1–16. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Cuperus, J.T.; Weigel, D.; Carrington, J.C. Regulation and Functional Specialization of Small RNA-Target Nodes during Plant Development. Curr. Opin. Plant Biol. 2009, 12, 622–627. [Google Scholar] [CrossRef]
- Zhu, Q.-H.; Helliwell, C.A. Regulation of Flowering Time and Floral Patterning by MiR172. J. Exp. Bot. 2011, 62, 487–495. [Google Scholar] [CrossRef]
- Wang, J.-W.; Czech, B.; Weigel, D. MiR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The Stem Cell Population of Arabidopsis Shoot Meristems in Maintained by a Regulatory Loop between the CLAVATA and WUSCHEL Genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Pi, L.; Aichinger, E.; van der Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-Derived WOX5 Signal Maintains Root Columella Stem Cells through Chromatin-Mediated Repression of CDF4 Expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef]
- Gancheva, M.S.; Malovichko, Y.V.; Poliushkevich, L.O.; Dodueva, I.E.; Lutova, L.A. Plant Peptide Hormones. Russ. J. Plant Physiol. 2019, 66, 171–189. [Google Scholar] [CrossRef]
- Dodueva, I.; Lebedeva, M.; Lutova, L. Dialog between Kingdoms: Enemies, Allies and Peptide Phytohormones. Plants 2021, 10, 2243. [Google Scholar] [CrossRef]
- Lebedeva, M.A.; Dodueva, I.E.; Gancheva, M.S.; Tvorogova, V.E.; Kuznetsova, K.A.; Lutova, L.A. The Evolutionary Aspects of Flowering Control: Florigens and Anti-Florigens. Russ. J. Genet. 2020, 56, 1323–1344. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, R.; Devisetty, U.K.; Maloof, J.N.; Zuo, Y.; Li, J.; Shen, Y.; Zhao, J.; Bao, M.; Ning, G. The Divergence of Flowering Time Modulated by FT/TFL1 Is Independent to Their Interaction and Binding Activities. Front. Plant Sci. 2017, 8, 697. [Google Scholar] [CrossRef]
- Gaarslev, N.; Swinnen, G.; Soyk, S. Meristem Transitions and Plant Architecture-Learning from Domestication for Crop Breeding. Plant Physiol. 2021, 187, 1045–1056. [Google Scholar] [CrossRef]
- Eserman, L.A.; Jarret, R.L.; Leebens-Mack, J.H. Parallel Evolution of Storage Roots in Morning Glories (Convolvulaceae). BMC Plant Biol. 2018, 18, 95. [Google Scholar] [CrossRef]
- Van den Berg, C.; Willemsen, V.; Hendriks, G.; Weisbeek, P.; Scheres, B. Short-Range Control of Cell Differentiation in the Arabidopsis Root Meristem. Nature 1997, 390, 287–289. [Google Scholar] [CrossRef]
- Stahl, Y.; Simon, R. Plant Primary Meristems: Shared Functions and Regulatory Mechanisms. Curr. Opin. Plant Biol. 2010, 13, 53–58. [Google Scholar] [CrossRef]
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Sánchez Rodríguez, F.; Wu, M.-F.; Solé-Gil, A.; Leal Gavarrón, M.; Siligato, R.; Miyashima, S.; et al. High Levels of Auxin Signalling Define the Stem-Cell Organizer of the Vascular Cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Shi, D.; Lebovka, I.; López-Salmerón, V.; Sanchez, P.; Greb, T. Bifacial Cambium Stem Cells Generate Xylem and Phloem during Radial Plant Growth. Development 2019, 146, dev171355. [Google Scholar] [CrossRef]
- Murray, J.A.H.; Jones, A.; Godin, C.; Traas, J. Systems Analysis of Shoot Apical Meristem Growth and Development: Integrating Hormonal and Mechanical Signaling. Plant Cell 2012, 24, 3907–3919. [Google Scholar] [CrossRef]
- Benková, E.; Hejátko, J. Hormone Interactions at the Root Apical Meristem. Plant Mol. Biol. 2009, 69, 383–396. [Google Scholar] [CrossRef]
- Su, Y.-H.; Liu, Y.-B.; Zhang, X.-S. Auxin-Cytokinin Interaction Regulates Meristem Development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Bürglin, T.R.; Affolter, M. Homeodomain Proteins: An Update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Fletcher, J.C.; Brand, U.; Running, M.P.; Simon, R.; Meyerowitz, E.M. Signaling of Cell Fate Decisions by CLAVATA3 in Arabidopsis Shoot Meristems. Science 1999, 283, 1911–1914. [Google Scholar] [CrossRef]
- Ogawa, M.; Shinohara, H.; Sakagami, Y.; Matsubayashi, Y. Arabidopsis CLV3 Peptide Directly Binds CLV1 Ectodomain. Science 2008, 319, 294. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL Protein Movement Mediates Stem Cell Homeostasis in the Arabidopsis Shoot Apex. Genes. Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef]
- Lenhard, M.; Laux, T. Stem Cell Homeostasis in the Arabidopsis Shoot Meristem Is Regulated by Intercellular Movement of CLAVATA3 and Its Sequestration by CLAVATA1. Development 2003, 130, 3163–3173. [Google Scholar] [CrossRef]
- Busch, W.; Miotk, A.; Ariel, F.D.; Zhao, Z.; Forner, J.; Daum, G.; Suzaki, T.; Schuster, C.; Schultheiss, S.J.; Leibfried, A.; et al. Transcriptional Control of a Plant Stem Cell Niche. Dev. Cell 2010, 18, 849–861. [Google Scholar] [CrossRef]
- Nimchuk, Z.L.; Tarr, P.T.; Ohno, C.; Qu, X.; Meyerowitz, E.M. Plant Stem Cell Signaling Involves Ligand-Dependent Trafficking of the CLAVATA1 Receptor Kinase. Curr. Biol. 2011, 21, 345–352. [Google Scholar] [CrossRef]
- Ito, Y.; Nakanomyo, I.; Motose, H.; Iwamoto, K.; Sawa, S.; Dohmae, N.; Fukuda, H. Dodeca-CLE Peptides as Suppressors of Plant Stem Cell Differentiation. Science 2006, 313, 842–845. [Google Scholar] [CrossRef]
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS Homeobox-Containing (WOX) Protein Family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Hazak, O.; Hardtke, C.S. CLAVATA 1-Type Receptors in Plant Development. J. Exp. Bot. 2016, 67, 4827–4833. [Google Scholar] [CrossRef]
- Bleckmann, A.; Weidtkamp-Peters, S.; Seidel, C.A.M.; Simon, R. Stem Cell Signaling in Arabidopsis Requires CRN to Localize CLV2 to the Plasma Membrane. Plant Physiol. 2010, 152, 166–176. [Google Scholar] [CrossRef]
- Mizuno, S.; Osakabe, Y.; Maruyama, K.; Ito, T.; Osakabe, K.; Sato, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Receptor-like Protein Kinase 2 (RPK 2) Is a Novel Factor Controlling Anther Development in Arabidopsis Thaliana. Plant J. 2007, 50, 751–766. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Bickle, K.L.; Schrage, K.J.; Muskett, P.; Patel, K.; Clark, S.E. The CLAVATA1-Related BAM1, BAM2 and BAM3 Receptor Kinase-like Proteins Are Required for Meristem Function in Arabidopsis. Plant J. 2006, 45, 1–16. [Google Scholar] [CrossRef]
- Shinohara, H.; Matsubayashi, Y. Reevaluation of the CLV3-Receptor Interaction in the Shoot Apical Meristem: Dissection of the CLV3 Signaling Pathway from a Direct Ligand-Binding Point of View. Plant J. 2015, 82, 328–336. [Google Scholar] [CrossRef]
- Schlegel, J.; Denay, G.; Wink, R.; Pinto, K.G.; Stahl, Y.; Schmid, J.; Blümke, P.; Simon, R.G. Control of Arabidopsis Shoot Stem Cell Homeostasis by Two Antagonistic CLE Peptide Signalling Pathways. eLife 2021, 10, e70934. [Google Scholar] [CrossRef]
- Hu, C.; Zhu, Y.; Cui, Y.; Cheng, K.; Liang, W.; Wei, Z.; Zhu, M.; Yin, H.; Zeng, L.; Xiao, Y.; et al. A Group of Receptor Kinases Are Essential for CLAVATA Signalling to Maintain Stem Cell Homeostasis. Nat. Plants 2018, 4, 205–211. [Google Scholar] [CrossRef]
- Shimizu, N.; Ishida, T.; Yamada, M.; Shigenobu, S.; Tabata, R.; Kinoshita, A.; Yamaguchi, K.; Hasebe, M.; Mitsumasu, K.; Sawa, S. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 Constitute a Signaling Pathway and Modulate CLE Peptide-Triggered Growth Inhibition in Arabidopsis Root. New Phytol. 2015, 208, 1104–1113. [Google Scholar] [CrossRef]
- Chen, S.; Mitchum, M.G.; Wang, X. Characterization and Response of Two Potato Receptor-like Kinases to Cyst Nematode Infection. Plant Signal Behav. 2022, 17, 2148372. [Google Scholar] [CrossRef]
- Jun, J.; Fiume, E.; Roeder, A.H.K.; Meng, L.; Sharma, V.K.; Osmont, K.S.; Baker, C.; Ha, C.M.; Meyerowitz, E.M.; Feldman, L.J.; et al. Comprehensive Analysis of CLE Polypeptide Signaling Gene Expression and Overexpression Activity in Arabidopsis. Plant Physiol. 2010, 154, 1721–1736. [Google Scholar] [CrossRef]
- Meng, L.; Ruth, K.C.; Fletcher, J.C.; Feldman, L. The Roles of Different CLE Domains in Arabidopsis CLE Polypeptide Activity and Functional Specificity. Mol. Plant 2010, 3, 760–772. [Google Scholar] [CrossRef]
- Hobe, M.; Müller, R.; Grünewald, M.; Brand, U.; Simon, R. Loss of CLE40, a Protein Functionally Equivalent to the Stem Cell Restricting Signal CLV3, Enhances Root Waving in Arabidopsis. Dev. Genes Evol. 2003, 213, 371–381. [Google Scholar] [CrossRef]
- Adibi, M.; Yoshida, S.; Weijers, D.; Fleck, C. Centering the Organizing Center in the Arabidopsis thaliana Shoot Apical Meristem by a Combination of Cytokinin Signaling and Self-Organization. PLoS ONE 2016, 11, e0147830. [Google Scholar] [CrossRef]
- Wang, J.; Tian, C.; Zhang, C.; Shi, B.; Cao, X.; Zhang, T.-Q.; Zhao, Z.; Wang, J.-W.; Jiao, Y. Cytokinin Signaling Activates WUSCHEL Expression during Axillary Meristem Initiation. Plant Cell 2017, 29, 1373–1387. [Google Scholar] [CrossRef]
- Zhang, F.; May, A.; Irish, V.F. Type-B Arabidopsis Response Regulators Directly Activate WUSCHEL. Trends Plant Sci. 2017, 22, 815–817. [Google Scholar] [CrossRef]
- Snipes, S.A.; Rodriguez, K.; DeVries, A.E.; Miyawaki, K.N.; Perales, M.; Xie, M.; Reddy, G.V. Cytokinin Stabilizes WUSCHEL by Acting on the Protein Domains Required for Nuclear Enrichment and Transcription. PLoS Genet. 2018, 14, e1007351. [Google Scholar] [CrossRef]
- Han, P.; Zhu, Y.-X. BARD1 May Be Renamed ROW1 Because It Functions Mainly as a REPRESSOR OF WUSCHEL1. Plant Signal Behav. 2009, 4, 52–54. [Google Scholar] [CrossRef]
- Yu, L.P.; Miller, A.K.; Clark, S.E. POLTERGEIST Encodes a Protein Phosphatase 2C That Regulates CLAVATA Pathways Controlling Stem Cell Identity at Arabidopsis Shoot and Flower Meristems. Curr. Biol. 2003, 13, 179–188. [Google Scholar] [CrossRef]
- Bommert, P.; Je, B.I.; Goldshmidt, A.; Jackson, D. The Maize Gα Gene COMPACT PLANT2 Functions in CLAVATA Signalling to Control Shoot Meristem Size. Nature 2013, 502, 555–558. [Google Scholar] [CrossRef]
- Trotochaud, A.E.; Hao, T.; Wu, G.; Yang, Z.; Clark, S.E. The CLAVATA1 Receptor-like Kinase Requires CLAVATA3 for Its Assembly into a Signaling Complex That Includes KAPP and a Rho-Related Protein. Plant Cell 1999, 11, 393–406. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Takahashi, F.; Kinoshita, A.; Miwa, H.; Shinozaki, K.; Fukuda, H.; Sawa, S. Mitogen-Activated Protein Kinase Regulated by the CLAVATA Receptors Contributes to Shoot Apical Meristem Homeostasis. Plant Cell Physiol. 2011, 52, 14–29. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A Molecular Link between Stem Cell Regulation and Floral Patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef]
- Perales, M.; Rodriguez, K.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. Threshold-Dependent Transcriptional Discrimination Underlies Stem Cell Homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, E6298–E6306. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL Controls Meristem Function by Direct Regulation of Cytokinin-Inducible Response Regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Ohno, C.; Heisler, M.; Girke, T.; Jönsson, H.; Reddy, G.V. Plant Stem Cell Maintenance Involves Direct Transcriptional Repression of Differentiation Program. Mol. Syst. Biol. 2013, 9, 654. [Google Scholar] [CrossRef]
- Ma, Y.; Miotk, A.; Šutiković, Z.; Ermakova, O.; Wenzl, C.; Medzihradszky, A.; Gaillochet, C.; Forner, J.; Utan, G.; Brackmann, K.; et al. WUSCHEL Acts as an Auxin Response Rheostat to Maintain Apical Stem Cells in Arabidopsis. Nat. Commun. 2019, 10, 5093. [Google Scholar] [CrossRef]
- Daum, G.; Medzihradszky, A.; Suzaki, T.; Lohmann, J.U. A Mechanistic Framework for Noncell Autonomous Stem Cell Induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14619–14624. [Google Scholar] [CrossRef]
- Rodriguez, K.; Perales, M.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. DNA-Dependent Homodimerization, Sub-Cellular Partitioning, and Protein Destabilization Control WUSCHEL Levels and Spatial Patterning. Proc. Natl. Acad. Sci. USA 2016, 113, E6307–E6315. [Google Scholar] [CrossRef]
- Chickarmane, V.S.; Gordon, S.P.; Tarr, P.T.; Heisler, M.G.; Meyerowitz, E.M. Cytokinin Signaling as a Positional Cue for Patterning the Apical-Basal Axis of the Growing Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2012, 109, 4002–4007. [Google Scholar] [CrossRef]
- Schuetz, M.; Berleth, T.; Mattsson, J. Multiple MONOPTEROS-Dependent Pathways Are Involved in Leaf Initiation. Plant Physiol. 2008, 148, 870–880. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL Confines CLAVATA3 Expression to the Outer Apical Meristem Layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A Member of the KNOTTED Class of Homeodomain Proteins Encoded by the STM Gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Reiser, L.; Hake, S. Shoot Meristem Size Is Dependent on Inbred Background and Presence of the Maize Homeobox Gene, Knotted1. Development 2000, 127, 3161–3172. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of Pluripotency Pathways Regulates Stem Cell Maintenance in the Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef]
- Lopes, F.L.; Galvan-Ampudia, C.; Landrein, B. WUSCHEL in the Shoot Apical Meristem: Old Player, New Tricks. J. Exp. Bot. 2021, 72, 1527–1535. [Google Scholar] [CrossRef]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved Factors Regulate Signalling in Arabidopsis thaliana Shoot and Root Stem Cell Organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef]
- Richards, S.; Wink, R.H.; Simon, R. Mathematical Modelling of WOX5- and CLE40-Mediated Columella Stem Cell Homeostasis in Arabidopsis. J. Exp. Bot. 2015, 66, 5375–5384. [Google Scholar] [CrossRef]
- Gallois, J.-L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL Induces Shoot Stem Cell Activity and Developmental Plasticity in the Root Meristem. Genes. Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef]
- Lee, K.; Won, J.H.; Seo, P.J. Overexpression of the WOX5 Gene Inhibits Shoot Development. Plant Signal Behav. 2022, 17, 2050095. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiao, Y.; Liu, Z.; Zhu, Y.-X. ROW1 Maintains Quiescent Centre Identity by Confining WOX5 Expression to Specific Cells. Nat. Commun. 2015, 6, 6003. [Google Scholar] [CrossRef]
- Ding, Z.; Friml, J. Auxin Regulates Distal Stem Cell Differentiation in Arabidopsis Roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Shimotohno, A.; Heidstra, R.; Blilou, I.; Scheres, B. Root Stem Cell Niche Organizer Specification by Molecular Convergence of PLETHORA and SCARECROW Transcription Factor Modules. Genes Dev. 2018, 32, 1085–1100. [Google Scholar] [CrossRef]
- Guillotin, B.; Birnbaum, K.D. Just Passing through: The Auxin Gradient of the Root Meristem. Curr. Top. Dev. Biol. 2020, 137, 433–454. [Google Scholar] [CrossRef]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An Auxin-Dependent Distal Organizer of Pattern and Polarity in the Arabidopsis Root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN Auxin Efflux Facilitator Network Controls Growth and Patterning in Arabidopsis Roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Tian, H.; Wabnik, K.; Niu, T.; Li, H.; Yu, Q.; Pollmann, S.; Vanneste, S.; Govaerts, W.; Rolcík, J.; Geisler, M.; et al. WOX5-IAA17 Feedback Circuit-Mediated Cellular Auxin Response Is Crucial for the Patterning of Root Stem Cell Niches in Arabidopsis. Mol. Plant 2014, 7, 277–289. [Google Scholar] [CrossRef]
- Hu, X.; Xu, L. Transcription Factors WOX11/12 Directly Activate WOX5/7 to Promote Root Primordia Initiation and Organogenesis1. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.-S.; Amasino, R.; Scheres, B. The PLETHORA Genes Mediate Patterning of the Arabidopsis Root Stem Cell Niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA Proteins as Dose-Dependent Master Regulators of Arabidopsis Root Development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef]
- Mähönen, A.P.; Ten Tusscher, K.; Siligato, R.; Smetana, O.; Díaz-Triviño, S.; Salojärvi, J.; Wachsman, G.; Prasad, K.; Heidstra, R.; Scheres, B. PLETHORA Gradient Formation Mechanism Separates Auxin Responses. Nature 2014, 515, 125–129. [Google Scholar] [CrossRef]
- Scheres, B.; Krizek, B.A. Coordination of Growth in Root and Shoot Apices by AIL/PLT Transcription Factors. Curr. Opin. Plant Biol. 2018, 41, 95–101. [Google Scholar] [CrossRef]
- Santuari, L.; Sanchez-Perez, G.F.; Luijten, M.; Rutjens, B.; Terpstra, I.; Berke, L.; Gorte, M.; Prasad, K.; Bao, D.; Timmermans-Hereijgers, J.L.P.M.; et al. The PLETHORA Gene Regulatory Network Guides Growth and Cell Differentiation in Arabidopsis Roots. Plant Cell 2016, 28, 2937–2951. [Google Scholar] [CrossRef]
- Burkart, R.C.; Strotmann, V.I.; Kirschner, G.K.; Akinci, A.; Czempik, L.; Dolata, A.; Maizel, A.; Weidtkamp-Peters, S.; Stahl, Y. PLETHORA-WOX5 Interaction and Subnuclear Localization Control Arabidopsis Root Stem Cell Maintenance. EMBO Rep. 2022, 23, e54105. [Google Scholar] [CrossRef]
- Sabatini, S.; Heidstra, R.; Wildwater, M.; Scheres, B. SCARECROW Is Involved in Positioning the Stem Cell Niche in the Arabidopsis Root Meristem. Genes Dev. 2003, 17, 354–358. [Google Scholar] [CrossRef]
- Cruz-Ramírez, A.; Díaz-Triviño, S.; Wachsman, G.; Du, Y.; Arteága-Vázquez, M.; Zhang, H.; Benjamins, R.; Blilou, I.; Neef, A.B.; Chandler, V.; et al. A SCARECROW-RETINOBLASTOMA Protein Network Controls Protective Quiescence in the Arabidopsis Root Stem Cell Organizer. PLoS Biol. 2013, 11, e1001724. [Google Scholar] [CrossRef]
- Danisman, S.; van Dijk, A.D.J.; Bimbo, A.; van der Wal, F.; Hennig, L.; de Folter, S.; Angenent, G.C.; Immink, R.G.H. Analysis of Functional Redundancies within the Arabidopsis TCP Transcription Factor Family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef]
- Czyzewicz, N.; Nikonorova, N.; Meyer, M.R.; Sandal, P.; Shah, S.; Vu, L.D.; Gevaert, K.; Rao, A.G.; De Smet, I. The Growing Story of (ARABIDOPSIS) CRINKLY 4. J. Exp. Bot. 2016, 67, 4835–4847. [Google Scholar] [CrossRef]
- Stahl, Y.; Grabowski, S.; Bleckmann, A.; Kühnemuth, R.; Weidtkamp-Peters, S.; Pinto, K.G.; Kirschner, G.K.; Schmid, J.B.; Wink, R.H.; Hülsewede, A.; et al. Moderation of Arabidopsis Root Stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 Receptor Kinase Complexes. Curr. Biol. 2013, 23, 362–371. [Google Scholar] [CrossRef]
- Stahl, Y.; Simon, R. Gated Communities: Apoplastic and Symplastic Signals Converge at Plasmodesmata to Control Cell Fates. J. Exp. Bot. 2013, 64, 5237–5241. [Google Scholar] [CrossRef]
- Berckmans, B.; Kirschner, G.; Gerlitz, N.; Stadler, R.; Simon, R. CLE40 Signaling Regulates Root Stem Cell Fate1. Plant Physiol. 2020, 182, 1776–1792. [Google Scholar] [CrossRef]
- Müller, R.; Bleckmann, A.; Simon, R. The Receptor Kinase CORYNE of Arabidopsis Transmits the Stem Cell–Limiting Signal CLAVATA3 Independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof Family of Plant Transcription Factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Savina, M.S.; Pasternak, T.; Omelyanchuk, N.A.; Novikova, D.D.; Palme, K.; Mironova, V.V.; Lavrekha, V.V. Cell Dynamics in WOX5-Overexpressing Root Tips: The Impact of Local Auxin Biosynthesis. Front. Plant Sci. 2020, 11, 560169. [Google Scholar] [CrossRef]
- Boniotti, M.B.; Gutierrez, C. A Cell-Cycle-Regulated Kinase Activity Phosphorylates Plant Retinoblastoma Protein and Contains, in Arabidopsis, a CDKA/Cyclin D Complex. Plant J. 2001, 28, 341–350. [Google Scholar] [CrossRef]
- Forzani, C.; Aichinger, E.; Sornay, E.; Willemsen, V.; Laux, T.; Dewitte, W.; Murray, J.A.H. WOX5 Suppresses CYCLIN D Activity to Establish Quiescence at the Center of the Root Stem Cell Niche. Curr. Biol. 2014, 24, 1939–1944. [Google Scholar] [CrossRef]
- Romeiro Motta, M.; Zhao, X.; Pastuglia, M.; Belcram, K.; Roodbarkelari, F.; Komaki, M.; Harashima, H.; Komaki, S.; Kumar, M.; Bulankova, P.; et al. B1-Type Cyclins Control Microtubule Organization during Cell Division in Arabidopsis. EMBO Rep. 2022, 23, e53995. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The Main Auxin Biosynthesis Pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and Function of Nuclear Bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef]
- Ji, J.; Strable, J.; Shimizu, R.; Koenig, D.; Sinha, N.; Scanlon, M.J. WOX4 Promotes Procambial Development. Plant Physiol. 2010, 152, 1346–1356. [Google Scholar] [CrossRef]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 Act Downstream of the PXY Receptor Kinase to Regulate Plant Vascular Proliferation Independently of Any Role in Vascular Organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF Peptide Signaling Regulates Vascular Stem Cell Proliferation via the WOX4 Homeobox Gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-Cell-Autonomous Control of Vascular Stem Cell Fate by a CLE Peptide/Receptor System. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 Receptor Ligand Pair Defines a Multifunctional Pathway That Controls the Rate and Orientation of Vascular Cell Division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef]
- Etchells, J.P.; Smit, M.E.; Gaudinier, A.; Williams, C.J.; Brady, S.M. A Brief History of the TDIF-PXY Signalling Module: Balancing Meristem Identity and Differentiation during Vascular Development. New Phytol. 2016, 209, 474–484. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Shirasu, K.; Fukuda, H. Plant GSK3 Proteins Regulate Xylem Cell Differentiation Downstream of TDIF-TDR Signalling. Nat. Commun. 2014, 5, 3504. [Google Scholar] [CrossRef]
- Saito, M.; Kondo, Y.; Fukuda, H. BES1 and BZR1 Redundantly Promote Phloem and Xylem Differentiation. Plant Cell Physiol. 2018, 59, 590–600. [Google Scholar] [CrossRef]
- Smit, M.E.; McGregor, S.R.; Sun, H.; Gough, C.; Bågman, A.-M.; Soyars, C.L.; Kroon, J.T.; Gaudinier, A.; Williams, C.J.; Yang, X.; et al. A PXY-Mediated Transcriptional Network Integrates Signaling Mechanisms to Control Vascular Development in Arabidopsis. Plant Cell 2020, 32, 319–335. [Google Scholar] [CrossRef]
- Fisher, K.; Turner, S. PXY, a Receptor-like Kinase Essential for Maintaining Polarity during Plant Vascular-Tissue Development. Curr. Biol. 2007, 17, 1061–1066. [Google Scholar] [CrossRef]
- Zhang, H.; Lin, X.; Han, Z.; Wang, J.; Qu, L.-J.; Chai, J. SERK Family Receptor-like Kinases Function as Co-Receptors with PXY for Plant Vascular Development. Mol. Plant 2016, 9, 1406–1414. [Google Scholar] [CrossRef]
- Gursanscky, N.R.; Jouannet, V.; Grünwald, K.; Sanchez, P.; Laaber-Schwarz, M.; Greb, T. MOL1 Is Required for Cambium Homeostasis in Arabidopsis. Plant J. 2016, 86, 210–220. [Google Scholar] [CrossRef]
- Ren, S.-C.; Song, X.-F.; Chen, W.-Q.; Lu, R.; Lucas, W.J.; Liu, C.-M. CLE25 Peptide Regulates Phloem Initiation in Arabidopsis through a CLERK-CLV2 Receptor Complex. J. Integr. Plant Biol. 2019, 61, 1043–1061. [Google Scholar] [CrossRef]
- Qian, P.; Song, W.; Zaizen-Iida, M.; Kume, S.; Wang, G.; Zhang, Y.; Kinoshita-Tsujimura, K.; Chai, J.; Kakimoto, T. A Dof-CLE Circuit Controls Phloem Organization. Nat. Plants 2022, 8, 817–827. [Google Scholar] [CrossRef]
- Bonke, M.; Thitamadee, S.; Mähönen, A.P.; Hauser, M.-T.; Helariutta, Y. APL Regulates Vascular Tissue Identity in Arabidopsis. Nature 2003, 426, 181–186. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Regan, S.; Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN Transcription Factor Family Are Involved in the Regulation of Secondary Growth in Populus. Plant Cell 2010, 22, 3662–3677. [Google Scholar] [CrossRef]
- Wang, R.; Ji, Y.; Wang, J.; Yang, S.; Song, Y. Vascular Expression of Populus LRR-RLK Genes and the Effects of Their Overexpression on Wood Formation. Mol. Breed. 2015, 35, 220. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, D.; Zhang, R.; Luo, L.; Cao, S.; Huang, C.; Sun, J.; Gui, J.; Li, L. A Xylem-produced Peptide PtrCLE20 Inhibits Vascular Cambium Activity in Populus. Plant Biotechnol. J. 2020, 18, 195–206. [Google Scholar] [CrossRef]
- Kucukoglu, M.; Chaabouni, S.; Zheng, B.; Mähönen, A.P.; Helariutta, Y.; Nilsson, O. Peptide Encoding Populus CLV3/ESR-RELATED 47 (PttCLE47) Promotes Cambial Development and Secondary Xylem Formation in Hybrid Aspen. New Phytol. 2020, 226, 75–85. [Google Scholar] [CrossRef]
- Mergaert, P.; Kereszt, A.; Kondorosi, E. Gene Expression in Nitrogen-Fixing Symbiotic Nodule Cells in Medicago truncatula and Other Nodulating Plants. Plant Cell 2020, 32, 42–68. [Google Scholar] [CrossRef]
- Oka-Kira, E.; Kawaguchi, M. Long-Distance Signaling to Control Root Nodule Number. Curr. Opin. Plant Biol. 2006, 9, 496–502. [Google Scholar] [CrossRef]
- Searle, I.R.; Men, A.E.; Laniya, T.S.; Buzas, D.M.; Iturbe-Ormaetxe, I.; Carroll, B.J.; Gresshoff, P.M. Long-Distance Signaling in Nodulation Directed by a CLAVATA1-like Receptor Kinase. Science 2003, 299, 109–112. [Google Scholar] [CrossRef]
- Schnabel, E.; Journet, E.-P.; de Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The Medicago truncatula SUNN Gene Encodes a CLV1-like Leucine-Rich Repeat Receptor Kinase That Regulates Nodule Number and Root Length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod Factor/Nitrate-Induced CLE Genes That Drive HAR1-Mediated Systemic Regulation of Nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’haeseleer, K.; Holsters, M.; Goormachtig, S. CLE Peptides Control Medicago truncatula Nodulation Locally and Systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef]
- Nishida, H.; Handa, Y.; Tanaka, S.; Suzaki, T.; Kawaguchi, M. Expression of the CLE-RS3 Gene Suppresses Root Nodulation in Lotus japonicus. J. Plant Res. 2016, 129, 909–919. [Google Scholar] [CrossRef]
- Hastwell, A.H.; de Bang, T.C.; Gresshoff, P.M.; Ferguson, B.J. Author Correction: CLE Peptide-Encoding Gene Families in Medicago truncatula and Lotus japonicus, Compared with Those of Soybean, Common Bean and Arabidopsis. Sci. Rep. 2017, 7, 15474. [Google Scholar] [CrossRef]
- Lebedeva, M.; Azarakhsh, M.; Yashenkova, Y.; Lutova, L. Nitrate-Induced CLE Peptide Systemically Inhibits Nodulation in Medicago truncatula. Plants 2020, 9, 1456. [Google Scholar] [CrossRef]
- Mens, C.; Hastwell, A.H.; Su, H.; Gresshoff, P.M.; Mathesius, U.; Ferguson, B.J. Characterisation of Medicago truncatula CLE34 and CLE35 in Nitrate and Rhizobia Regulation of Nodulation. New Phytol. 2021, 229, 2525–2534. [Google Scholar] [CrossRef]
- Moreau, C.; Gautrat, P.; Frugier, F. Nitrate-Induced CLE35 Signaling Peptides Inhibit Nodulation through the SUNN Receptor and MiR2111 Repression. Plant Physiol. 2021, 185, 1216–1228. [Google Scholar] [CrossRef]
- Osipova, M.A.; Mortier, V.; Demchenko, K.N.; Tsyganov, V.E.; Tikhonovich, I.A.; Lutova, L.A.; Dolgikh, E.A.; Goormachtig, S. WUSCHEL-RELATED HOMEOBOX5 Gene Expression and Interaction of CLE Peptides with Components of the Systemic Control Add Two Pieces to the Puzzle of Autoregulation of Nodulation. Plant Physiol. 2012, 158, 1329–1341. [Google Scholar] [CrossRef]
- Liu, C.-W.; Breakspear, A.; Guan, D.; Cerri, M.R.; Jackson, K.; Jiang, S.; Robson, F.; Radhakrishnan, G.V.; Roy, S.; Bone, C.; et al. NIN Acts as a Network Hub Controlling a Growth Module Required for Rhizobial Infection. Plant Physiol. 2019, 179, 1704–1722. [Google Scholar] [CrossRef]
- Liu, J.; Bisseling, T. Evolution of NIN and NIN-like Genes in Relation to Nodule Symbiosis. Genes. 2020, 11, 777. [Google Scholar] [CrossRef]
- Azarakhsh, M.; Lebedeva, M.A.; Lutova, L.A. Identification and Expression Analysis of Medicago truncatula Isopentenyl Transferase Genes (IPTs) Involved in Local and Systemic Control of Nodulation. Front. Plant Sci. 2018, 9, 304. [Google Scholar] [CrossRef]
- Serra, O.; Mähönen, A.P.; Hetherington, A.J.; Ragni, L. The Making of Plant Armor: The Periderm. Annu. Rev. Plant Biol. 2022, 73, 405–432. [Google Scholar] [CrossRef]
- Xiao, W.; Molina, D.; Wunderling, A.; Ripper, D.; Vermeer, J.E.M.; Ragni, L. Pluripotent Pericycle Cells Trigger Different Growth Outputs by Integrating Developmental Cues into Distinct Regulatory Modules. Curr. Biol. 2020, 30, 4384–4398.e5. [Google Scholar] [CrossRef]
- Wunderling, A.; Ripper, D.; Barra-Jimenez, A.; Mahn, S.; Sajak, K.; Targem, M.B.; Ragni, L. A Molecular Framework to Study Periderm Formation in Arabidopsis. New Phytol. 2018, 219, 216–229. [Google Scholar] [CrossRef]
- Fernández-Piñán, S.; Boher, P.; Soler, M.; Figueras, M.; Serra, O. Transcriptomic Analysis of Cork during Seasonal Growth Highlights Regulatory and Developmental Processes from Phellogen to Phellem Formation. Sci. Rep. 2021, 11, 12053. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Rymen, B.; Lambolez, A.; Kojima, M.; Takebayashi, Y.; Heyman, J.; Watanabe, S.; Seo, M.; De Veylder, L.; et al. Wounding Triggers Callus Formation via Dynamic Hormonal and Transcriptional Changes. Plant Physiol. 2017, 175, 1158–1174. [Google Scholar] [CrossRef]
- Radhakrishnan, D.; Shanmukhan, A.P.; Kareem, A.; Aiyaz, M.; Varapparambathu, V.; Toms, A.; Kerstens, M.; Valsakumar, D.; Landge, A.N.; Shaji, A.; et al. A Coherent Feed-Forward Loop Drives Vascular Regeneration in Damaged Aerial Organs of Plants Growing in a Normal Developmental Context. Development 2020, 147, dev185710. [Google Scholar] [CrossRef]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis Regeneration from Multiple Tissues Occurs via a Root Development Pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 Are Involved in the First-Step Cell Fate Transition during de Novo Root Organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Lee, K.; Kim, J.H.; Park, O.-S.; Jung, Y.J.; Seo, P.J. Ectopic Expression of WOX5 Promotes Cytokinin Signaling and de Novo Shoot Regeneration. Plant Cell Rep. 2022, 41, 2415–2422. [Google Scholar] [CrossRef]
- Kyo, M.; Maida, K.; Nishioka, Y.; Matsui, K. Coexpression of WUSCHEL Related Homeobox (WOX) 2 with WOX8 or WOX9 Promotes Regeneration from Leaf Segments and Free Cells in Nicotiana tabacum L. Plant Biotechnol. 2018, 35, 23–30. [Google Scholar] [CrossRef]
- Tvorogova, V.E.; Krasnoperova, E.Y.; Kudriashov, A.A.; Kuznetsova, K.A.; Potsenkovskaya, E.A.; Fedorova, Y.A.; Lutova, L.A. Transcriptomic Analysis of Medicago truncatula Calli with MtWOX9-1 Overexpression. Vestn. VOGiS 2019, 23, 691–699. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Ito, T.; Tanaka, H.; Favero, D.S.; Kawamura, A.; Sakamoto, S.; Wakazaki, M.; Tameshige, T.; Fujii, H.; et al. Wound-Inducible WUSCHEL-RELATED HOMEOBOX 13 Is Required for Callus Growth and Organ Reconnection. Plant Physiol. 2021, 188, 425–441. [Google Scholar] [CrossRef]
- Kadri, A.; Grenier De March, G.; Guerineau, F.; Cosson, V.; Ratet, P. WUSCHEL Overexpression Promotes Callogenesis and Somatic Embryogenesis in Medicago truncatula Gaertn. Plants 2021, 10, 715. [Google Scholar] [CrossRef]
- Kang, J.; Wang, X.; Ishida, T.; Grienenberger, E.; Zheng, Q.; Wang, J.; Zhang, Y.; Chen, W.; Chen, M.; Song, X.-F.; et al. A Group of CLE Peptides Regulates de Novo Shoot Regeneration in Arabidopsis thaliana. New Phytol. 2022, 235, 2300–2312. [Google Scholar] [CrossRef]
- Kudriashov, A.A.; Zlydneva, N.S.; Efremova, E.P.; Tvorogova, V.E.; Lutova, L.A. MtCLE08, MtCLE16, and MtCLE18 Transcription Patterns and Their Possible Functions in the Embryogenic Calli of Medicago truncatula. Plants 2023, 12, 435. [Google Scholar] [CrossRef]
- Dodueva, I.E.; Lebedeva, M.A.; Kuznetsova, K.A.; Gancheva, M.S.; Paponova, S.S.; Lutova, L.L. Plant Tumors: A Hundred Years of Study. Planta 2020, 251, 82. [Google Scholar] [CrossRef]
- Lebedeva (Osipova), M.A.; Tvorogova, V.E.; Vinogradova, A.P.; Gancheva, M.S.; Azarakhsh, M.; Ilina, E.L.; Demchenko, K.N.; Dodueva, I.E.; Lutova, L.A. Initiation of Spontaneous Tumors in Radish (Raphanus sativus): Cellular, Molecular and Physiological Events. J. Plant Physiol. 2015, 173, 97–104. [Google Scholar] [CrossRef]
- Wang, J.; Lee, C.; Replogle, A.; Joshi, S.; Korkin, D.; Hussey, R.; Baum, T.J.; Davis, E.L.; Wang, X.; Mitchum, M.G. Dual Roles for the Variable Domain in Protein Trafficking and Host-Specific Recognition of Heterodera glycines CLE Effector Proteins. New Phytol. 2010, 187, 1003–1017. [Google Scholar] [CrossRef]
- Wang, J.; Replogle, A.; Hussey, R.; Baum, T.; Wang, X.; Davis, E.L.; Mitchum, M.G. Identification of Potential Host Plant Mimics of CLAVATA3/ESR (CLE)-like Peptides from the Plant-Parasitic Nematode Heterodera schachtii. Mol. Plant Pathol. 2011, 12, 177–186. [Google Scholar] [CrossRef]
- Replogle, A.; Wang, J.; Paolillo, V.; Smeda, J.; Kinoshita, A.; Durbak, A.; Tax, F.E.; Wang, X.; Sawa, S.; Mitchum, M.G. Synergistic Interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in Cyst Nematode Parasitism of Arabidopsis. Mol. Plant Microbe Interact. 2013, 26, 87–96. [Google Scholar] [CrossRef]
- Guo, X.; Wang, J.; Gardner, M.; Fukuda, H.; Kondo, Y.; Etchells, J.P.; Wang, X.; Mitchum, M.G. Identification of Cyst Nematode B-Type CLE Peptides and Modulation of the Vascular Stem Cell Pathway for Feeding Cell Formation. PLoS Pathog. 2017, 13, e1006142. [Google Scholar] [CrossRef]
- Patel, N.; Hamamouch, N.; Li, C.; Hussey, R.; Mitchum, M.; Baum, T.; Wang, X.; Davis, E.L. Similarity and Functional Analyses of Expressed Parasitism Genes in Heterodera schachtii and Heterodera glycines. J. Nematol. 2008, 40, 299–310. [Google Scholar]
- Haegeman, A.; Mantelin, S.; Jones, J.T.; Gheysen, G. Functional Roles of Effectors of Plant-Parasitic Nematodes. Gene 2012, 492, 19–31. [Google Scholar] [CrossRef]
- Nemec-Venza, Z.; Madden, C.; Stewart, A.; Liu, W.; Novák, O.; Pěnčík, A.; Cuming, A.C.; Kamisugi, Y.; Harrison, C.J. CLAVATA Modulates Auxin Homeostasis and Transport to Regulate Stem Cell Identity and Plant Shape in a Moss. New Phytol. 2022, 234, 149–163. [Google Scholar] [CrossRef]
- Gheysen, G.; Mitchum, M.G. Phytoparasitic Nematode Control of Plant Hormone Pathways. Plant Physiol. 2019, 179, 1212–1226. [Google Scholar] [CrossRef]
- Taoka, K.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 Proteins Act as Intracellular Receptors for Rice Hd3a Florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef]
- Pnueli, L.; Gutfinger, T.; Hareven, D.; Ben-Naim, O.; Ron, N.; Adir, N.; Lifschitz, E. Tomato SP-Interacting Proteins Define a Conserved Signaling System That Regulates Shoot Architecture and Flowering. Plant Cell 2001, 13, 2687–2702. [Google Scholar] [CrossRef]
- De Boer, A.H.; van Kleeff, P.J.M.; Gao, J. Plant 14-3-3 Proteins as Spiders in a Web of Phosphorylation. Protoplasma 2013, 250, 425–440. [Google Scholar] [CrossRef]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of Spatial and Temporal Information during Floral Induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef]
- Kawamoto, N.; Sasabe, M.; Endo, M.; Machida, Y.; Araki, T. Calcium-Dependent Protein Kinases Responsible for the Phosphorylation of a BZIP Transcription Factor FD Crucial for the Florigen Complex Formation. Sci. Rep. 2015, 5, 8341. [Google Scholar] [CrossRef]
- Collani, S.; Neumann, M.; Yant, L.; Schmid, M. FT Modulates Genome-Wide DNA-Binding of the BZIP Transcription Factor FD. Plant Physiol. 2019, 180, 367–380. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Wigge, P.A. FT Protein Acts as a Long-Range Signal in Arabidopsis. Curr. Biol. 2007, 17, 1050–1054. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a Chrysanthemum FLOWERING LOCUS T-like Gene, Is a Key Regulator of Photoperiodic Flowering in Chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation Tagging of the Floral Inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef]
- Chailakhyan, M.K. Gormonal’naya Teoriya Razvitiya Rastenii (Hormonal Theory of Plant Development); Akad Nauk SSSR: Moscow, Russia, 1937. [Google Scholar]
- Matsoukas, I.G. Florigens and Antiflorigens: A Molecular Genetic Understanding. Essays Biochem. 2015, 58, 133–149. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Hou, X.; Xi, W.; Shen, L.; Tao, Z.; Wang, Y.; Yu, H. FTIP1 Is an Essential Regulator Required for Florigen Transport. PLoS Biol. 2012, 10, e1001313. [Google Scholar] [CrossRef]
- Liu, L.; Li, C.; Song, S.; Teo, Z.W.N.; Shen, L.; Wang, Y.; Jackson, D.; Yu, H. FTIP-Dependent STM Trafficking Regulates Shoot Meristem Development in Arabidopsis. Cell Rep. 2018, 23, 1879–1890. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, L.; Shen, L.; Yu, H. NaKR1 Regulates Long-Distance Movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2016, 2, 16075. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP Gene Family in Plants: Functional Diversification in Seed Plant Evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef]
- Yu, X.; Liu, H.; Sang, N.; Li, Y.; Zhang, T.; Sun, J.; Huang, X. Identification of Cotton MOTHER OF FT AND TFL1 Homologs, GhMFT1 and GhMFT2, Involved in Seed Germination. PLoS ONE 2019, 14, e0215771. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 Regulates Seed Germination through a Negative Feedback Loop Modulating ABA Signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Vaistij, F.E.; Barros-Galvão, T.; Cole, A.F.; Gilday, A.D.; He, Z.; Li, Y.; Harvey, D.; Larson, T.R.; Graham, I.A. MOTHER-OF-FT-AND-TFL1 Represses Seed Germination under Far-Red Light by Modulating Phytohormone Responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, 8442–8447. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Yang, K.-Z.; Wei, X.-X.; Wang, X.-Q. Revisiting the Phosphatidylethanolamine-Binding Protein (PEBP) Gene Family Reveals Cryptic FLOWERING LOCUS T Gene Homologs in Gymnosperms and Sheds New Light on Functional Evolution. New Phytol. 2016, 212, 730–744. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Kobayashi, Y.; Goto, K.; Abe, M.; Araki, T. TWIN SISTER OF FT (TSF) Acts as a Floral Pathway Integrator Redundantly with FT. Plant Cell Physiol. 2005, 46, 1175–1189. [Google Scholar] [CrossRef]
- Kinoshita, T.; Ono, N.; Hayashi, Y.; Morimoto, S.; Nakamura, S.; Soda, M.; Kato, Y.; Ohnishi, M.; Nakano, T.; Inoue, S.; et al. FLOWERING LOCUS T Regulates Stomatal Opening. Curr. Biol. 2011, 21, 1232–1238. [Google Scholar] [CrossRef]
- Huang, N.-C.; Jane, W.-N.; Chen, J.; Yu, T.-S. Arabidopsis thaliana CENTRORADIALIS Homologue (ATC) Acts Systemically to Inhibit Floral Initiation in Arabidopsis. Plant J. 2012, 72, 175–184. [Google Scholar] [CrossRef]
- Goretti, D.; Silvestre, M.; Collani, S.; Langenecker, T.; Méndez, C.; Madueño, F.; Schmid, M. TERMINAL FLOWER1 Functions as a Mobile Transcriptional Cofactor in the Shoot Apical Meristem. Plant Physiol. 2020, 182, 2081–2095. [Google Scholar] [CrossRef]
- Conti, L.; Bradley, D. TERMINAL FLOWER1 Is a Mobile Signal Controlling Arabidopsis Architecture. Plant Cell 2007, 19, 767–778. [Google Scholar] [CrossRef]
- Baumann, K.; Venail, J.; Berbel, A.; Domenech, M.J.; Money, T.; Conti, L.; Hanzawa, Y.; Madueno, F.; Bradley, D. Changing the Spatial Pattern of TFL1 Expression Reveals Its Key Role in the Shoot Meristem in Controlling Arabidopsis Flowering Architecture. J. Exp. Bot. 2015, 66, 4769–4780. [Google Scholar] [CrossRef]
- Shannon, S.; Meeks-Wagner, D.R. A Mutation in the Arabidopsis TFL1 Gene Affects Inflorescence Meristem Development. Plant Cell 1991, 3, 877–892. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Money, T.; Bradley, D. A Single Amino Acid Converts a Repressor to an Activator of Flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A Divergent External Loop Confers Antagonistic Activity on Floral Regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early Evolution of the MFT-like Gene Family in Plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Shen, Y.; Chang, H.-C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The Flowering Time Regulator CONSTANS Is Recruited to the FLOWERING LOCUS T Promoter via a Unique Cis-Element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for MiR156-SPL Module in Arabidopsis Thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Wood, C.C.; Robertson, M.; James Peacock, W.; Dennis, E.S. The Arabidopsis FLC Protein Interacts Directly in Vivo with SOC1 and FT Chromatin and Is Part of a High-Molecular-Weight Protein Complex. Plant J. 2006, 46, 183–192. [Google Scholar] [CrossRef]
- Tsuji, H.; Taoka, K.; Shimamoto, K. Regulation of Flowering in Rice: Two Florigen Genes, a Complex Gene Network, and Natural Variation. Curr. Opin. Plant Biol. 2011, 14, 45–52. [Google Scholar] [CrossRef]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1,a CONSTANS Ortholog in Rice, Functions as an Ehd1 Repressor through Interaction with Monocot-Specific CCT-Domain Protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef]
- Woods, D.P.; McKeown, M.A.; Dong, Y.; Preston, J.C.; Amasino, R.M. Evolution of VRN2/Ghd7-Like Genes in Vernalization-Mediated Repression of Grass Flowering. Plant Physiol. 2016, 170, 2124–2135. [Google Scholar] [CrossRef]
- Pin, P.A.; Zhang, W.; Vogt, S.H.; Dally, N.; Büttner, B.; Schulze-Buxloh, G.; Jelly, N.S.; Chia, T.Y.P.; Mutasa-Göttgens, E.S.; Dohm, J.C.; et al. The Role of a Pseudo-Response Regulator Gene in Life Cycle Adaptation and Domestication of Beet. Curr. Biol. 2012, 22, 1095–1101. [Google Scholar] [CrossRef]
- Sun, J.; Wang, H.; Ren, L.; Chen, S.; Chen, F.; Jiang, J. CmFTL2 Is Involved in the Photoperiod- and Sucrose-Mediated Control of Flowering Time in Chrysanthemum. Hortic. Res. 2017, 4, 17001. [Google Scholar] [CrossRef]
- Nakano, Y.; Takase, T.; Takahashi, S.; Sumitomo, K.; Higuchi, Y.; Hisamatsu, T. Chrysanthemum Requires Short-Day Repeats for Anthesis: Gradual CsFTL3 Induction through a Feedback Loop under Short-Day Conditions. Plant Sci. 2019, 283, 247–255. [Google Scholar] [CrossRef]
- Böhlenius, H.; Huang, T.; Charbonnel-Campaa, L.; Brunner, A.M.; Jansson, S.; Strauss, S.H.; Nilsson, O. CO/FT Regulatory Module Controls Timing of Flowering and Seasonal Growth Cessation in Trees. Science 2006, 312, 1040–1043. [Google Scholar] [CrossRef]
- Mohamed, R.; Wang, C.-T.; Ma, C.; Shevchenko, O.; Dye, S.J.; Puzey, J.R.; Etherington, E.; Sheng, X.; Meilan, R.; Strauss, S.H.; et al. Populus CEN/TFL1 Regulates First Onset of Flowering, Axillary Meristem Identity and Dormancy Release in Populus. Plant J. 2010, 62, 674–688. [Google Scholar] [CrossRef]
- Brunner, A.M.; Evans, L.M.; Hsu, C.-Y.; Sheng, X. Vernalization and the Chilling Requirement to Exit Bud Dormancy: Shared or Separate Regulation? Front. Plant Sci. 2014, 5, 732. [Google Scholar] [CrossRef]
- Niwa, M.; Daimon, Y.; Kurotani, K.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 Interacts with FLOWERING LOCUS T to Repress the Floral Transition of the Axillary Meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef]
- Higuchi, Y.; Hisamatsu, T. CsTFL1, a Constitutive Local Repressor of Flowering, Modulates Floral Initiation by Antagonising Florigen Complex Activity in Chrysanthemum. Plant Sci. 2015, 237, 1–7. [Google Scholar] [CrossRef]
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of Flowering and Storage Organ Formation in Potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef]
- Teo, C.-J.; Takahashi, K.; Shimizu, K.; Shimamoto, K.; Taoka, K.-I. Potato Tuber Induction Is Regulated by Interactions between Components of a Tuberigen Complex. Plant Cell Physiol. 2017, 58, 365–374. [Google Scholar] [CrossRef]
- González-Schain, N.D.; Díaz-Mendoza, M.; Zurczak, M.; Suárez-López, P. Potato CONSTANS Is Involved in Photoperiodic Tuberization in a Graft-Transmissible Manner. Plant J. 2012, 70, 678–690. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Cruz-Oró, E.; Franco-Zorrilla, J.M.; Prat, S. Potato StCONSTANS-Like1 Suppresses Storage Organ Formation by Directly Activating the FT-like StSP5G Repressor. Curr. Biol. 2016, 26, 872–881. [Google Scholar] [CrossRef]
- Lehretz, G.G.; Sonnewald, S.; Hornyik, C.; Corral, J.M.; Sonnewald, U. Post-Transcriptional Regulation of FLOWERING LOCUS T Modulates Heat-Dependent Source-Sink Development in Potato. Curr. Biol. 2019, 29, 1614–1624.e3. [Google Scholar] [CrossRef]
- Morris, W.L.; Ducreux, L.J.M.; Morris, J.; Campbell, R.; Usman, M.; Hedley, P.E.; Prat, S.; Taylor, M.A. Identification of TIMING OF CAB EXPRESSION 1 as a Temperature-Sensitive Negative Regulator of Tuberization in Potato. J. Exp. Bot. 2019, 70, 5703–5714. [Google Scholar] [CrossRef]
- Gebregwergis, F.; Gebremicheal, M.; Gebremedhin, H.; Asefa, A. The Effects of Flower Removal and Earthing up on Tuber Yield and Quality of Potato (Solanum tuberosum L.). J. Agric. Sci. BGD 2021, 66, 121–137. [Google Scholar] [CrossRef]
- Gao, K.; Zhang, Z.; Zhu, T.; Coulter, J.A. The Influence of Flower Removal on Tuber Yield and Biomass Characteristics of Helianthus tuberosus L. in a Semi-Arid Area. Ind. Crops Prod. 2020, 150, 112374. [Google Scholar] [CrossRef]
- Plantenga, F.D.M.; Bergonzi, S.; Abelenda, J.A.; Bachem, C.W.B.; Visser, R.G.F.; Heuvelink, E.; Marcelis, L.F.M. The Tuberization Signal StSP6A Represses Flower Bud Development in Potato. J. Exp. Bot. 2019, 70, 937–948. [Google Scholar] [CrossRef]
- Lee, R.; Baldwin, S.; Kenel, F.; McCallum, J.; Macknight, R. FLOWERING LOCUS T Genes Control Onion Bulb Formation and Flowering. Nat. Commun. 2013, 4, 2884. [Google Scholar] [CrossRef]
- Atif, M.J.; Ahanger, M.A.; Amin, B.; Ghani, M.I.; Ali, M.; Cheng, Z. Mechanism of Allium Crops Bulb Enlargement in Response to Photoperiod: A Review. Int. J. Mol. Sci. 2020, 21, 1325. [Google Scholar] [CrossRef]
- Shalit-Kaneh, A.; Eviatar-Ribak, T.; Horev, G.; Suss, N.; Aloni, R.; Eshed, Y.; Lifschitz, E. The Flowering Hormone Florigen Accelerates Secondary Cell Wall Biogenesis to Harmonize Vascular Maturation with Reproductive Development. Proc. Natl. Acad. Sci. USA 2019, 116, 16127–16136. [Google Scholar] [CrossRef]
- Guo, R.; Li, W.; Wang, X.; Chen, B.; Huang, Z.; Liu, T.; Chen, X.; XuHan, X.; Lai, Z. Effect of Photoperiod on the Formation of Cherry Radish Root. Sci. Hortic. 2019, 244, 193–199. [Google Scholar] [CrossRef]
- Natarajan, B.; Kondhare, K.R.; Hannapel, D.J.; Banerjee, A.K. Mobile RNAs and Proteins: Prospects in Storage Organ Development of Tuber and Root Crops. Plant Sci. 2019, 284, 73–81. [Google Scholar] [CrossRef]
- Susila, H.; Purwestri, Y.A. PEBP Signaling Network in Tubers and Tuberous Root Crops. Plants 2023, 12, 264. [Google Scholar] [CrossRef]
- Chen, S.; Wang, S. GLABRA2, A Common Regulator for Epidermal Cell Fate Determination and Anthocyanin Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4997. [Google Scholar] [CrossRef]
- Khusnutdinov, E.; Sukhareva, A.; Panfilova, M.; Mikhaylova, E. Anthocyanin Biosynthesis Genes as Model Genes for Genome Editing in Plants. Int. J. Mol. Sci. 2021, 22, 8752. [Google Scholar] [CrossRef]
- Chakraborty, S.; Nguyen, B.; Wasti, S.D.; Xu, G. Plant Leucine-Rich Repeat Receptor Kinase (LRR-RK): Structure, Ligand Perception, and Activation Mechanism. Molecules 2019, 24, 3081. [Google Scholar] [CrossRef]
- Ruprecht, C.; Vaid, N.; Proost, S.; Persson, S.; Mutwil, M. Beyond Genomics: Studying Evolution with Gene Coexpression Networks. Trends Plant Sci. 2017, 22, 298–307. [Google Scholar] [CrossRef]
- Bowman, J.L.; Briginshaw, L.N.; Florent, S.N. Evolution and Co-Option of Developmental Regulatory Networks in Early Land Plants. Curr. Top. Dev. Biol. 2019, 131, 35–53. [Google Scholar] [CrossRef]
- Kohchi, T.; Yamato, K.T.; Ishizaki, K.; Yamaoka, S.; Nishihama, R. Development and Molecular Genetics of Marchantia polymorpha. Annu. Rev. Plant Biol. 2021, 72, 677–702. [Google Scholar] [CrossRef]
- Clowes, F.A.L. Pattern in Root Meristem Development in Angiosperms. New Phytol. 2000, 146, 83–94. [Google Scholar] [CrossRef]
- Fujinami, R.; Yamada, T.; Nakajima, A.; Takagi, S.; Idogawa, A.; Kawakami, E.; Tsutsumi, M.; Imaichi, R. Root Apical Meristem Diversity in Extant Lycophytes and Implications for Root Origins. New Phytol. 2017, 215, 1210–1220. [Google Scholar] [CrossRef]
- Imaichi, R.; Moritoki, N.; Solvang, H.K. Evolution of Root Apical Meristem Structures in Vascular Plants: Plasmodesmatal Networks. Am. J. Bot. 2018, 105, 1453–1468. [Google Scholar] [CrossRef]
- Imaichi, R.; Hiratsuka, R. Evolution of Shoot Apical Meristem Structures in Vascular Plants with Respect to Plasmodesmatal Network. Am. J. Bot. 2007, 94, 1911–1921. [Google Scholar] [CrossRef]
- Tomescu, A.M.F.; Groover, A.T. Mosaic Modularity: An Updated Perspective and Research Agenda for the Evolution of Vascular Cambial Growth. New Phytol. 2019, 222, 1719–1735. [Google Scholar] [CrossRef]
- Tomescu, A.M.F. The Stele—A Developmental Perspective on the Diversity and Evolution of Primary Vascular Architecture. Biol. Rev. Camb. Philos. Soc. 2021, 96, 1263–1283. [Google Scholar] [CrossRef]
- Jura-Morawiec, J.; Oskolski, A.; Simpson, P. Revisiting the Anatomy of the Monocot Cambium, a Novel Meristem. Planta 2021, 254, 6. [Google Scholar] [CrossRef]
- Nardmann, J.; Werr, W. The Invention of WUS-like Stem Cell-Promoting Functions in Plants Predates Leptosporangiate Ferns. Plant Mol. Biol. 2012, 78, 123–134. [Google Scholar] [CrossRef]
- Nardmann, J.; Werr, W. Symplesiomorphies in the WUSCHEL Clade Suggest That the Last Common Ancestor of Seed Plants Contained at Least Four Independent Stem Cell Niches. New Phytol. 2013, 199, 1081–1092. [Google Scholar] [CrossRef]
- Wu, C.-C.; Li, F.-W.; Kramer, E.M. Large-Scale Phylogenomic Analysis Suggests Three Ancient Superclades of the WUSCHEL-RELATED HOMEOBOX Transcription Factor Family in Plants. PLoS ONE 2019, 14, e0223521. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem Cell Regulation by Arabidopsis WOX Genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef]
- Frank, M.H.; Edwards, M.B.; Schultz, E.R.; McKain, M.R.; Fei, Z.; Sørensen, I.; Rose, J.K.C.; Scanlon, M.J. Dissecting the Molecular Signatures of Apical Cell-Type Shoot Meristems from Two Ancient Land Plant Lineages. New Phytol. 2015, 207, 893–904. [Google Scholar] [CrossRef]
- Ge, Y.; Liu, J.; Zeng, M.; He, J.; Qin, P.; Huang, H.; Xu, L. Identification of WOX Family Genes in Selaginella Kraussiana for Studies on Stem Cells and Regeneration in Lycophytes. Front. Plant Sci. 2016, 7, 93. [Google Scholar] [CrossRef]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 2017, 171, 287–304.e15. [Google Scholar] [CrossRef]
- Youngstrom, C.E.; Withers, K.A.; Irish, E.E.; Cheng, C.-L. Vascular Function of the T3/Modern Clade WUSCHEL-Related HOMEOBOX Transcription Factor Genes Predate Apical Meristem-Maintenance Function. BMC Plant Biol. 2022, 22, 210. [Google Scholar] [CrossRef]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential Expression of WOX Genes Mediates Apical-Basal Axis Formation in the Arabidopsis Embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef]
- Nardmann, J.; Reisewitz, P.; Werr, W. Discrete Shoot and Root Stem Cell-Promoting WUS/WOX5 Functions Are an Evolutionary Innovation of Angiosperms. Mol. Biol. Evol. 2009, 26, 1745–1755. [Google Scholar] [CrossRef]
- Hedman, H.; Zhu, T.; von Arnold, S.; Sohlberg, J.J. Analysis of the WUSCHEL-RELATED HOMEOBOX Gene Family in the Conifer Picea Abies Reveals Extensive Conservation as Well as Dynamic Patterns. BMC Plant Biol. 2013, 13, 89. [Google Scholar] [CrossRef]
- Frank, M.H.; Scanlon, M.J. Transcriptomic Evidence for the Evolution of Shoot Meristem Function in Sporophyte-Dominant Land Plants through Concerted Selection of Ancestral Gametophytic and Sporophytic Genetic Programs. Mol. Biol. Evol. 2015, 32, 355–367. [Google Scholar] [CrossRef]
- Hirakawa, Y. Evolution of Meristem Zonation by CLE Gene Duplication in Land Plants. Nat. Plants 2022, 8, 735–740. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Uchida, N.; Yamaguchi, Y.L.; Tabata, R.; Ishida, S.; Ishizaki, K.; Nishihama, R.; Kohchi, T.; Sawa, S.; Bowman, J.L. Control of Proliferation in the Haploid Meristem by CLE Peptide Signaling in Marchantia polymorpha. PLoS Genet. 2019, 15, e1007997. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Fujimoto, T.; Ishida, S.; Uchida, N.; Sawa, S.; Kiyosue, T.; Ishizaki, K.; Nishihama, R.; Kohchi, T.; Bowman, J.L. Induction of Multichotomous Branching by CLAVATA Peptide in Marchantia polymorpha. Curr. Biol. 2020, 30, 3833–3840.e4. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, X.-X.; Li, R.-Q.; Zhao, X.; Liu, Y.; Li, M.-H.; Zwaenepoel, A.; Ma, H.; Goffinet, B.; Guan, Y.-L.; et al. The Hornwort Genome and Early Land Plant Evolution. Nat. Plants 2020, 6, 107–118. [Google Scholar] [CrossRef]
- Hirakawa, Y. CLAVATA3, a Plant Peptide Controlling Stem Cell Fate in the Meristem. Peptides 2021, 142, 170579. [Google Scholar] [CrossRef]
- Whitewoods, C.D.; Cammarata, J.; Nemec Venza, Z.; Sang, S.; Crook, A.D.; Aoyama, T.; Wang, X.Y.; Waller, M.; Kamisugi, Y.; Cuming, A.C.; et al. CLAVATA Was a Genetic Novelty for the Morphological Innovation of 3D Growth in Land Plants. Curr. Biol. 2018, 28, 2365–2376.e5. [Google Scholar] [CrossRef]
- Furumizu, C.; Sawa, S. The RGF/GLV/CLEL Family of Short Peptides Evolved Through Lineage-Specific Losses and Diversification and Yet Conserves Its Signaling Role between Vascular Plants and Bryophytes. Front. Plant Sci. 2021, 12, 703012. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Bowman, J.L. A Role of TDIF Peptide Signaling in Vascular Cell Differentiation Is Conserved among Euphyllophytes. Front. Plant Sci. 2015, 6, 1048. [Google Scholar] [CrossRef]
- Strabala, T.J.; Phillips, L.; West, M.; Stanbra, L. Bioinformatic and Phylogenetic Analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE Signal Peptide Genes in the Pinophyta. BMC Plant Biol. 2014, 14, 47. [Google Scholar] [CrossRef]
- Galibina, N.A.; Moshchenskaya, Y.L.; Tarelkina, T.V.; Nikerova, K.M.; Korzhenevskii, M.A.; Serkova, A.A.; Afoshin, N.V.; Semenova, L.I.; Ivanova, D.S.; Guljaeva, E.N.; et al. Identification and Expression Profile of CLE41/44-PXY-WOX Genes in Adult Trees Pinus sylvestris L. Trunk Tissues during Cambial Activity. Plants 2023, 12, 835. [Google Scholar] [CrossRef]
- Suzaki, T.; Yoshida, A.; Hirano, H.-Y. Functional Diversification of CLAVATA3-Related CLE Proteins in Meristem Maintenance in Rice. Plant Cell 2008, 20, 2049–2058. [Google Scholar] [CrossRef]
- Li, X.; Yang, H.; Wang, C.; Yang, S.; Wang, J. Distinct Transgenic Effects of Poplar TDIF Genes on Vascular Development in Arabidopsis. Plant Cell Rep. 2018, 37, 799–808. [Google Scholar] [CrossRef]
- Goad, D.M.; Zhu, C.; Kellogg, E.A. Comprehensive Identification and Clustering of CLV3/ESR-Related (CLE) Genes in Plants Finds Groups with Potentially Shared Function. New Phytol. 2017, 216, 605–616. [Google Scholar] [CrossRef]
- Geng, Y.; Guo, L.; Han, H.; Liu, X.; Banks, J.A.; Wisecaver, J.H.; Zhou, Y. Conservation and Diversification of HAIRY MERISTEM Gene Family in Land Plants. Plant J. 2021, 106, 366–378. [Google Scholar] [CrossRef]
- Brouwer, P.; Bräutigam, A.; Külahoglu, C.; Tazelaar, A.O.E.; Kurz, S.; Nierop, K.G.J.; van der Werf, A.; Weber, A.P.M.; Schluepmann, H. Azolla Domestication towards a Biobased Economy? New Phytol. 2014, 202, 1069–1082. [Google Scholar] [CrossRef]
- Vallée, B.S.; Coadou, G.; Labbé, H.; Sy, D.; Vovelle, F.; Schoentgen, F. Peptides Corresponding to the N- and C-Terminal Parts of PEBP Are Well-Structured in Solution: New Insights into Their Possible Interaction with Partners In Vivo. J. Pept. Res. 2003, 61, 47–57. [Google Scholar] [CrossRef]
- Eklund, D.M.; Kanei, M.; Flores-Sandoval, E.; Ishizaki, K.; Nishihama, R.; Kohchi, T.; Lagercrantz, U.; Bhalerao, R.P.; Sakata, Y.; Bowman, J.L. An Evolutionarily Conserved Abscisic Acid Signaling Pathway Regulates Dormancy in the Liverwort Marchantia polymorpha. Curr. Biol. 2018, 28, 3691–3699.e3. [Google Scholar] [CrossRef]
- Hou, C.-J.; Yang, C.-H. Comparative Analysis of the Pteridophyte Adiantum MFT Ortholog Reveals the Specificity of Combined FT/MFT C and N Terminal Interaction with FD for the Regulation of the Downstream Gene AP1. Plant Mol. Biol. 2016, 91, 563–579. [Google Scholar] [CrossRef]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef]
- Harig, L.; Beinecke, F.A.; Oltmanns, J.; Muth, J.; Müller, O.; Rüping, B.; Twyman, R.M.; Fischer, R.; Prüfer, D.; Noll, G.A. Proteins from the FLOWERING LOCUS T-like Subclade of the PEBP Family Act Antagonistically to Regulate Floral Initiation in Tobacco. Plant J. 2012, 72, 908–921. [Google Scholar] [CrossRef]
- Pin, P.A.; Benlloch, R.; Bonnet, D.; Wremerth-Weich, E.; Kraft, T.; Gielen, J.J.L.; Nilsson, O. An Antagonistic Pair of FT Homologs Mediates the Control of Flowering Time in Sugar Beet. Science 2010, 330, 1397–1400. [Google Scholar] [CrossRef]
- Zhai, H.; Lü, S.; Liang, S.; Wu, H.; Zhang, X.; Liu, B.; Kong, F.; Yuan, X.; Li, J.; Xia, Z. GmFT4, a Homolog of FLOWERING LOCUS T, Is Positively Regulated by E1 and Functions as a Flowering Repressor in Soybean. PLoS ONE 2014, 9, e89030. [Google Scholar] [CrossRef]
- Blackman, B.K.; Strasburg, J.L.; Raduski, A.R.; Michaels, S.D.; Rieseberg, L.H. The Role of Recently Derived FT Paralogs in Sunflower Domestication. Curr. Biol. 2010, 20, 629–635. [Google Scholar] [CrossRef]
- Hiraoka, K.; Yamaguchi, A.; Abe, M.; Araki, T. The Florigen Genes FT and TSF Modulate Lateral Shoot Outgrowth in Arabidopsis thaliana. Plant Cell Physiol. 2013, 54, 352–368. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Lee, H.-J.; Seo, P.J.; Jung, J.-H.; Ahn, J.H.; Park, C.-M. The Arabidopsis Floral Repressor BFT Delays Flowering by Competing with FT for FD Binding under High Salinity. Mol. Plant 2014, 7, 377–387. [Google Scholar] [CrossRef]
- Armenta-Medina, A.; Gillmor, C.S.; Gao, P.; Mora-Macias, J.; Kochian, L.V.; Xiang, D.; Datla, R. Developmental and Genomic Architecture of Plant Embryogenesis: From Model Plant to Crops. Plant Commun. 2021, 2, 100136. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, F.; Jackson, D. All Together Now, a Magical Mystery Tour of the Maize Shoot Meristem. Curr. Opin. Plant Biol. 2018, 45, 26–35. [Google Scholar] [CrossRef]
- Je, B.I.; Gruel, J.; Lee, Y.K.; Bommert, P.; Arevalo, E.D.; Eveland, A.L.; Wu, Q.; Goldshmidt, A.; Meeley, R.; Bartlett, M.; et al. Signaling from Maize Organ Primordia via FASCIATED EAR3 Regulates Stem Cell Proliferation and Yield Traits. Nat. Genet. 2016, 48, 785–791. [Google Scholar] [CrossRef]
- Bommert, P.; Lunde, C.; Nardmann, J.; Vollbrecht, E.; Running, M.; Jackson, D.; Hake, S.; Werr, W. Thick Tassel Dwarf1 Encodes a Putative Maize Ortholog of the Arabidopsis CLAVATA1 Leucine-Rich Repeat Receptor-like Kinase. Development 2005, 132, 1235–1245. [Google Scholar] [CrossRef]
- Je, B.I.; Xu, F.; Wu, Q.; Liu, L.; Meeley, R.; Gallagher, J.P.; Corcilius, L.; Payne, R.J.; Bartlett, M.E.; Jackson, D. The CLAVATA Receptor FASCIATED EAR2 Responds to Distinct CLE Peptides by Signaling through Two Downstream Effectors. eLife 2018, 7, e35673. [Google Scholar] [CrossRef]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing Grain-Yield-Related Traits by CRISPR-Cas9 Promoter Editing of Maize CLE Genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef]
- Kloosterman, B.; Abelenda, J.A.; Gomez, M.d.M.C.; Oortwijn, M.; de Boer, J.M.; Kowitwanich, K.; Horvath, B.M.; van Eck, H.J.; Smaczniak, C.; Prat, S.; et al. Naturally Occurring Allele Diversity Allows Potato Cultivation in Northern Latitudes. Nature 2013, 495, 246–250. [Google Scholar] [CrossRef]
- Gutaker, R.M.; Weiß, C.L.; Ellis, D.; Anglin, N.L.; Knapp, S.; Luis Fernández-Alonso, J.; Prat, S.; Burbano, H.A. The Origins and Adaptation of European Potatoes Reconstructed from Historical Genomes. Nat. Ecol. Evol. 2019, 3, 1093–1101. [Google Scholar] [CrossRef]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the Flowering Gene SELF PRUNING 5G Promotes Day-Neutrality and Early Yield in Tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Zhang, S.; Jiao, Z.; Liu, L.; Wang, K.; Zhong, D.; Li, S.; Zhao, T.; Xu, X.; Cui, X. Enhancer-Promoter Interaction of SELF PRUNING 5G Shapes Photoperiod Adaptation. Plant Physiol. 2018, 178, 1631–1642. [Google Scholar] [CrossRef]
- Carmel-Goren, L.; Liu, Y.S.; Lifschitz, E.; Zamir, D. The SELF-PRUNING Gene Family in Tomato. Plant Mol. Biol. 2003, 52, 1215–1222. [Google Scholar] [CrossRef]
- Semeniuk, P. Inheritance of Recurrent and Nonrecurrent Blooming in ‘Goldilocks’ X Rosa Wichuraiana Progeny. J. Heredity 1971, 62, 319–320. [Google Scholar] [CrossRef]
- Brown, T.; Wareing, P.F. The Genetical Control of Everbearing Habit and Three Other Characters in Varieties of Fragaria vesca. Euphytica 1965, 14, 97–112. [Google Scholar] [CrossRef]
- Iwata, H.; Gaston, A.; Remay, A.; Thouroude, T.; Jeauffre, J.; Kawamura, K.; Oyant, L.H.-S.; Araki, T.; Denoyes, B.; Foucher, F. The TFL1 Homologue KSN Is a Regulator of Continuous Flowering in Rose and Strawberry. Plant J. 2012, 69, 116–125. [Google Scholar] [CrossRef]
- Mouhu, K.; Kurokura, T.; Koskela, E.A.; Albert, V.A.; Elomaa, P.; Hytönen, T. The Fragaria vesca Homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 Represses Flowering and Promotes Vegetative Growth. Plant Cell 2013, 25, 3296–3310. [Google Scholar] [CrossRef]
- Koskela, E.A.; Mouhu, K.; Albani, M.C.; Kurokura, T.; Rantanen, M.; Sargent, D.J.; Battey, N.H.; Coupland, G.; Elomaa, P.; Hytönen, T. Mutation in TERMINAL FLOWER1 Reverses the Photoperiodic Requirement for Flowering in the Wild Strawberry Fragaria vesca. Plant Physiol. 2012, 159, 1043–1054. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Tränkner, C.; Hanke, M.-V. The MdTFL1 Gene of Apple (Malus × Domestica Borkh.) Reduces Vegetative Growth and Generation Time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, K.; Efremova, E.; Dodueva, I.; Lebedeva, M.; Lutova, L. Functional Modules in the Meristems: “Tinkering” in Action. Plants 2023, 12, 3661. https://doi.org/10.3390/plants12203661
Kuznetsova K, Efremova E, Dodueva I, Lebedeva M, Lutova L. Functional Modules in the Meristems: “Tinkering” in Action. Plants. 2023; 12(20):3661. https://doi.org/10.3390/plants12203661
Chicago/Turabian StyleKuznetsova, Ksenia, Elena Efremova, Irina Dodueva, Maria Lebedeva, and Ludmila Lutova. 2023. "Functional Modules in the Meristems: “Tinkering” in Action" Plants 12, no. 20: 3661. https://doi.org/10.3390/plants12203661





